Abstract
The SET and MYND domain-containing (SMYD) family of lysine methyltransferases are essential in several mammalian developmental pathways. Although predominantly expressed in the heart, the role of SMYD2 in heart development has yet to be fully elucidated and has even been shown to be dispensable in a murine Nkx2-5-associated conditional knockout. Additionally, SMYD2 was recently shown to be necessary not only for lymphocyte development but also for the viability of hematopoietic leukemias. Based on the broad expression pattern of SMYD2 in mammalian tissues, it is likely that it plays pivotal roles in a host of additional normal and pathological processes. In this brief review, we consider what is currently known about the normal and pathogenic functions of SMYD2 and propose specific future directions for characterizing its role in embryogenesis.
1. Introduction
The 5-member SET and MYND domain-containing (SMYD) family of lysine methyltransferases are broadly expressed throughout the embryo and are essential for a wide range of developmental pathways [1,2,3,4]. These enzymes alter the epigenetic landscapes of cells by catalyzing the transfer of methyl groups to histone lysine residues, thereby modifying chromatin architecture and controlling gene expression during development. In addition, SMYD proteins catalyze methylation of non-histone proteins, thus altering signaling cascades and gene transcription. These multi-domain enzymes catalyze methylation by binding specific enzyme targets in a pocket formed predominantly by the i-SET and core-SET domains and bringing the target into close proximity with the methyl donor s-adenosylmethionine (SAM) in an adjacent pocket (Figure 1c). The second member of the SMYD family, SMYD2, was initially characterized in mice cardiomyocytes during embryogenesis [5]. However, the bulk of research involving SMYD2 has centered on its role in carcinogenesis because of its detected overexpression in a wide range of human cancers (Table 1). Due to its identification as an oncogene, studies investigating SMYD2 in embryogenesis are lacking. We previously detected clear SMYD2 expression in the hypothalamus, liver, kidneys, thymus, vomeronasal organ, and ovaries during development in mice (Figure 1a,b) [5], but the roles of SMYD2 in these organ systems have yet to be fully elucidated and merit further study. In this review, we overview what is currently known about SMYD2 in cancer and embryogenesis and hypothesize new areas for studying SMYD2 in embryogenesis based on preliminary data and analysis of what is currently known. Specifically, we: (1) hypothesize that SMYD2 is essential for the development of mesendoderm and several of its downstream organs (heart, kidney, liver, thymus, and ovaries) in part via its role in WNT signaling; (2) highlight the recently discovered role of SMYD2 in hematopoiesis and leukemias; and (3) suggest a possible role of SMYD2 in the migration of gonadotropin-releasing hormone (GnRH) neurons during the development of the vomeronasal organ (VNO).
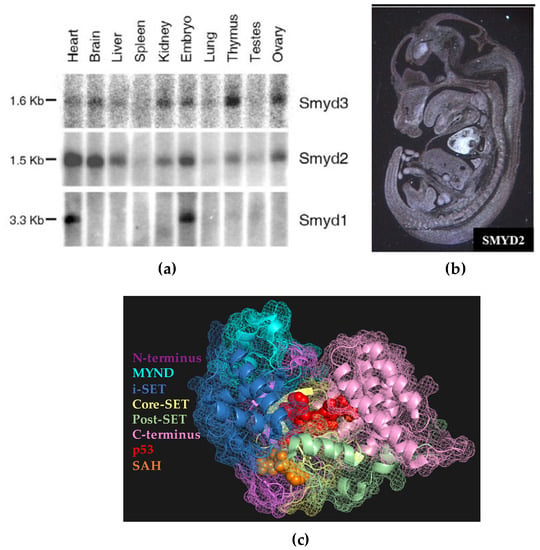
Figure 1.
Expression patterns of SMYD2. (a) Northern blot displaying tissue-specific mRNA expression levels of SMYD1, SMYD2, and SMYD3 during mouse development. Middle panel: SMYD2 is widely expressed in the heart, brain, liver, kidney, thymus, and ovary; (b) Murine E13.5 sagittal section with an anti-sense SMYD2 label depicting broad SMYD2 expression in the developing embryo, especially in the heart, hypothalamus, and vomeronasal organ; (c) the color-coded domains of SMYD2, common to all five SMYD proteins. Panel (a) used with permission from Ref. [5] according to Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0). Panel (c) created using PyMol software and the protein data bank file 3TG5 [6].

Table 1.
SMYD2 target proteins, functions, and associated cancers.
2. SMYD2 in Cancer
SMYD2 was initially characterized in mouse cardiomyocytes [5,6,7,8], where it was shown to methylate histone H3 at K4 and K36 [9]. However, SMYD2 has received the bulk of its research attention because of its overexpression in several cancers and its interactions with known oncogenes. Overexpression of SMYD2 has been observed in breast, pancreatic, colorectal, esophageal, blood, lung, bladder, and hepatocellular cancers (see refs. in Table 1), and SMYD2-positive tumors are often correlated with poor patient outcomes. These studies have revealed two principal mechanisms by which SMYD2 contributes to carcinogenesis and cancer progression. First, aberrant histone methylation has been associated with many cancer types and results in altered expression levels of several oncogenes. Second, in addition to histone targets, SMYD2 has been shown to directly modulate the activity of cell cycle regulators via post-translational methylation. Studies using human cells both in vitro and in immunodeficient mice demonstrated that SMYD2 represses the activity of tumor suppressers p53 (apoptosis) and RB1 (cell-cycle arrest) via mono-methylation at p53:K370 and RB1:K860 and K810 [8,10,11]. The methylation of AHNAK and AHNAK2—two proteins involved in cell migration and invasion—may also contribute to the role of SMYD2 in carcionogenesis [12]. Other SMYD2 methylation targets include HSP90AB1, ERα, PARP1, PTEN, BMPR2, and β-Catenin [13,14,15,16,17], which is discussed further in Section 3. The known targets of SMYD2, the tissue(s) that are involved, and observed correlations with cancers are summarized in Table 1.
An improved understanding of the specific mechanisms by which SMYD2 contributes to carcinogenesis is likely to reveal new targets for clinical intervention Tremendous efforts by universities and pharmaceutical companies have identified several effective and specific small molecule SMYD2 inhibitors, including AZ505, AZ506, EPZ033294, A-893, BAY-598, LLY-507 [18,19,20,21,22,23,24]. These compounds are promising for the treatment of a wide range of cancers and may also prove to be useful for further investigation of SMYD2 function. While our understanding of the role of SMYD2 in cancers is certainly incomplete and requires further investigation, its function in embryogenesis is even less understood. Further study into the normal roles of SMYD2 in development is necessary for a more-complete understanding of organ development, stem cell differentiation, epigenetics, and SMYD2-mediated pathogenic pathways.
3. SMYD2 in Embryogenesis
While our understanding of the role of SMYD2 in cancers remains incomplete, its function in embryogenesis is even less understood. Further study into the normal roles of SMYD2 in development is necessary for a more-complete understanding of organ development, stem cell differentiation, epigenetics, and SMYD2-mediated pathogenic pathways. Towards this understanding, several SMYD2 knockout studies have been conducted both in vitro and in vivo (summarized in Table 2). Of particular note, global SMYD2-knockouts (KO) in zebrafish have revealed severe developmental defects [25,40]; however, mice lacking SMYD2 exhibited no severe developmental phenotypes or life-span defects and in fact gained a reduced susceptibility to leukemias [41]. Our group developed conditional SMYD2 knockouts in mice to study the embryonic role of SMYD2 in heart development [7] and hematopoiesis [35]. These studies are elaborated below in Section 3.1 and Section 3.2, respectively, and serve as a foundation for future investigation of the role of SMYD2 in the development of specific organ systems.

Table 2.
SMYD2 knockout studies and observed developmental phenotypes.
3.1. SMYD2 in the WNT Pathway and Mesendodermal Differentiation
SMYD2 expression has been detected in embryonic tissues derived from all three germ layers, including muscle (mesoderm), liver (endoderm), and brain (ectoderm). It is predominantly expressed in the heart during development, however its role in cardiac development remains unclear. Donlin and colleagues [26] demonstrated that SMYD2 forms a complex with the cytoplasmic protein chaperone Hsp90 and the abundant sarcomeric protein titin. Their work revealed that a SMYD2 deficiency resulted in impaired titin stability and altered muscle function. Unexpectedly, however, an Nkx2.5-conditional Smyd2-knockout (cardiac-specific) in mice did not impact heart development [7], suggesting that SMYD2 is not essential for heart development or that its role manifests prior to Nkx2.5 expression in cardiac progenitor cells. Further investigation into the latter conclusion led us to studies revealing a role for SMYD2 prior to Nkx2.5 expression. SMYD2 methylates β-Catenin, which is essential for its nuclear translocation and subsequent activation of WNT signaling [13]. Activation of WNT signaling drives pluripotent stem cell commitment to mesendoderm lineages [40] and is the first step in the differentiation of human pluripotent stem cells (hPSC) to cardiomyocytes in vitro [27,42]. In addition to direct activation of β-Catenin, SMYD2-knockout hPSC lines demonstrated a remarkable reduction of H3K4me1 and H3K36me2 levels at the transcriptional start sites of several signature mesendoderm genes (T, EOMES, MIXL1, and GSC) during differentiation to mesoderm and endoderm [30]. Therefore, we hypothesize that the principal role of SMYD2 in heart development occurs prior to Nkx2.5 expression, which begins 3–4 days after WNT activation via β-Catenin translocation [43]. The role of SMYD2 in the WNT signaling pathway may also explain its presence and importance in other mesendodermal tissues during development, including the liver, kidney, and reproductive systems. Future studies should investigate the specific roles of SMYD2 in the development of these organs using tissue-specific conditional knockout models that allow for ablation of SMYD2 expression at various timepoints during development. Finally, dysregulation of the WNT pathway has been observed in a wide range of cancers, including colorectal, hepatocellular, and breast carcinomas [36,44,45]. The overlap between SMYD2 and the WNT pathway, particularly given their independent associations with such a wide range of cancers, merits immediate further investigation.
3.2. SMYD2 in Hematopoiesis and Leukemia
SMYD2 overexpression has been observed in several blood cancers, including B-ALL, T-ALL, CML, MLLr-B-ALL, AML and additional hematopoietic lesions, including CLL and DLBCL [46,47,48,49]. Despite this knowledge, the role of SMYD2 in hematopoeisis was unclear prior to our recent work investigating the effects of SMYD2 deletion in hematopoietic stem cells (HSCs) and their downstream progenitor pools [35]. We discovered that a murine HSC-specific Smyd2 conditional knockout (CKO) yielded a significant decrease in HSC numbers as well as a decrease in some, but not all, downstream myeloid and lymphoid lineages. Deeper investigation revealed that these effects are mediated at least in part by the induction of apoptosis in blood progenitor cells of Smyd2-CKO animals. Additionally, we found that Smyd2-CKO mice exhibited disrupted STAT3 and WNT/β-Catenin signaling in HSC lineages, agreeing with previous studies describing the interactions between SMYD2 and these two proliferation-inducing signaling pathways [13,29].
3.3. SMYD2 in the Hypothalamus and Vomeronasal Organ (VNO)
In our initial characterization of SMYD2 in 2006 [5], we sought to determine which embryonic tissues express SMYD2 during development. We performed whole-mount in situ hybridization using murine embryos at day 13.5 with a probe specific to Smyd2 (Figure 1b). While the most pronounced expression was observed in the heart, we also observed clear SMYD2 expression in the hypothalamus and in the vicinity of the vomeronasal organ (VNO). To our knowledge, a possible developmental role of SMYD2 in this region has never been investigated following these initial observations.
Herein, we propose that SMYD2 may play a role in the origin and/or migration of gonadotropin-releasing hormone (GnRH) neurons from the VNO, which are known to emerge from the VNO around E11 and migrate into the basal forebrain between E12–E17 in mice [50,51]. GnRH is released from GnRH neurons located in the hypothalamus in a pulsatile fashion during adolescence, and is essential for proper reproductive function. The release of GnRH represents the initial step in the hypothalamic-pituitary-gonadal signaling axis. Due to the strong SMYD2 signal in the VNO and the hypothalamus during GnRH neural migration between the two developing organs, it is plausible that SMYD2 plays a role in proper neuronal differentiation and/or networking during development. Several factors are implicated in proper GnRH neural migration, including growth factors, extracellular matrix/adhesion molecules, neuro-transmitters, G-protein-coupled receptors, and transcription factors [26,42]. Studies that investigate the roles of histone methylation at transcriptional start sites or direct post-transcriptional methylation in the expression or activity of these factors may elucidate a role for SMYD2 in VNO and hypothalamic development. Besides SMYD2-mediated histone methylation, thorough epigenetic analysis in these regions is likely to further clarify the specific mechanisms that drive VNO development, GnRH neuron migration, and neural networking in the hypothalamus.
4. Conclusions and Future Directions
Methyltransferase enzymes have recently been recognized as key players in modulating gene expression and protein activity in a wide range of physiologic processes. The SMYD family of methyltransferase enzymes has been shown to be particularly important during embryogenesis via histone methylation, and have also been shown to regulate the expression and activity of several known oncogenes. We previously demonstrated that the second member of the SMYD family, SMYD2, is widely expressed during normal embryogenesis but also in several cancers. To further elucidate the role of SMYD2 in development and carcinogenesis, we propose four areas for future study.
First, the purpose of the robust expression of SMYD2 in the heart during development has yet to be uncovered. It is known that SMYD2 expression is dispensable in early cardiac progenitor cells expressing Nkx2.5 [7], which may be explained by compensation from SMYD1 or by an earlier role for SMYD2 during heart development. Experimentation seeking to elucidate an earlier role for SMYD2 in heart development would be valuable in answering this question.
Second, SMYD2-WNT-β-catenin signaling merits deeper investigation in two contexts. First, WNT signaling is essential for mesendoderm commitment, which could explain the role of SMYD2 in the development of a wide range of downstream organs (heart, kidneys, thymus, liver, and ovaries) and represents a reasonable explanation for the role of SMYD2 in heart development prior to Nkx2.5 expression. Second, this signaling axis is essential for hematopoetic stem cell (HSC) renewal and is disrupted in a wide range of HSC-derived blood cancers. Targeting the SMYD2-WNT-β-catenin pathway in HSCs may provide a successful approach for future cancer therapies.
Third, while our recent work established SMYD2 as an important mediator in leukemias [35], the specific mechanisms by which it directs apoptosis in HSCs and downstream progenitor pools remain unclear. Further study into the roles of SMYD2 in normal and pathogenic blood cell renewal and differentiation is likely to reveal important transcriptional leukemic targets for clinical intervention.
Fourth, SMYD2 is clearly expressed near the vomeronasal organ (VNO) during development but its role has yet to be studied in this organ system [5], Figure 1b. Due to the parallel expression in the hypothalamus during GnRH neuron migration from the VNO, we postulate that SMYD2 may play a key role in the proper migration and networking of these neurons from the VNO to the hypothalamus.
The broad expression patterns of SMYD2 in embryogenesis and carcinogenesis have yet to be completely explained. Further investigation of the functions of SMYD2 in these contexts will expand our understanding of mammalian development and improve our clinical management of cancer.
Funding
This research was funded by the NSF-GRFP grant number DGE-1553798 (awarded to D.K.J.) and by NSF Grants 1060548 and 1930417 (awarded to M.A.B.).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Doughan, M.; Spellmon, N.; Li, C.; Yang, Z. SMYD proteins in immunity: Dawning of a new era. AIMS Biophys 2016, 3, 450–455. [Google Scholar] [CrossRef] [PubMed]
- Du, S.J.; Tan, X.; Zhang, J. SMYD Proteins: Key Regulators in Skeletal and Cardiac Muscle Development and Function. Anat. Rec. (Hoboken) 2014, 297, 1650–1662. [Google Scholar] [CrossRef]
- Leinhart, K.; Brown, M. SET/MYND Lysine Methyltransferases Regulate Gene Transcription and Protein Activity. Genes (Basel) 2011, 2, 210–218. [Google Scholar] [CrossRef]
- Spellmon, N.; Holcomb, J.; Trescott, L.; Sirinupong, N.; Yang, Z. Structure and function of SET and MYND domain-containing proteins. Int. J. Mol. Sci. 2015, 16, 1406–1428. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.A.; Sims, R.J., 3rd; Gottlieb, P.D.; Tucker, P.W. Identification and characterization of Smyd2: A split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Mol. Cancer 2006, 5, 26. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, L.; Zhang, H.; Luo, X.; Dai, J.; Zhou, S.; Gu, J.; Zhu, J.; Atadja, P.; Lu, C.; et al. Structure of human SMYD2 protein reveals the basis of p53 tumor suppressor methylation. J. Biol. Chem. 2011, 286, 38725–38737. [Google Scholar] [CrossRef] [PubMed]
- Diehl, F.; Brown, M.A.; van Amerongen, M.J.; Novoyatleva, T.; Wietelmann, A.; Harriss, J.; Ferrazzi, F.; Bottger, T.; Harvey, R.P.; Tucker, P.W.; et al. Cardiac deletion of Smyd2 is dispensable for mouse heart development. PLoS ONE 2010, 5, e9748. [Google Scholar] [CrossRef]
- Huang, J.; Perez-Burgos, L.; Placek, B.J.; Sengupta, R.; Richter, M.; Dorsey, J.A.; Kubicek, S.; Opravil, S.; Jenuwein, T.; Berger, S.L. Repression of p53 activity by Smyd2-mediated methylation. Nature 2006, 444, 629–632. [Google Scholar] [CrossRef]
- Clappier, E.; Gerby, B.; Sigaux, F.; Delord, M.; Touzri, F.; Hernandez, L.; Ballerini, P.; Baruchel, A.; Pflumio, F.; Soulier, J. Clonal selection in xenografted human T cell acute lymphoblastic leukemia recapitulates gain of malignancy at relapse. J. Exp. Med. 2011, 208, 653–661. [Google Scholar] [CrossRef]
- Cho, H.S.; Hayami, S.; Toyokawa, G.; Maejima, K.; Yamane, Y.; Suzuki, T.; Dohmae, N.; Kogure, M.; Kang, D.; Neal, S.; et al. RB1 methylation by SMYD2 enhances cell cycle progression through an increase of RB1 phosphorylation. Neoplasia 2012, 14, 476–486. [Google Scholar] [CrossRef]
- Saddic, L.A.; West, L.E.; Aslanian, A.; Yates, J.R., 3rd; Rubin, S.M.; Gozani, O.; Sage, J. Methylation of the retinoblastoma tumor suppressor by SMYD2. J. Biol. Chem. 2010, 285, 37733–37740. [Google Scholar] [CrossRef] [PubMed]
- Olsen, J.B.; Cao, X.J.; Han, B.; Chen, L.H.; Horvath, A.; Richardson, T.I.; Campbell, R.M.; Garcia, B.A.; Nguyen, H. Quantitative Profiling of the Activity of Protein Lysine Methyltransferase SMYD2 Using SILAC-Based Proteomics. Mol. Cell Proteomics 2016, 15, 892–905. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Hamamoto, R.; Vougiouklakis, T.; Wang, R.; Yoshioka, Y.; Suzuki, T.; Dohmae, N.; Matsuo, Y.; Park, J.H.; Nakamura, Y. Critical roles of SMYD2-mediated beta-catenin methylation for nuclear translocation and activation of Wnt signaling. Oncotarget 2017, 8, 55837–55847. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Wang, Z.; Wang, W.; Hu, X.; Chen, P.; Li, J.; Feng, X.; Wong, J.; Du, J.X. The lysine methyltransferase SMYD2 methylates the kinase domain of type II receptor BMPR2 and stimulates bone morphogenetic protein signaling. J. Biol. Chem. 2017, 292, 12702–12712. [Google Scholar] [CrossRef]
- Nakakido, M.; Deng, Z.; Suzuki, T.; Dohmae, N.; Nakamura, Y.; Hamamoto, R. Dysregulation of AKT Pathway by SMYD2-Mediated Lysine Methylation on PTEN. Neoplasia 2015, 17, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Piao, L.; Kang, D.; Suzuki, T.; Masuda, A.; Dohmae, N.; Nakamura, Y.; Hamamoto, R. The histone methyltransferase SMYD2 methylates PARP1 and promotes poly(ADP-ribosyl)ation activity in cancer cells. Neoplasia 2014, 16, 257–264.e2. [Google Scholar] [CrossRef]
- Zhang, X.; Tanaka, K.; Yan, J.; Li, J.; Peng, D.; Jiang, Y.; Yang, Z.; Barton, M.C.; Wen, H.; Shi, X. Regulation of estrogen receptor alpha by histone methyltransferase SMYD2-mediated protein methylation. Proc. Natl. Acad. Sci. USA 2013, 110, 17284–17289. [Google Scholar] [CrossRef] [PubMed]
- Cowen, S.D.; Russell, D.; Dakin, L.A.; Chen, H.; Larsen, N.A.; Godin, R.; Throner, S.; Zheng, X.; Molina, A.; Wu, J.; et al. Design, Synthesis, and Biological Activity of Substrate Competitive SMYD2 Inhibitors. J. Med. Chem. 2016, 59, 11079–11097. [Google Scholar] [CrossRef] [PubMed]
- Eggert, E.; Hillig, R.C.; Koehr, S.; Stockigt, D.; Weiske, J.; Barak, N.; Mowat, J.; Brumby, T.; Christ, C.D.; Ter Laak, A.; et al. Discovery and Characterization of a Highly Potent and Selective Aminopyrazoline-Based in Vivo Probe (BAY-598) for the Protein Lysine Methyltransferase SMYD2. J. Med. Chem. 2016, 59, 4578–4600. [Google Scholar] [CrossRef] [PubMed]
- Li, L.X.; Zhou, J.X.; Calvet, J.P.; Godwin, A.K.; Jensen, R.A.; Li, X. Lysine methyltransferase SMYD2 promotes triple negative breast cancer progression. Cell Death Dis. 2018, 9, 326. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.; Allali-Hassani, A.; Antonysamy, S.; Chang, S.; Chen, L.H.; Curtis, C.; Emtage, S.; Fan, L.; Gheyi, T.; Li, F.; et al. LLY-507, a Cell-active, Potent, and Selective Inhibitor of Protein-lysine Methyltransferase SMYD2. J. Biol. Chem. 2015, 290, 13641–13653. [Google Scholar] [CrossRef] [PubMed]
- Sweis, R.F.; Wang, Z.; Algire, M.; Arrowsmith, C.H.; Brown, P.J.; Chiang, G.G.; Guo, J.; Jakob, C.G.; Kennedy, S.; Li, F.; et al. Discovery of A-893, A New Cell-Active Benzoxazinone Inhibitor of Lysine Methyltransferase SMYD2. ACS Med. Chem. Lett. 2015, 6, 695–700. [Google Scholar] [CrossRef] [PubMed]
- Thomenius, M.J.; Totman, J.; Harvey, D.; Mitchell, L.H.; Riera, T.V.; Cosmopoulos, K.; Grassian, A.R.; Klaus, C.; Foley, M.; Admirand, E.A.; et al. Small molecule inhibitors and CRISPR/Cas9 mutagenesis demonstrate that SMYD2 and SMYD3 activity are dispensable for autonomous cancer cell proliferation. PLoS ONE 2018, 13, e0197372. [Google Scholar] [CrossRef] [PubMed]
- Fabini, E.; Manoni, E.; Ferroni, C.; Rio, A.D.; Bartolini, M. Small-molecule inhibitors of lysine methyltransferases SMYD2 and SMYD3: Current trends. Future Med. Chem. 2019, 11, 901–921. [Google Scholar] [CrossRef] [PubMed]
- Voelkel, T.; Andresen, C.; Unger, A.; Just, S.; Rottbauer, W.; Linke, W.A. Lysine methyltransferase Smyd2 regulates Hsp90-mediated protection of the sarcomeric titin springs and cardiac function. Biochim. Biophys. Acta 2013, 1833, 812–822. [Google Scholar] [CrossRef]
- Donlin, L.T.; Andresen, C.; Just, S.; Rudensky, E.; Pappas, C.T.; Kruger, M.; Jacobs, E.Y.; Unger, A.; Zieseniss, A.; Dobenecker, M.W.; et al. Smyd2 controls cytoplasmic lysine methylation of Hsp90 and myofilament organization. Genes Dev. 2012, 26, 114–119. [Google Scholar] [CrossRef]
- Lian, X.; Hsiao, C.; Wilson, G.; Zhu, K.; Hazeltine, L.B.; Azarin, S.M.; Raval, K.K.; Zhang, J.; Kamp, T.J.; Palecek, S.P. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. USA 2012, 109, E1848–E1857. [Google Scholar] [CrossRef]
- Hamamoto, R.; Toyokawa, G.; Nakakido, M.; Ueda, K.; Nakamura, Y. SMYD2-dependent HSP90 methylation promotes cancer cell proliferation by regulating the chaperone complex formation. Cancer Lett. 2014, 351, 126–133. [Google Scholar] [CrossRef]
- Li, L.X.; Fan, L.X.; Zhou, J.X.; Grantham, J.J.; Calvet, J.P.; Sage, J.; Li, X. Lysine methyltransferase SMYD2 promotes cyst growth in autosomal dominant polycystic kidney disease. J. Clin. Investig. 2017, 127, 2751–2764. [Google Scholar] [CrossRef]
- Bai, H.J.; Zhang, P.; Ma, L.; Liang, H.; Wei, G.; Yang, H.T. SMYD2 Drives Mesendodermal Differentiation of Human Embryonic Stem Cells Through Mediating the Transcriptional Activation of Key Mesendodermal Genes. Stem Cells 2019, 37, 1401–1415. [Google Scholar] [CrossRef]
- Komatsu, S.; Ichikawa, D.; Hirajima, S.; Nagata, H.; Nishimura, Y.; Kawaguchi, T.; Miyamae, M.; Okajima, W.; Ohashi, T.; Konishi, H.; et al. Overexpression of SMYD2 relates to tumor cell proliferation and malignant outcome of esophageal squamous cell carcinoma. Carcinogenesis 2009, 30, 1139–1146. [Google Scholar] [CrossRef] [PubMed]
- Reynoird, N.; Mazur, P.K.; Stellfeld, T.; Flores, N.M.; Lofgren, S.M.; Carlson, S.M.; Brambilla, E.; Hainaut, P.; Kaznowska, E.B.; Arrowsmith, K.; et al. Coordination of stress signals by the lysine methyltransferase SMYD2 promotes pancreatic cancer. Genes Dev. 2016, 30, 772–785. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Ichikawa, D.; Hirajima, S.; Nagata, H.; Nishimura, Y.; Kawaguchi, T.; Miyamae, M.; Okajima, W.; Ohashi, T.; Konishi, H.; et al. Overexpression of SMYD2 contributes to malignant outcome in gastric cancer. Br. J. Cancer 2015, 112, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Abu-Farha, M.; Lambert, J.P.; Al-Madhoun, A.S.; Elisma, F.; Skerjanc, I.S.; Figeys, D. The tale of two domains: Proteomics and genomics analysis of SMYD2, a new histone methyltransferase. Mol. Cell Proteomics 2008, 7, 560–572. [Google Scholar] [CrossRef]
- Brown, M.A.; Edwards, M.A.; Alshiraihi, I.; Geng, H.; Dekker, J.D.; Tucker, H.O. The lysine methyltransferase SMYD2 is required for normal lymphocyte development and survival of hematopoietic leukemias. Genes Immun. 2020, 21, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Nusse, R.; Clevers, H. Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017, 169, 985–999. [Google Scholar] [CrossRef]
- Sakamoto, L.H.; Andrade, R.V.; Felipe, M.S.; Motoyama, A.B.; Pittella Silva, F. SMYD2 is highly expressed in pediatric acute lymphoblastic leukemia and constitutes a bad prognostic factor. Leuk. Res. 2014, 38, 496–502. [Google Scholar] [CrossRef]
- Ou, H.; Chen, Z.; Xiang, L.; Fang, Y.; Xu, Y.; Liu, Q.; Hu, Z.; Li, X.; Huang, Y.; Yang, D. Frizzled 2-induced epithelial-mesenchymal transition correlates with vasculogenic mimicry, stemness, and Hippo signaling in hepatocellular carcinoma. Cancer Sci. 2019, 110, 1169–1182. [Google Scholar] [CrossRef]
- Wang, R.; Deng, X.; Yoshioka, Y.; Vougiouklakis, T.; Park, J.H.; Suzuki, T.; Dohmae, N.; Ueda, K.; Hamamoto, R.; Nakamura, Y. Effects of SMYD2-mediated EML4-ALK methylation on the signaling pathway and growth in non-small-cell lung cancer cells. Cancer Sci. 2017, 108, 1203–1209. [Google Scholar] [CrossRef]
- Sese, B.; Barrero, M.J.; Fabregat, M.C.; Sander, V.; Izpisua Belmonte, J.C. SMYD2 is induced during cell differentiation and participates in early development. Int. J. Dev. Biol. 2013, 57, 357–364. [Google Scholar] [CrossRef]
- Bagislar, S.; Sabo, A.; Kress, T.R.; Doni, M.; Nicoli, P.; Campaner, S.; Amati, B. Smyd2 is a Myc-regulated gene critical for MLL-AF9 induced leukemogenesis. Oncotarget 2016, 7, 66398–66415. [Google Scholar] [CrossRef]
- Burridge, P.W.; Matsa, E.; Shukla, P.; Lin, Z.C.; Churko, J.M.; Ebert, A.D.; Lan, F.; Diecke, S.; Huber, B.; Mordwinkin, N.; et al. Chemically defined generation of human cardiomyocytes. Nat. Methods 2014, 11, 855–860. [Google Scholar] [CrossRef]
- Zhang, J.Z.; Termglinchan, V.; Shao, N.Y.; Itzhaki, I.; Liu, C.; Ma, N.; Tian, L.; Wang, V.Y.; Chang, A.C.Y.; Guo, H.; et al. A Human iPSC Double-Reporter System Enables Purification of Cardiac Lineage Subpopulations with Distinct Function and Drug Response Profiles. Cell Stem Cell 2019, 24, 802–811.e5. [Google Scholar] [CrossRef]
- Krishnamurthy, N.; Kurzrock, R. Targeting the Wnt/beta-catenin pathway in cancer: Update on effectors and inhibitors. Cancer Treat. Rev. 2018, 62, 50–60. [Google Scholar] [CrossRef]
- Taciak, B.; Pruszynska, I.; Kiraga, L.; Bialasek, M.; Krol, M. Wnt signaling pathway in development and cancer. J. Physiol. Pharmacol. 2018, 69. [Google Scholar]
- Bagger, F.O.; Sasivarevic, D.; Sohi, S.H.; Laursen, L.G.; Pundhir, S.; Sonderby, C.K.; Winther, O.; Rapin, N.; Porse, B.T. BloodSpot: A database of gene expression profiles and transcriptional programs for healthy and malignant haematopoiesis. Nucleic Acids Res. 2016, 44, D917–D924. [Google Scholar] [CrossRef]
- Barrett, T.; Troup, D.B.; Wilhite, S.E.; Ledoux, P.; Rudnev, D.; Evangelista, C.; Kim, I.F.; Soboleva, A.; Tomashevsky, M.; Edgar, R. NCBI GEO: Mining tens of millions of expression profiles—Database and tools update. Nucleic Acids Res. 2007, 35, D760–D765. [Google Scholar] [CrossRef]
- Geng, H.; Brennan, S.; Milne, T.A.; Chen, W.Y.; Li, Y.; Hurtz, C.; Kweon, S.M.; Zickl, L.; Shojaee, S.; Neuberg, D.; et al. Integrative epigenomic analysis identifies biomarkers and therapeutic targets in adult B-acute lymphoblastic leukemia. Cancer Discov. 2012, 2, 1004–1023. [Google Scholar] [CrossRef]
- Juric, D.; Lacayo, N.J.; Ramsey, M.C.; Racevskis, J.; Wiernik, P.H.; Rowe, J.M.; Goldstone, A.H.; O’Dwyer, P.J.; Paietta, E.; Sikic, B.I. Differential gene expression patterns and interaction networks in BCR-ABL-positive and -negative adult acute lymphoblastic leukemias. J. Clin. Oncol. 2007, 25, 1341–1349. [Google Scholar] [CrossRef]
- Tobet, S.A.; Schwarting, G.A. Minireview: Recent progress in gonadotropin-releasing hormone neuronal migration. Endocrinology 2006, 147, 1159–1165. [Google Scholar] [CrossRef]
- Wierman, M.E.; Kiseljak-Vassiliades, K.; Tobet, S. Gonadotropin-releasing hormone (GnRH) neuron migration: Initiation, maintenance and cessation as critical steps to ensure normal reproductive function. Front. Neuroendocrinol. 2011, 32, 43–52. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).