Abstract
Akkermansia muciniphila is a mucin-degrading bacterium that has shown the potential to provide anti-inflammatory and anti-obesity effects in mouse and man. We here focus on companion animals, specifically cats and dogs, and evaluate the microbial degradation of mucus and its health impact in the context of the worldwide epidemic of pet obesity. A literature survey revealed that the two presently known Akkermansia spp., A. muciniphila and A. glycaniphila, as well as other members of the phylum of Verrucomicrobia seem to be neither very prevalent nor abundant in the digestive tract of cats and dog. While this may be due to methodological aspects, it suggests that bacteria related to Akkermansia are not the major mucus degraders in these pets and hence other mucus-utilizing taxa may deserve attention. Hence, we will discuss the potential of these endogenous mucus utilizers and dietary interventions to boost these as well as the use of Akkermansia spp. related bacteria or their components as strategies to target feline and canine obesity.
1. Introduction
Obesity is a complex disease that is closely related to host genetics, environmental factors, and the microorganisms inhabiting the digestive tract (gut microbiota). According to the World Health Organization, globally, there are more than 1 billion overweight adults, and at least 300 million of these clinically obese [1]. Obesity is associated with additional medical problems, such as cardiovascular, musculoskeletal, and psychological disorders, diabetes, and an overall lower quality of life.
According to the American Veterinary Medical Association (AVMA), there are approximately 76 million dogs and 58 million cats in the US, and approximately 60% of these animals are overweight or obese [2]. Obesity is one of the most common nutritional imbalances in companion animals [3,4] and carries similar medical problems as in people. Traditional therapies to treat obese cats and dogs include exercise as well as changes in diet and dietary patterns [5]. More recently, the use of beneficial microorganisms has been considered as a feasible alternative solution in obese people [6]. However, the development of effective bacterial therapies with weight-loss applications in obese cats and dogs has shown to be challenging [7], in part due to the wide inter-individual variation in the gut microbiota of these animal species.
Akkermansia muciniphila is a mucin-degrading bacterium that has shown the potential to provide anti-inflammatory and anti-obesity effects in mouse and man by reducing insulin resistance, glucose intolerance, and gut permeability. We here focus on microbial degradation of intestinal mucus and its health impact in the context of the worldwide epidemic of feline and canine obesity.
2. The Gut Microbiota
From birth, and perhaps even before that, the digestive tract is inhabited by a complex mixture of microorganisms, including bacteriophages and other viruses. The density of microbial life varies widely throughout the gut. The stomach and the small intestine contain small numbers of bacteria (<1 × 104 bacteria/mL, mostly attached to the mucosal layer with little luminal microbes), while the large intestine is much more densely populated (>1 × 1011 microbial cells/g of intestinal content) [8]. Despite the high microbial density, in normal circumstances, the gut microbiota lives in equilibrium with the immune system from the host [9]. However, in the event of physiological or physical alterations, for example when the mucosal barrier is compromised, the host–microbiota relationship is no longer in balance, and a process of inflammation may develop [10,11,12]. This is an area of great interest particularly for clinicians because the manipulation of the host–microbiota milieu inside the gut, such as bacteria marketed as probiotics, can lead to better strategies to maintain and restore health.
In human beings, it has been shown that each individual harbors a highly individualized microbiota inside the gut [13], a finding that has been reproduced at large scale that showed even personalization at the strain level [14]. While a core microbiome can be identified, the microbial signature in each individual may be better considered as an ‘evolving fingerprint’ since the gut microbiota also shows pronounced temporal variability across months, weeks, and even days [15,16]. This uniqueness of the gut microbiota is also true for pet animal species as shown by a growing number of studies in cats and dogs (see “The feline and canine gut microbiota” below). While an evolving individualized microbiota profile may be difficult to visualize, this makes sense when considering the individuality and evolution of microorganisms and host–microbe interactions [17].
The gut microbiota, particularly that in the large intestine, is closely related to energy homeostasis, fat metabolism, and obesity. One seminal study showed that colonization of adult germ-free mice with a distal gut microbiota harvested from conventionally raised mice produced a remarkable increase in body fat within two weeks, despite an associated decrease in food consumption [18]. On the other hand, certain diets in mice can induce obesity and this phenomenon is linked to marked alterations in the distal gut microbiome, although specific microbial manipulations may limit weight gain [19,20]. While these and other studies have shed light into the complex host–microbiota relation in obesity, a causal relationship has been found difficult to establish [21].
The gut microbiota is composed of hundreds of different types of microbes with wildly different ecophysiologies [22]. Within this complex and unique assortment of microorganisms, some microbes have apparently evolved to live in the mucus, a heavily glycosylated protein that is continuously produced to cover and protect the inner epithelial layer of the digestive tract. Some of these mucus-degraders have specialized in surviving on host-produced mucus rather than diet-derived compounds that feed most of the colonic bacteria. It has been even suggested that bacteria that live in the mucosa do not compete with the microbiota present in the lumen and, therefore, do not depend on dietary nutrients deriving from host food consumption [23,24]. This is a very important corollary that will be discussed again in the last section of this article (see “Implications of microbial mucus degradation for health in cats and dogs” below).
3. Gastrointestinal (GI) Mucus
The digestive tract of humans and other animals is a muscular tube covered on the inside by epithelium containing mucus-secreting goblet cells [25,26,27]. This mucus has important protective, lubricative, and immune functions that are relevant to preserve health [28,29,30,31]. Studies in laboratory animals have shown that the stomach and the small intestine are covered by one layer of mucus (~100 μm), while the large intestine has a two-layered mucus (~150 μm) that differs in biochemical composition, structure, and relationship with the microorganisms inhabiting the gut [32,33,34,35,36].
Biochemical studies showed that the mucus layer is composed of two layers, a sterile inner layer that protects the enterocytes and an outer layer that is colonized and provides nutrients and attachment sites for the intestinal bacteria [37]. In this context, it should be noted that mucus production in the animal intestinal tract is high and may account for approximately half of the carbon present in the colon and hence is an important substrate for bacteria that are fed directly by the host [38]. In addition, it was found that development in the colon of mice takes several weeks, and that is a phenomenon with a close dependence on the presence and metabolic activities of different groups of microorganisms [39]. These findings are relevant, especially in a context of GI diseases that involve destruction of mucus and mucosal epithelium, for example inflammatory bowel disease (IBD, [40,41]). The unbalanced destruction of mucus has several negative consequences for the host, including a diminished defense of enterocytes, an increased presence of mucus-associated bacteria in the lumen, and more nutrients to luminal bacteria that usually do not feed on these substrates. These consequences may lead to a more unbalanced state during GI disease, and it has been proposed that this leads to biofilms that are often observed in patients suffering from IBD or colorectal cancer [11,42,43].
The production and release of mucus is a phenomenon affected primarily by anatomical location, host genetics, and pathogenic infections, as well as the gut microbiota and their metabolites (e.g., short-chain fatty acids (SCFAs), acetate, propionate, and butyrate). One early study showed that SCFAs did not influence the number of cells staining for mucin in the rat colon, but solutions of low osmolality (i.e., with a low number of dissolved particles) caused a considerable release of mucin from goblet cells [44]. Other, more recent, studies have shown that certain dietary ingredients can lead to enhanced production of SCFA or lactic acid, which in turn may have stimulating effects on mucus production and affect epithelial barrier integrity in mouse and man [45,46]. On the other hand, each animal species is unique in terms of dietary and physiological characteristics, and therefore, it is expected that each species also possesses a distinctive mechanism of mucus production as well as different mucus structure and biochemical composition.
Several studies have investigated the physicochemical and antigenic characteristics of GI mucus in health and disease in humans [47,48,49,50], while others have focused on mice [28,51,52,53]. To our knowledge, there have been no comparisons of mucus physicochemical properties among animal species, in part due to the inherent complications of analyzing mucus separately of intestinal contents. An early study used a radioimmunoassay for the quantitative measurement of human goblet cell mucin and showed that human goblet cell mucin cross-reacted with dog, monkey, and rabbit mucins, but not with mucins of rat, pig, toad, and oyster [54].
A. muciniphila and Microbial Degradation of Mucus
The gut microbiota affects the whole-body metabolism by affecting the energy balance and gut permeability, particularly in the large intestine. A. muciniphila was first isolated in 2004 at Wageningen University from a healthy subject as an abundant specialist growing on animal and human mucus [55]. Later on, it was demonstrated that A. muciniphila cells are related to energy storage and whole-body metabolism by improving insulin sensitivity, reducing insulinemia and plasma total cholesterol, increasing the intestinal levels of endocannabinoids that control inflammation, the gut barrier function, and gut peptide secretion in mouse and man [20,56]. It remains to be seen whether these properties could also be linked to other mucus-degrading bacteria. However, these findings help clarifying, at least partly, the relationship between microbial degradation of mucus and obesity.
Presently, only two recognized species of the genus Akkermansia have been described. A. muciniphila, found in man and many other species, and A. glycaniphila isolated from a Burmese python [57,58]. A recent genomic study of 70 human strains suggested the presence of four species-level groups, although the 16S rRNA sequences of all isolates showed more than 97% identity [59,60]. Of interest, two of these new isolates could produce vitamin B12 and contained corrin ring biosynthesis-associated genes, similar to A. glycaniphila [59]. Several review papers have summarized the state-of-the-art knowledge on mucin–bacterial interactions and A. muciniphila [24,58,61,62,63]. Hence, we focused here on particular aspects of this bacterium that have not been discussed and are relevant for this work. Interestingly, A. muciniphila has been investigated for its effect on systemic markers of GI permeability and epithelial damage following antimicrobial administration in dogs [64].
Verrucomicrobium spinosum was the first isolated organism of a new phylum that is now known as the Verrucomicrobia and was described in 1987 [65]. This bacterium was given such name because of the presence of bundles of fimbriae originating from the tips of the prosthecae. Different types of microbes are currently catalogued within the Verrucomicrobia, including some without the fimbriae in V. spinosum. The phylogenetic relationship of Verrucomicrobia with other bacterial groups is constantly evolving. For example, it has been suggested that Verrucomicrobia, Planctomycetes, and other groups form a monophyletic group referred to as the PVC superphylum [66].
It has been suggested that Akkermansia-like organisms are universally distributed in the intestines of the animal kingdom [24]. However, it was also observed that Akkermansia spp. were not detected in all taxa within the order of the Mammalia, which at that moment was explained by the possibility that Akkermansia sequences might have been overlooked because the sequence depth was limited. Others have also suggested a global distribution of A. muciniphila in the mammalian gut microbiota but have only used data from feces of humans, mice, and pigs [67]. In an early massive study of the microbiota in mammals, it was shown that most Verrucomicrobia 16S rRNA sequences belonged to Akkermansia but also to the Class Subdivision 5 (1054 sequences in the Ribosomal Database Project, RDP) [68]. Moreover, following the characterization of A. muciniphila type strain at the genome level and using this in human metagenomic libraries, it was found that there may be new and uncultured Akkermansia species, since metagenomic sequences with 84–88% average nucleotide identity were detected [69]. This study also indicated that there could be at least eight different representative species within the Akkermansia genus based on the 16S rRNA gene sequences when a cut off of 98% identity was used and suggested that some individuals may be colonized by different species of Akkermansia [69]. The latter makes sense when considering the individuality in bacteria [17] and was later confirmed experimentally [70,71]. It is also important to remember that identical 16S rRNA gene sequences have been found in microorganisms with highly divergent ecophysiologies [72], a highly relevant example being the Escherichia coli K12, an innocent gut isolate, and E. coli O157, responsible for many disease outbreaks [73]. This indicates that the metabolic potential cannot always be deduced from the taxonomic position of an Akkermansia-like isolate. However, microbial taxonomy will always be based on cultured and deposited representatives and is still a polyphasic approach including the use of phenotypic, chemotaxonomic, and genotypic data [74].
To help shed light onto the nature and distribution of Akkermansia spp., we depicted the phylogenetic relationship of all 1872 Akkermansia-like 16S rRNA gene sequences (>1200 nucleotides, common gaps removed) retrieved from the RDP database (Figure 1; analysis done by December 2019) using iTOL [75]. Akkermansia is one of the best represented genera within the Verrucomicrobia in RDP, accounting for ~17% of all 10,731 16S rRNA sequences of Verrucomicrobia. From the Akkermansia-like sequences, a subset of 250 sequences (~13% of all Akkermansia sequences in RDP) belong to the study of fecal microbial communities from 60 mammalian species from 13 taxonomic orders [68]. Note that RDP contains a total of 16,763 16S rRNA gene sequences from this study, most of which belong to Firmicutes (10,697 sequences), Bacteroidetes (2636 sequences), and Proteobacteria (1579 sequences) (these three taxa account for 89% of all sequences from Ley et al. [68]). Interestingly, 219 sequences from this study also belong to Verrucomicrobia but were catalogued within the Class Subdivision 5, including taxa that we did not discuss here but may deserve attention in future studies.
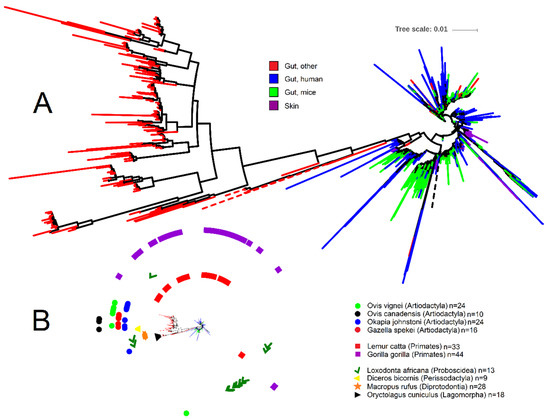
Figure 1.
Phylogenetic portrayal of all 1872 16S rRNA gene sequences classified as Akkermansia from the Ribosomal Database Project. (A) Circular phylogenetic tree showing the relationship of 16S rRNA gene sequences (note that most sequences belonging to the category of other animal species clustered together—“Gut, other” in red; other sequences from this category clustered with human and mice samples but are hard to visualize due to sequence overlap). The tree was generated using FastTree [76] in QIIME [77] 1.8. (B) Miniaturized circular tree from Figure 1A showing symbols for each animal species from the study by Ley et al. [68]. Akkermansia sequences clustered closely together for several animal species (Oryctolagus cuniculus, Diceros bicornis, and all animal species within the order Artiodactyla), suggesting host specialization. The most dispersed Akkermansia sequences belonged to the order of Primates (Lemur catta and Gorilla gorilla) and Proboscidea (Loxodonta africana). Note that a total of eight sequences from the order Perissodactyla (three sequences from Equus equus, one sequence from Equus grevyi, four sequences from Equus asinus), 17 sequences from the order Artiodactyla (three sequences from Budorcas taxicolor, four sequences from Sus cebifrons, six sequences from Babyrousa babyrussa, one sequence from Ovis ammon, one sequence from Giraffa camelopardalis reticulata, and two sequences from Antidorcas marsupialis), four sequences from the order Carnivora (Acinonyx jubatus), one sequence from the order Chiroptera (Pteropus giganteus), and two sequences from the order Rodentia (Callosciurus prevosti) are not shown for sake of visual clarity. Tree scale refers to sequence dissimilarity (the longer the scale the bigger the dissimilarity).
The analysis of Akkermansia-like16S rRNA gene sequences from RDP offered several important insights. First, the majority of Akkermansia-like 16S rRNA sequences belonging to non-human and non-mice gut contents clustered together (bootstrap support 0.862); of these, most belonged to the study of zoo animals (Figure 1). Second, this cluster showed a much stronger sequence dissimilarity compared to the rest of the sequences, as evidenced by the longer tree scale (Figure 1). Third, Akkermansia-like 16S rRNA sequences were present in only nine out of the 13 orders sampled by Ley et al. [68] (Figure 1). While this may suggest that Akkermansia-like bacteria is not universally distributed across the mammalian kingdom, as indicated above, this may also be due to limited sequence depth or absence in the animal species sampled. Fourth, Akkermansia-like 16S rRNA sequences from a given animal species sometimes clustered together (for example for the European rabbit Oryctolagus cuniculus and Black rhinoceros Diceros bicornis), thus suggesting host speciation even though different individuals from each animal species were sampled. However, one should keep in mind that the vast majority of samples derived from zoo animals that may have been housed together or exposed to the human species. This can be exemplified from the genomes of A. muciniphila isolated from laboratory mice that are almost identical to that of the A. muciniphila human type strain. On the other hand, several Akkermansia 16S rRNA sequences from other animal species (e.g., samples from Primates) were more dispersed in the tree and clustered together with human and mice sequences (Figure 1), thus suggesting that host-speciation is different among animal species. Finally, several sequences from two different studies [78,79] were obtained from skin, thus confirming the presence of this organism in this environment [80]. However, several of these sequences derived from breasts of women that may have been lactating and it is known that A. muciniphila is present in breast milk and can grow well on human milk oligosaccharides [71,81]. Overall, these insights suggest either a) that different animal species are colonized by different types of Akkermansia-like bacteria, or b) that the same types of Akkermansia-like bacteria colonize different hosts. This latter hypothesis has also been suggested by others [24], but it is very well possible that both options are feasible and not necessarily exclude each other.
Lastly, we and others suggest that 16S rRNA sequences from Akkermansia-like and other low abundant bacterial groups may not be detected even from massive sequencing surveys due to inadequate sequence depth (see “The feline and canine gut microbiota” below). Therefore, it is important to briefly discuss the numbers of A. muciniphila. Relative abundance of A. muciniphila in the colon of healthy humans is approximately 3% [63] (the minimum reported is often 0.1%, and the maximum reported is up to 85% [69,82]). Now, relative percentages of 16S rRNA gene reads cannot be accurately translated into cell numbers, but in an environment of 1 × 1011 bacteria/g of contents in the large intestine, these percentages would imply approximately 1 × 108 to 3 × 109 bacteria/g of contents. These hypothetical numbers of cells based on sequence reads agree with other studies that have looked at the actual numbers of A. muciniphila. Collado et al. [83] showed 1 × 108 cells/g in feces, and Derrien et al. [84] showed that A. muciniphila accounted for more than 1% of the total fecal cells, which in an environment of 1 × 1011 bacteria/g of contents would represent 1 × 109 cells. Another study showed that one single dose of 109 A. muciniphila to germ-free mice yielded 1.9 × 108 cells/g of ileal contents, 3.1 × 1010 cells/g cecal content, and 1.7 × 109 cells/g colonic content [23]. Another more recent study has also shown that viable A. muciniphila cells can be recovered from mice cecal and colon contents in a concentration of up to 1 × 1010 cells/g [70]. Overall, these studies agree with a possible concentration of 1 × 108–1 × 1010 in large intestinal contents. Now, in a regular sequencing survey using, for example, the Illumina MiSeq platform, an approximate 30,000 16S rRNA reads are analyzed, and 0.1–4% represents only 30–120 sequences. These estimations, albeit imprecise, strongly suggest that in the case that Akkermansia or other microbes are present with an abundance lower than 0.1% in sequencing surveys of 30,000 sequences/sample, we should not overlook taxa with less than 100 sequences, as done by others [85]. However, an important point to make here is that the presence of an Akkermansia-like 16S rRNA sequence in DNA isolated from a given sample does not necessarily supports the presence of Akkermansia-like bacteria in the habitat where that samples are taken from. Many artefacts related to cross-sample contaminations have been reported, notably with the use of sensitive PCR methods as well as 454 and Illumina sequencing approaches. Hence, relying on a limited number of sequences has a risk, and with the present new platforms, such as the Illumina NovaSeq, it may be possible to have very deep analyses of fecal and other samples, amounting to more than millions of reads.
4. The Feline and Canine Gut Microbiota
Similar to humans and other mammals, cats and dogs harbor a complex assemblage of different types of microorganisms inside their digestive tracts. Also, each section of the GI tract has different physiological conditions, including complex defense mechanisms [86] that in turn help develop different microbial communities [87,88,89,90,91].
As indicated above, the development of microbial therapies with weight-loss applications in obese cats and dogs is challenging [7], in part due to the little we know about microbes and obesity [92,93] in these animal species and the wide inter-individual variation in the gut microbiome. This variation in the gut microbiome in pets is associated with the passing of time and aging [94,95,96,97,98], environmental factors, including diet [99], microbes in surrounding people [100], breed and other host genetics factors [101,102], clinical and subclinical conditions [91,93,103], gender [104], and behavior [105]. Here, we discuss the literature on feline and canine gut microbiota with regards to Verrucomicrobia- and Akkermansia-like 16S rRNA sequences (Table 1).

Table 1.
Summary of studies on the feline and canine gut microbiota based on molecular methods 1.
4.1. Oral Cavity
Verrucomicrobia has not been detected in the oral microbiota of cats [115,116,118]. An early molecular description of the canine oral microbiota (5958 full-length 16S rRNA gene sequences) did not find any sequence belonging to Verrucomicrobia in this environment [112]. Other studies of the canine oral microbiota have also failed to show the presence of Verrucomicrobia [147,148]. Similarly, Verrucomicrobia have not been detected in the human oral cavity [58,149,150,151,152], although a single report describes the presence of Akkermansia-like sequences in the oral cavity of a choledocholithiasis patient [153]. The Human Oral Microbiome Database [154,155] also does not contain any information about Verrucomicrobia. These studies strongly suggest that Akkermansia is absent from the healthy oral cavity of humans, cats, and dogs, something that could be explained by its strict anaerobic nature as described by Derrien et al. [55] although the type strain can tolerate and use small amounts of oxygen [156]. However, several anaerobes have been described and isolated from the human oral microbiota, but these often include pathogens that are colonizing deep and anerobic pockets or are in specific biofilms [43,157]. However, recent data indicate that A. muciniphila can be used to prevent some pathogens like the anaerobic Poryphyromonas gingivalis to produce gingivitis and cause bone loss in mouse models [158].
4.2. Stomach
The studies of the feline gastric microbiota have focused on specific bacteria such as Helicobacter [159] and no Verrucomicrobia was detected in the stomach of healthy dogs using high-throughput 16S rRNA sequencing [104]. Similarly, Verrucomicrobia has not been detected in some studies of the human gastric microbiota [160,161]. This makes sense at least when considering that the original A. muciniphila strain MucT did not grow below pH 5.5 or above pH 8 [55] while A. glycaniphila grew best between pH 5.0 and pH 7.5 [57]. However, it remains possible that other and novel Akkermansia species may grow at more extreme pH values.
4.3. Intestinal Tract
Verrucomicrobia has been detected in very low abundance (0.1%, [110]) or not at all [41,108] in the canine duodenum. Moreover, an analysis of almost all sections of the digestive tract of cats [106] and dogs [107] did not detect Verrucomicrobia. More recent studies of the small and large mucosal microbiota have also failed to detect Verrucomicrobia in dogs [91] or not attempted to look for this group at all [162]. These and other studies (Table 1) strongly suggest that Akkermansia has a very low prevalence and is not an abundant member of the microbiota in the small intestine of cats and dogs. Possible explanations for this include the fact that the mucosal layer in the small intestine is thinner and less dense. This fact and other biochemical conditions (e.g., presence of bile) may limit microbial life in this section of the GI tract. An alternative explanation can be the fact that dogs, and to some extent also cats, are highly inbred and their microbiota may be dependent on the breeders. It has been shown that laboratory mice derived from different suppliers have a greatly variable microbiome, including a highly varying level of Akkermansia spp. [163].
A great number of studies have analyzed the fecal microbiota in cats and dogs, and most have failed to detect Verrucomicrobia. Exceptions include the study by Handl et al. [111] where Akkermansia was not detected in any fecal sample from 12 healthy pet dogs and in only 1 fecal sample from 12 healthy pet cats, albeit in very low proportion (0.01%) (in relation to the numbers discussed above, it is interesting that this study used 454-pyrosequencing and generated only ~5000 sequences per sample, and 0.01% of these equals approximately one sequence only). In another study comparing the fecal microbiota between lean (n = 21) and obese (n = 22) dogs (also using low-throughput pyrosequencing), Verrucomicrobia were detected in only one lean dog and in very low abundance (<0.01% [92]). A recent study of household pet cats (n = 46) and dogs (n = 192) showed that the abundance of fecal Verrucomicrobia was 0.11% in cats and 0.02% in dogs [138]. Similarly, some studies of the human fecal microbiota have not detected this group either [164] and others have detected it in high levels in only a few sections of the digestive tract [165], suggesting that either Akkermansia is not that universally distributed or that its abundance in some individuals is simply undetectable. One more recent study used a FISH probe developed previously [84] to look for the first time into the abundance of Akkermansia in colonic mucosa of healthy dogs and dogs with chronic enteropathy [136]. Interestingly, the abundance of Akkermansia was similar to other microbes such as Faecalibacterium, and it was statistically higher in healthy dogs. This topic indeed deserves more attention with regards to prevalence and abundance, for instance one study of non-human primates showed that Verrucomicrobia was relatively highly abundant in feces (0.3–2.6%) but common to all nine subjects from three different primate species [166].
4.4. Verrucomicrobia in Other Anatomical Areas
While A. muciniphila is often regarded as part of the human colonic mucus-associated microbiota, recently, the intriguing possibility was discussed of having Akkermansia in other anatomical areas such as the pancreas and gallbladder [58]. However, only few studies have analyzed bacteria and other microbes in these organs in cats or dogs [167]. To our knowledge, Akkermansia and other members of Verrucomicrobia have not been searched or detected in these sites. Moreover, Verrucomicrobia has not been detected in the urinary microbiome of healthy dogs [168], the microbiota in bronchoalveolar fluid and blood of healthy cats [169], or the upper and lower airway microbiota of healthy dogs [170]. Verrucomicrobia have not been detected in the skin microbiome of healthy and allergic cats and dogs [171,172,173], and this deserves special attention because Verrucomicrobia has been found in human skin [80].
5. Why is Akkermansia So Rare in the Digestive Tract of Cats and Dogs?
The presence or absence of certain types of microorganisms in specific ecological niches is intriguing and has important implications in several fields of science [174,175]. In the case of the GI tract of humans and animals, this topic becomes especially relevant in a context of health and disease. On one extreme, we have the abundant and prevalent microbes, sometimes referred to as the core microbiota, a continuously revisited concept [176]. Several efforts have been directed to identify a possible relationship between this core microbiota with intestinal symptoms of disease [15] or metabolic diseases such as obesity [177]. In this context of core microbiota, it is interesting that Akkermansia has been shown to co-occur with two of the three main enterotypes in humans as previously described based on metagenomic information [178].
On the other extreme, we have the rare and less prevalent microbes, which have been detected only in a few individuals and in very low abundance or not at all. Unfortunately, we know little about the contribution of these low abundant groups to overall gut homeostasis [179]; in fact, some researchers suggest and promote the removal of low abundant microbes from analysis in order to detect “true” phylotypes [180]. This is not a trivial topic, at least in other environments such as soil it has been shown that the low-abundance bacteria play a fundamental biological role [181] and may, in fact, be keystone species regulating the function of different microbial environments, including host-associated microbiomes [182].
With few exemptions, the literature search found no evidence to believe that Akkermansia is present in high prevalence or abundance in any portion of the digestive tract of cats and dogs. Therefore, we suggest that this bacterium (and in fact the whole Verrucomicrobia) is not part of the core feline or canine gut microbiota. But why not? One technical reason may relate to sequencing depth and this has also been suggested by others [24]; in fact, based on our calculations described above, a relative abundance lower than 0.1% would be difficult to detect when having up to 30,000 sequences per sample. In our own experience, we have detected high levels (up to 5%) of Akkermansia in fecal samples from mice [183] using the same sequencing depth (minimum 60,000 sequences per sample) as some of our more recent studies in cats and dogs. Another reason may be related to the use of 16S rRNA primers and probes. Members of Verrucomicrobia are deeply rooted and some new species may not be detected with the canonical universal primers. This is particularly important when using other techniques such as FISH because the 16S rRNA sequence of A. muciniphila contains two mismatches with the traditional universal bacterial probe EUB 338 [84]. Another reason for not having Akkermansia may be related to the reference sequence database used to select OTUs (for example, the one from GreenGenes). However, even the earliest version of this database (August 2012) already contained sequences belonging to Akkermansia spp. and other taxa within the Verrucomicrobia. Another possibility is that the Akkermansia-like sequences found in the studies of the cat and dog microbiota were very divergent compared to the reference A. muciniphila or A. glycaniphila sequences and hence did not match with the reference sequences (the fate of these sequences will depend on the OTU clustering method, some methods, such as the pick_closed_reference_otus.py in QIIME1, discard the most divergent sequences to be able to detect only “true” phylotypes). This is indeed a possibility especially when considering, for example, the existence of non-Akkermansia-like sequences in databases such as the Class Subdivision 5 in RDP from the same samples published previously [68]. It should also be noted that some studies have shown a high abundance of Verrucomicrobia (~4%) in feces of horses [184], while it was shown that Akkermansia spp. were present in only a few samples of horses or other members of the order Perissodactyla [68]. Indeed, as indicated previously, the nature and divergence of 16S rRNA genes of Akkermansia-related organisms is worth investigating [24].
If we assume the obligatory existence of bacteria that degrade mucin and other host-compounds, then other bacteria must be playing the function of Akkermansia spp. in cats and dogs. In fact, using an elegant approach, it was shown that Bacteroides acidifaciens was also an important host-compound forager in vivo, and A. muciniphila and B. acidifaciens received more attention in this study only because they were the more abundant ones (other microorganisms such as members of Ruminococcaceae, Lactobacillaceae, Enterococcaceae, and Mucispirillum were also enriched in host-secreted proteins) [185]. Indeed, mucin degradation is phylogenetically widespread [186], and actually, the outer mucus layer contains bacteria without specialized mucolytic capabilities [187]. This has been elegantly explained by the fact that the structural heterogeneity of the substrates and the associated enzymatic diversity required for complete mucus degradation needs an assortment of many different microorganisms [188]. In dogs, different microorganisms have been shown in mucosal samples of healthy dogs (e.g., Bacteroidaceae, Prevotellaceae, Clostridiales, Faecalibacterium, and the Fusobacteria phylum [41,136], and these groups may in fact represent some of the native mucus degraders in these animal species.
6. Implications of Microbial Mucus Degradation for Health in Cats and Dogs
Microbial mucus degradation is an important phenomenon to consider in feline and canine gut health. Several lines of research can emerge from not having Akkermansia spp. in cats and dogs in high prevalence or abundance.
More research is needed to investigate the type of mucins produced and secreted by the GI tract of cats and dogs. For example, more than 20 genes encoding mucins have been identified in humans, and different mucins have been found in the different sections, including the stomach [26]. Further, more research is necessary to reveal the identities and quantities of the most predominant mucus-degraders as well as the extent of host-compound foraging in cats and dogs. For this end, one can use a similarly elegant but complex approach as used previously with high-resolution secondary ion mass spectrometry (NanoSIMS) combined with fluorescence in situ hybridization to determine the isotope content of individual cells hybridized with specific phylogenetic probes [185]. Note that whether A. muciniphila is also a major host-compound forager in humans has not been proven in such studies that use labeled amino acids.
Obesity is a growing problem in cats and dogs and several lines of research point out a potential role of the gut microbiota in this disorder [189,190,191,192,193]; therefore, several efforts have been directed to help these patients but mostly centered around dietary therapies [3]. A. muciniphila is currently considered a beneficial microbe that could help in the treatment of obesity, diabetes, and associated disorders in humans when administered orally, either alive or even pasteurized [56,194,195]. Given the increased prevalence of obesity in cats and dogs, the question remains as to whether Akkermansia spp. could also be considered as an option for a therapy for obesity in these animal species. Note that probiotics do not necessarily need to be isolated from the same animal species, or even from the same environment, in order to survive and colonize the intestinal tract temporarily [196]. Therefore, even if Akkermansia spp. are not considered native to the feline and canine gut, both A. muciniphila or A. glycaniphila could nonetheless be considered for these animal species. Another option to consider is the use of a purified membrane protein from A. muciniphila that interacts with Toll-like receptor (TLR) 2 and recapitulates the beneficial effects of the bacterium [195]. This correlation with TLR2 is important, because of its involvement in maintaining intestinal homeostasis, bacterial recognition, and host metabolism [197]. The signaling to TLR2 is also particularly relevant in cases of IBD in veterinary medicine [198].
Dietary strategies can also be developed to target native mucus degraders in cats and dogs. For example, prebiotic administration helps increase the levels of Akkermansia likely due to an increase in the number of goblet cells and mucus layer thickness [20]. Although the overall effect of prebiotics on mucus foragers remains to be investigated in cats and dogs, it was recently shown that cherry powder (high in prebiotics and polyphenolics) led to a five-fold difference in relative abundance of Akkermansia in feces of obese db/db mice compared to both lean and obese controls [183]. It is likely that at least some prebiotics strengthen gut health through an indirect effect on mucus microbial degraders in cats and dogs [199]. Other dietary alternatives to boost local mucus degraders include the use of polyphenols [200]. Finally, the growing conditions of Akkermansia and other microbes can be modified to obtain a different phenotype [201], but few data exist regarding the biochemical composition of intestinal mucus in cats and dogs to hypothesize what culture condition may lead to potentially advantageous down- or up-regulation of specific genes.
Another important line of research worth investigating is whether the presence of Akkermansia spp. and other mucus degraders in the mucus correlate with its abundance in luminal contents and feces, or if there is variation in the mucus adhesion properties as to open the possibility that the shed bacteria are different from those that are fixed into the mucosal layer. This may explain the apparent low concentration of Akkermansia in feces of cats and dogs because these bacteria may be closely attached to the mucosa, thus reducing its loss in feces. Interestingly, with other microbes such as in some strains of Lactobacillus, there is great variation in mucin adhesion properties and structures [202]. Therefore, it is feasible to hypothesize that each Akkermansia species or strain displays variation in their behavior in vivo, a possibility that has been proven in silico [67]. Feces of cats and dogs may contain a high degree of intestinal mucus as shown by studies in humans [40]. In this regard, it was reported that differences between lumen and mucosal communities may be quite small as a consequence of extensive mucus shedding and mixing in the lumen [175]. This is particularly important in light of new evidence suggesting the presence of potentially undiscovered A. muciniphila (see above).
Finally, it has been suggested that bacteria that live in the mucosa do not compete with the microbiota present in the lumen and therefore do not depend on dietary nutrients deriving from host food consumption [23,24]. However, the biochemical properties of mucin depend on the diet [203,204] and the glycan repertoire can select for distinct mucosa-associated bacteria [205]. Therefore, diet is also strongly, albeit indirectly, associated with the microbes attached to the mucosa. Moreover, it is unlikely that bacteria that are shed into the lumen cease all activities and die. In fact, ingested and salivary microbes may integrate into the native microbiome, although the alterations in the large intestine are mostly of limited extent [206,207].
7. Conclusions
In summary, microbial degradation of mucus is important for GI health and disease of cats and dogs, but this topic has received very little attention. Akkermansia is apparently not that universal across the animal kingdom, and thus far, there is no indication to believe that this taxon is prevalent or highly abundant in the digestive tract of cats and dogs. Other bacteria that have been detected in intestinal mucosa, such as members of Bacteroidaceae, Prevotellaceae, Clostridiales, Faecalibacterium, and the poorly studied Fusobacteria phylum, may deserve more attention as possible contributors to mucus degradation in the gut of cats and dogs. Remaining challenges include more research into the identities of mucus foragers in these companion animals, the search for dietary alternatives to boost these native mucus degraders, and the fate of mucosal bacteria shed into the lumen.
Author Contributions
All authors contributed equally to this review. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
The authors would like to thank Aashish Jha for revising the text and providing helpful comments and suggestions.
Conflicts of Interest
Jose F. Garcia-Mazcorro is an employee of MNA de Mexico, a company devoted to Animal Nutrition. Willem M de Vos is the founder and shareholder of A-mansia Biotech, a company developing Akkermansia spp. and their products.
References
- Waxman, A. World Health Assembly 2004. WHO global strategy on diet, physical activity and health. Food Nutr. Bull. 2004, 25, 292–302. [Google Scholar]
- Association for Pet Obesity Prevention. Available online: https://petobesityprevention.org/ (accessed on 26 November 2019).
- German, A.J. The growing problem of obesity in dogs and cats. J. Nutr. 2006, 136, 1940S–1946S. [Google Scholar] [CrossRef] [PubMed]
- McGreevy, P.D.; Thomson, P.C.; Pride, C.; Fawcett, A.; Grassi, T.; Jones, B. Prevalence of obesity in dogs examined by Australian veterinary practices and the risk factors involved. Vet. Rec. 2011, 156, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Crane, S.W. Occurrence and management of obesity in companion animals. J. Small Anim. Pract. 1991, 32, 275–282. [Google Scholar] [CrossRef]
- Brusaferro, A.; Cozzali, R.; Orabona, C.; Biscarini, A.; Farinelli, E.; Cavalli, E.; Grohmann, U.; Principi, N.; Esposito, S. Is it time to use probiotics to prevent or treat obesity? Nutrients 2018, 10, 1613. [Google Scholar] [CrossRef]
- Grześkowiak, Ł.; Endo, A.; Beasley, S.; Salminen, S. Microbiota and probiotics in canine and feline welfare. Anaerobe 2015, 34, 14–23. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef]
- Hill, D.A.; Artis, D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu. Rev. Immunol. 2010, 28, 623–667. [Google Scholar] [CrossRef]
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef]
- Ansaldo, E.; Slayden, L.C.; Ching, K.L.; Koch, M.A.; Wolf, N.K.; Plichta, D.R.; Brown, E.M.; Graham, D.B.; Xavier, R.J.; Moon, J.J.; et al. Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science 2019, 364, 1179–1184. [Google Scholar] [CrossRef]
- Million, M.; Tomas, J.; Wagner, C.; Lelouard, H.; Raoult, D.; Gorvel, J.P. New insights in gut microbiota and mucosal immunity of the small intestine. Hum. Microbiome J. 2018, 7, 23–32. [Google Scholar] [CrossRef]
- Zoetendal, E.G.; Akkermans, A.D.L.; de Vos, W.M. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 1998, 64, 3854–3859. [Google Scholar] [CrossRef] [PubMed]
- The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Jalanka-Tuovinen, J.; Salonen, A.; Nikkilä, J.; Immonen, O.; Kekkonen, R.; Lahti, L.; Palva, A.; de Vos, W.M. Intestinal microbiota in healthy adults: Temporal analysis reveals individual and common core and relation to intestinal symptoms. PLoS ONE 2011, 6, e23035. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Costello, E.K.; Berg-Lyons, D.; Gonzalez, A.; Stombaugh, J.; Knights, D.; Gajer, P.; Ravel, J.; Fierer, N.; et al. Moving pictures of the human microbiome. Genome Biol. 2011, 12, R50. [Google Scholar] [CrossRef] [PubMed]
- Davidson, C.J.; Surette, M.G. Individuality in bacteria. Annu. Rev. Genet. 2008, 42, 253–268. [Google Scholar] [CrossRef]
- Bäckhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Young Koh, G.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Bäckhed, F.; Fulton, L.; Gordon, J.I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 2008, 3, 213–223. [Google Scholar] [CrossRef]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef]
- Maruvada, P.; Leone, V.; Kaplan, L.M.; Chang, E.B. The human microbiome and obesity: Moving beyond associations. Cell Host Microbe 2017, 22, 589–599. [Google Scholar] [CrossRef]
- Pasolli, E.; Asnicar, F.; Manara, S.; Zolfo, M.; Karcher, N.; Armanini, F.; Beghini, F.; Manghi, P.; Tett, A.; Ghensi, P.; et al. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell 2019, 176, 649–662. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; van Baarlen, P.; Hooiveld, G.; Norin, E.; Muller, M.; de Vos, W.M. Modulation of mucosal immune response, tolerance and proliferation in mice colonized by the mucin-degrader Akkermansia muciniphila. Front. Cell Infect. Microbiol. 2011, 2, 166. [Google Scholar] [CrossRef] [PubMed]
- Belzer, C.; de Vos, W.M. Microbes inside—From diversity to function: The case of Akkermansia. ISME J. 2012, 6, 1449–1458. [Google Scholar] [CrossRef]
- Johansson, M.E.; Ambort, D.; Pelaseyed, T.; Schütte, A.; Gustafsson, J.K.; Ermund, A.; Subramani, D.B.; Holmén-Larsson, J.M.; Thomsson, K.A.; Bergström, J.H.; et al. Composition and functional role of the mucus layers in the intestine. Cell Mol. Life Sci. 2011, 68, 3635–3641. [Google Scholar] [CrossRef] [PubMed]
- Tailford, L.E.; Crost, E.H.; Kavanaugh, D.; Juge, N. Mucin glycan foraging in the human gut microbiome. Front. Genet. 2015, 6, 81. [Google Scholar] [CrossRef] [PubMed]
- Taherali, F.; Varum, F.; Basit, A.W. A slippery slope: On the origin, role and physiology of mucus. Adv. Drug Deliv. Rev. 2017, 124, 16–33. [Google Scholar] [CrossRef] [PubMed]
- Ermund, A.; Schütte, A.; Johansson, M.E.V.; Gustafsson, J.K.; Hansson, G.C. Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer’s patches. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 305, G341–G347. [Google Scholar] [CrossRef]
- Birchenough, G.M.H.; Johansson, M.E.V.; Gustafsson, J.K.; Bergström, J.H.; Hansson, G.C. New developments in goblet cell mucus secretion and function. Mucosal Immunol. 2015, 8, 712–719. [Google Scholar] [CrossRef]
- Johansson, M.E.; Hansson, G.C. Immunological aspects of intestinal mucus and mucins. Nat. Rev. Immunol. 2016, 16, 639–649. [Google Scholar] [CrossRef]
- Allaire, J.M.; Morampudi, V.; Crowley, S.M.; Stahl, M.; Yu, H.; Bhullar, K.; Knodler, L.A.; Bressler, B.; Jacobson, K.; Vallance, B.A. Frontline defenders: Goblet cell mediators dictate host-microbe interactions in the intestinal tract during health and disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2017, 314, G360–G377. [Google Scholar] [CrossRef]
- Atuma, C.; Strugula, V.; Allen, A.; Holm, L. The adherent gastrointestinal mucus gel layer: Thickness and physical state in vivo. Am. J. Physiol. 2001, 280, G922–G929. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.E.V.; Phillipson, M.; Petersson, J.; Velcich, A.; Holm, L.; Hansson, G.C. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 15064–15069. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.E.V.; Hansson, G.C. Mucus and the goblet cell. Dig. Dis. 2013, 31, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, H.E.; Rodriguez-Piñeiro, A.M.; Schütte, A.; Ermund, A.; Boysen, P.; Bemark, M.; Sommer, F.; Bäckhed, F.; Hansson, G.C.; Johansson, M.E. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep. 2015, 16, 164–177. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Piñeiro, A.M.; Johansson, M.E. The colonic mucus protection depends on the microbiota. Gut Microbes 2015, 6, 326–330. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.E.; Larsson, J.M.; Hansson, G.C. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc. Natl. Acad. Sci. USA 2011, 108, 4659–4665. [Google Scholar] [CrossRef] [PubMed]
- Ouwehand, A.C.; Derrien, M.M.N.; de Vos, W.M.; Tiihonen, K.; Rautonen, N. Prebiotics and other microbial substrates for gut functionality. Curr. Opin. Biotechnol. 2005, 16, 212–217. [Google Scholar] [CrossRef]
- Johansson, M.E.V.; Jakobsson, H.E.; Holmén-Larsson, J.; Schütte, A.; Ermund, A.; Rodríguez-Piñeiro, A.M.; Arike, L.; Wising, C.; Svensson, F.; Bäckhed, F.; et al. Normalization of host intestinal mucus layers requires long-term microbial colonization. Cell Host Microbe 2015, 18, 582–592. [Google Scholar] [CrossRef]
- Swidsinski, A.; Loening-Baucke, V.; Vaneechoutte, M.; Doerffel, Y. Active Chron’s disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflamm. Bowel Dis. 2008, 14, 147–161. [Google Scholar] [CrossRef]
- Suchodolski, J.S.; Dowd, S.E.; Wilke, V.; Steiner, J.M.; Jergens, A.E. 16S rRNA gene pyrosequencing reveals bacterial dysbiosis in the duodenum of dogs with idiopathic inflammatory bowel disease. PLoS ONE 2012, 7, e39333. [Google Scholar] [CrossRef]
- Capaldo, C.T.; Powell, D.N.; Kalman, D. Layered defense: How mucus and tight junctions seal the intestinal barrier. J. Mol. Med. 2017, 95, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Tytgat, H.L.P.; Nobrega, F.L.; van der Oost, J.; de Vos, W.M. Bowel biofilms: Tipping points between a healthy and compromised gut? Trends Microbiol. 2019, 27, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Sakata, T.; von Engelhardt, W. Influence of short-chain fatty acids and osmolality on mucin release in the rat colon. Cell Tissue Res. 1981, 219, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Ten Bruggencate, S.J.; Bovee-Oudenhoven, I.M.; Lettink-Wissink, M.L.; Katan, M.B.; van der Meer, R. Dietary fructooligosaccharides affect intestinal barrier function in healthy men. J. Nutr. 2006, 136, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Monk, J.M.; Lepp, D.; Wu, W.; Pauls, K.P.; Robinson, L.E.; Power, K.A. Navy and black bean supplementation primes the colonic mucosal microenvironment to improve gut health. J. Nutr. Biochem. 2017, 49, 89–199. [Google Scholar] [CrossRef]
- Culling, C.F.; Reid, P.E.; Dunn, W.L.; Clay, M.G. Histochemical comparison of the epithelial mucins in the ileum in Crohn’s disease and in normal controls. J. Clin. Pathol. 1977, 30, 1063–1067. [Google Scholar] [CrossRef]
- Filipe, M.I.; Fenger, C. Histochemical characteristics of mucins in the small intestine. A comparative study of normal mucosa, benign epithelial tumours and carcinoma. Histochem. J. 1979, 11, 277–287. [Google Scholar] [CrossRef]
- Mantle, M.; Forstner, G.G.; Forstner, J.F. Antigenic and structural features of goblet-cell mucin of human small intestine. Biochem. J. 1984, 217, 159–167. [Google Scholar] [CrossRef]
- Bansil, R.; Turner, B.S. The biology of mucus: Composition, synthesis and organization. Adv. Drug Deliv. Rev. 2017, 124, 3–15. [Google Scholar] [CrossRef]
- Forstner, J.F.; Jabbal, I.; Forstner, G.G. Goblet cell mucin of rat small intestine. Chemical and physical characterization. Can. J. Biochem. 1973, 51, 1154–1166. [Google Scholar] [CrossRef]
- Rodriguez-Piñeiro, A.M.; Bergström, J.H.; Ermund, A.; Gustafsson, J.K.; Schütte, A.; Johansson, M.E.V.; Hansson, G.C. Studies of mucus in mouse stomach, small intestine, and colon. II. Gastrointestinal mucus proteome reveals Muc2 and Muc5ac accompanied by a set of core proteins. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 305, G348–G356. [Google Scholar]
- Holmén Larsson, J.M.; Thomsson, K.A.; Rodríguez-Piñeiro, A.M.; Karlsson, H.; Hansson, G.C. Studies of mucus in mouse stomach, small intestine, and colon. III. Gastrointestinal Muc5ac and Muc2 mucin O-glycan patterns reveals a regiospecific distribution. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 305, G357–G363. [Google Scholar]
- Qureshi, R.; Forstner, G.G.; Forstner, J.F. Radioimmunoassay of human intestinal goblet cell mucin. Investigation of mucus from different organs and species. J. Clin. Investig. 1979, 64, 1149–1156. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; Vaughan, E.E.; Plugge, C.M.; de Vos, W.M. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 1469–1476. [Google Scholar] [CrossRef]
- Depommier, C.; Everard, A.; Druart, C.; Plovier, H.; Van Hul, M.; Vieira-Silva, S.; Falony, G.; Raes, J.; Maiter, D.; Delzenne, N.M.; et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: A proof-of-concept exploratory study. Nat. Med. 2019, 25, 1096–1103. [Google Scholar] [CrossRef]
- Ouwerkerk, J.P.; Aalvink, S.; Belzer, C.; de Vos, W.M. Akkermansia glycaniphila sp. nov.: An anaerobic mucin-degrading bacterium isolated from reticulated python faeces. Int. J. Syst. Evol. Microbiol. 2016, 66, 4614–4620. [Google Scholar] [CrossRef]
- Geerlings, S.Y.; Kostopoulos, I.; de Vos, W.M.; Belzer, C. Akkermansia muciniphila in the human gastrointestinal tract: When, where, and how? Microorganisms 2018, 6, 75. [Google Scholar] [CrossRef]
- Kirmiz, N.; Galindo, K.; Cross, K.L.; Luna, E.; Rhoades, N.; Podar, M.; Flores, G.E. Comparative genomics guides elucidation of vitamin B12 biosynthesis in novel human associated Akkermansia. Appl. Environ. Microbiol. 2020, 86, e02117–e02119. [Google Scholar] [CrossRef]
- Xing, J.; Li, X.; Sun, Y.; Zhao, J.; Miao, S.; Xiong, Q.; Zhang, Y.; Zhang, G. Comparative genomic and functional analysis of Akkermansia muciniphila and closely related species. Genes Genom. 2019, 41, 1253–1264. [Google Scholar] [CrossRef]
- Derrien, M.; van Passel, M.W.J.; van de Bovenkamp, J.H.B.; Schipper, R.G.; de Vos, W.M.; Dekker, J. Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut Microbes 2010, 1, 254–268. [Google Scholar] [CrossRef]
- Gómez-Gallego, C.; Pohl, S.; Salminen, S.; de Vos, W.M.; Kneifel, W. Akkermansia muciniphila: A novel functional microbe with probiotic properties. Benef. Microbes 2016, 7, 571–584. [Google Scholar] [CrossRef] [PubMed]
- De Vos, W.M. Microbe profile: Akkermansia muciniphila: A conserved intestinal symbiont that acts as the gatekeeper of our mucosa. Microbiology 2017, 163, 646–648. [Google Scholar] [CrossRef] [PubMed]
- Jugan, M.C.; Rudinsky, A.J.; Gordon, A.; Kramer, D.L.; Daniels, J.B.; Paliy, O.; Boyaka, P.; Gilor, C. Effects of oral Akkermansia muciniphila supplementation in healthy dogs following antimicrobial administration. Am. J. Vet. Res. 2018, 79, 884–892. [Google Scholar] [CrossRef] [PubMed]
- Schlesner, H. Verrucomicrobium spinosum gen. nov., sp. nov.: A fimbriated prosthecate Bacterium. System. Appl. Microbiol. 1987, 10, 54–56. [Google Scholar] [CrossRef]
- Wagner, M.; Horn, M. The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr. Opin. Biotechnol. 2006, 17, 241–249. [Google Scholar] [CrossRef]
- Guo, X.; Li, S.; Zhang, J.; Wu, F.; Li, X.; Wu, D.; Zhang, M.; Ou, Z.; Jie, Z.; Yan, Q.; et al. Genome sequencing of 39 Akkermansia muciniphila isolates reveals its population structure, genomic and functional diversity, and global distribution in mammalian gut microbiotas. BMC Genom. 2017, 18, 800. [Google Scholar] [CrossRef]
- Ley, R.E.; Hamady, M.; Lozupone, C.; Turnbaugh, P.; Ramey, R.R.; Bircher, J.S.; Schlegel, M.L.; Tucker, T.A.; Schrenzel, M.D.; Knight, R.; et al. Evolution of mammals and their gut microbes. Science 2008, 320, 1647–1651. [Google Scholar] [CrossRef]
- Van Passel, M.W.J.; Kant, R.; Zoetendal, E.G.; Plugge, C.M.; Derrien, M.; Malfatti, S.A.; Chain, P.S.G.; Woyke, T.; Palva, A.; de Vos, W.M.; et al. The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS ONE 2011, 6, e16876. [Google Scholar] [CrossRef]
- Ouwerkerk, J.P.; Aalvink, S.; Belzer, C.; de Vos, W.M. Preparation and preservation of viable Akkermansia muciniphila cells for therapeutic interventions. Benef. Microbes 2017, 8, 163–169. [Google Scholar] [CrossRef]
- Ottman, N.A. Host Immunostimulation and Substrate Utilization of the Gut Symbiont Akkermansia muciniphila. Ph.D. Thesis, Wageningen University, Wageningen, The Netherdlands, 2015. [Google Scholar]
- Jaspers, E.; Overmann, J. Ecological significance of microdiversity: Identical 16S rRNA gene sequences can be found in bacteria with highly divergent genomes and ecophysiologies. Appl. Environ. Microbiol. 2004, 70, 4831–4839. [Google Scholar] [CrossRef]
- Sharma, V.K.; Akavaram, S.; Schaut, R.G.; Bayles, D.O. Comparative genomics reveals structural and functional features specific to the genome of a foodborne Escherichia coli. BMC Genom. 2019, 20, 196. [Google Scholar] [CrossRef]
- Chun, J.; Rainey, F.A. Integrating genomics into the taxonomy and systematics of the Bacteria and Archaea. Int. J. Syst. Evol. Microbiol. 2014, 64, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; NISC Comparative Sequencing Program; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; et al. Topographical and temporal diversity of the human skin microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef]
- Kong, H.H.; Oh, J.; Deming, C. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012, 22, 850–859. [Google Scholar] [CrossRef]
- Urbaniak, C.; Cummins, J.; Brackstone, M.; MacKlaim, J.M.; Gloor, G.B.; Baban, C.K.; Scott, L.; O’Hanlon, D.M.; Burton, J.P.; Francis, K.P.; et al. Microbiota of human breast tissue. Appl. Environ. Microbiol. 2014, 80, 3007–3014. [Google Scholar] [CrossRef]
- Collado, M.C.; Laitinen, K.; Salminen, S.; Isolauri, E. Maternal weight and excessive weight gain during pregnancy modify the immunomodulatory potential of breast milk. Pediatr. Res. 2012, 72, 77–85. [Google Scholar] [CrossRef]
- Dubourg, G.; Lagier, J.C.; Armougom, F.; Robert, C.; Audoly, G.; Papazian, L.; Raoult, D. High-level colonisation of the human gut by Verrucomicrobia following broad-spectrum antibiotic treatment. Int. J. Antimicrob. Agents. 2013, 41, 149–155. [Google Scholar] [CrossRef]
- Collado, M.C.; Derrien, M.; Isolauri, E.; de Vos, W.M.; Salminen, S. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl. Environ. Microbiol. 2007, 73, 7767–7770. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; Collado, M.C.; Ben-Amor, K.; Salminen, S.; de Vos, W.M. The mucin degrader Akkermansia muciniphila is an abundant resident of the human intestinal tract. Appl. Environ. Microbiol. 2008, 74, 1646–1648. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Stombaugh, J.; Gonzalez, A.; Ackermann, G.; Wendel, D.; Vázquez-Baeza, Y.; Jansson, J.K.; Gordon, J.I.; Knight, R. Meta-analyses of studies of the human microbiota. Genome Res. 2013, 23, 1704–1714. [Google Scholar] [CrossRef] [PubMed]
- Stokes, C.; Waly, N. Mucosal defence along the gastrointestinal tract of cats and dogs. Vet. Res. 2006, 37, 281–293. [Google Scholar] [CrossRef][Green Version]
- Suchodolski, J.S. Intestinal microbiota of dogs and cats: A bigger world than we thought. Vet. Clin. North Am. Small Anim. Pract. 2011, 41, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Minamoto, Y.; Hooda, S.; Swanson, K.S.; Suchodolski, J.S. Feline gastrointestinal microbiota. Anim. Health Res. Rev. 2012, 13, 64–77. [Google Scholar] [CrossRef]
- Hooda, S.; Minamoto, Y.; Suchodolski, J.S.; Swanson, K.S. Current state of knowledge: The canine gastrointestinal microbiome. Anim. Health Res. Rev. 2012, 13, 78–88. [Google Scholar] [CrossRef]
- Redfern, A.; Suchodolski, J.S.; Jergens, A. Role of the gastrointestinal microbiota in small animal health and disease. Vet Rec. 2017, 181, 370. [Google Scholar] [CrossRef]
- Kalenyak, K.; Isaiah, A.; Heilmann, R.M.; Suchodolski, J.S.; Burgener, I.A. Comparison of the intestinal mucosal microbiota in dogs diagnosed with idiopathic inflammatory bowel disease and dogs with food-responsive diarrhea before and after treatment. FEMS Microbiol. Ecol. 2018, 94. [Google Scholar] [CrossRef]
- Handl, S.; German, A.J.; Holden, S.L.; Dowd, S.E.; Steiner, J.M.; Heilmann, R.M.; Grant, R.W.; Swanson, K.S.; Suchodolski, J.S. Faecal microbiota in lean and obese dogs. FEMS Microbiol. Ecol. 2013, 84, 332–343. [Google Scholar] [CrossRef]
- Forster, G.M.; Stockman, J.; Noyes, N.; Heuberger, A.L.; Broeckling, C.D.; Bantle, C.M.; Ryan, E.P. A comparative study of serum biochemistry, metabolome and microbiome parameters of clinically healthy, normal weight, overweight, and obese companion dogs. Top. Companion Anim. M. 2018, 33, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Mazcorro, J.F.; Dowd, S.E.; Poulsen, J.; Steiner, J.M.; Suchodolski, J.S. Abundance and short-term temporal variability of fecal microbiota in healthy dogs. MicrobiologyOpen 2012, 1, 340–347. [Google Scholar] [CrossRef]
- Deusch, O.; O’Flynn, C.; Colyer, A.; Swanson, K.S.; Allaway, D.; Morris, P. A longitudinal study of the feline faecal microbiome identifies changes into early adulthood irrespective of sexual development. PLoS ONE 2015, 10, e0144881. [Google Scholar] [CrossRef] [PubMed]
- Vilson, Å.; Ramadan, Z.; Li, Q.; Hedhammar, Å.; Reynolds, A.; Spears, J.; Labuda, J.; Pelker, R.; Björkstén, B.; Dicksved, J.; et al. Disentangling factors that shape the gut microbiota in German Shepherd dogs. PLoS ONE 2018, 13, e0193507. [Google Scholar] [CrossRef]
- Masuoka, H.; Shimada, K.; Kiyosue-Yasuda, T.; Kiyosue, M.; Oishi, Y.; Kimura, S.; Yamada, A.; Hirayama, K. Transition of the intestinal microbiota of dogs with age. Biosci. Microbiota Food Health 2016, 36, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Masuoka, H.; Shimada, K.; Kiyosue-Yasuda, T.; Kiyosue, M.; Oishi, Y.; Kimura, S.; Ohashi, Y.; Fujisawa, T.; Hotta, K.; Yamada, A.; et al. Transition of the intestinal microbiota of cats with age. PLoS ONE 2017, 12, e0181739. [Google Scholar] [CrossRef] [PubMed]
- Herstad, K.M.V.; Gajardo, K.; Bakke, A.M.; Moe, L.; Ludvigsen, J.; Rudi, K.; Rud, I.; Sekelja, M.; Skancke, E. A diet change from dry food to beef induces reversible changes on the faecal microbiota in healthy, adult client-owned dogs. BMC Vet. Res. 2017, 13, 147. [Google Scholar] [CrossRef] [PubMed]
- Song, S.J.; Lauber, C.; Costello, E.K.; Lozupone, C.A.; Humphrey, G.; Berg-Lyons, D.; Caporaso, J.G.; Knights, D.; Clemente, J.C.; Nakielny, S.; et al. Cohabiting family members share microbiota with one another and with their dogs. eLife 2013, 2, e00458. [Google Scholar] [CrossRef]
- Alessandri, G.; Milani, C.; Mancabelli, L.; Mangifesta, M.; Lugli, G.A.; Viappiani, A.; Duranti, S.; Turroni, F.; Ossiprandi, M.C.; van Sinderen, D.; et al. Metagenomic dissection of the canine gut microbiota: Insights into taxonomic, metabolic and nutritional features. Environ. Microbiol. 2019, 21, 1331–1343. [Google Scholar] [CrossRef]
- Reddy, K.E.; Kim, H.R.; Jeong, J.Y.; So, K.M.; Lee, S.; Ji, S.Y.; Kim, M.; Lee, H.J.; Lee, S.; Kim, K.H.; et al. Impact of breed on the fecal microbiome of dogs under the same dietary condition. J. Microbiol. Biotechnol. 2019, 29, 1947–1956. [Google Scholar] [CrossRef]
- Jeffery, N.D.; Barker, A.K.; Alcott, C.J.; Levine, J.M.; Meren, I.; Wengert, J.; Jergens, A.E.; Suchodolski, J.S. The association of specific constituents of the fecal microbiota with immune-mediated brain disease in dogs. PLoS ONE 2017, 12, e0170589. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Mazcorro, J.F.; Suchodolski, J.S.; Jones, K.R.; Clark-Price, S.C.; Dowd, S.E.; Minamoto, Y.; Markel, M.; Steiner, J.M.; Dossin, O. Effect of the proton pump inhibitor omeprazole on the gastrointestinal bacterial microbiota of healthy dogs. FEMS Microbiol. Ecol. 2012, 80, 624–636. [Google Scholar] [CrossRef]
- Kirchoff, N.S.; Udell, M.A.R.; Sharpton, T.J. The gut microbiome correlates with conspecific aggression in a small population of rescued dogs (Canis familiaris). Peer J. 2019, 7, e6103. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, L.E.; Steiner, J.M.; Suchodolski, J.S. Assessment of microbial diversity along the feline intestinal tract using 16S rRNA gene analysis. FEMS Microbiol. Ecol. 2008, 66, 590–598. [Google Scholar] [CrossRef] [PubMed]
- Suchodolski, J.S.; Camacho, J.; Steiner, J.M. Analysis of bacterial diversity in the canine duodenum, jejunum, ileum, and colon by comparative 16S rRNA gene analysis. FEMS Microbiol Ecol. 2008, 66, 567–578. [Google Scholar] [CrossRef]
- Xenoulis, P.G.; Palculict, B.; Allenspach, K.; Steiner, J.M.; Van House, A.M.; Suchodolski, J.S. Molecular-phylogenetic characterization of microbial communities imbalances in the small intestine of dogs with inflammatory bowel disease. FEMS Microbiol. Ecol. 2008, 66, 579–589. [Google Scholar] [CrossRef]
- Jia, J.; Frantz, N.; Khoo, C.; Gibson, G.R.; Rastall, R.A.; McCartney, A.L. Investigation of the faecal microbiota associated with canine chronic diarrhea. FEMS Microbiol. Ecol. 2010, 71, 304–312. [Google Scholar] [CrossRef]
- Suchodolski, J.S.; Xenoulis, P.G.; Paddock, C.G.; Steiner, J.M.; Jergens, A.E. Molecular analysis of the bacterial microbiota in duodenal biopsies from dogs with idiopathic inflammatory bowel disease. Vet. Microbiol. 2010, 142, 394–400. [Google Scholar] [CrossRef]
- Handl, S.; Dowd, S.E.; Garcia-Mazcorro, J.F.; Steiner, J.M.; Suchodolski, J.S. Massive parallel 16S rRNA gene pyrosequencing reveals highly diverse fecal bacterial and fungal communities in healthy dogs and cats. FEMS Microbiol. Ecol. 2011, 76, 301–310. [Google Scholar] [CrossRef]
- Dewhirst, F.E.; Klein, E.A.; Thompson, E.C.; Blanton, J.M.; Chen, T.; Milella, L.; Buckley, C.M.F.; Davis, I.J.; Bennett, M.L.; Marshall-Jones, Z.V. The canine oral microbiome. PLoS ONE 2012, 7, e36067. [Google Scholar] [CrossRef]
- Suchodolski, J.S.; Markel, M.E.; Garcia-Mazcorro, J.F.; Unterer, S.; Heilmann, R.M.; Dowd, S.E.; Kachroo, P.; Ivanov, I.; Minamoto, Y.; Dillman, E.M.; et al. The fecal microbiome in dogs with acute diarrhea and idiopathic inflammatory bowel disease. PLoS ONE 2012, 7, e51907. [Google Scholar] [CrossRef] [PubMed]
- Hand, D.; Wallis, C.; Colyer, A.; Penn, C.W. Pyrosequencing the canine faecal microbiota: Breadth and depth of biodiversity. PLoS ONE 2013, 8, e53115. [Google Scholar] [CrossRef] [PubMed]
- Sturgeon, A.; Pinder, S.L.; Costa, M.C.; Weese, J.S. Characterization of the oral microbiota of healthy cats using next-generation sequencing. Vet. J. 2014, 201, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Dewhirst, F.E.; Klein, E.A.; Bennett, M.L.; Croft, J.M.; Harris, S.J.; Marshall-Jones, Z.V. The feline oral microbiome: A provisional 16S rRNA gene based taxonomy with full-length reference sequences. Vet. Microbiol. 2015, 175, 294–303. [Google Scholar] [CrossRef] [PubMed]
- Guard, B.C.; Barr, J.W.; Reddivari, L.; Klemashevich, C.; Jayaraman, A.; Steiner, J.M.; Vanamala, J.; Suchodolski, J.S. Characterization of microbial dysbiosis and metabolomic changes in dogs with acute diarrhea. PLoS ONE 2015, 10, e0127259. [Google Scholar] [CrossRef] [PubMed]
- Harris, S.; Croft, J.; O’Flynn, C.; Deusch, O.; Colyer, A.; Allsopp, J.; Milella, L.; Davis, I.J. A pyrosequencing investigation of differences in the feline subgingival microbiota in health, gingivitis and mild periodontitis. PLoS ONE 2015, 10, e0136986. [Google Scholar] [CrossRef]
- Minamoto, Y.; Otoni, C.C.; Steelman, S.M.; Büyüklebebici, O.; Steiner, J.M.; Jergens, A.E.; Suchodolski, J.S. Alteration of the fecal microbiota and serum metabolite profiles in dogs with idiopathic inflammatory bowel disease. Gut Microbes 2015, 6, 33–47. [Google Scholar] [CrossRef]
- Wallis, C.; Marshall, M.; Colyer, A.; O’Flynn, C.; Deusch, O.; Harris, S. A longitudinal assessment of changes in bacterial community composition associated with the development of periodontal disease in dogs. Vet. Microbiol. 2015, 181, 271–282. [Google Scholar] [CrossRef]
- Kasiraj, A.C.; Harmoinen, J.; Isaiah, A.; Westermarck, E.; Steiner, J.M.; Spillmann, T.; Suchodolski, J.S. The effects of feeding and withholding food on the canine small intestinal microbiota. FEMS Microbiol. Ecol. 2016, 92, fiw085. [Google Scholar] [CrossRef]
- Sabbioni, A.; Ferrario, C.; Milani, C.; Mancabelli, L.; Riccardi, E.; Di Ianni, F.; Beretti, V.; Superchi, P.; Ossiprandi, M.C. Modulation of the Bifidobacterial communities of the dog microbiota by zeolite. Front. Microbiol. 2016, 7, 1491. [Google Scholar] [CrossRef]
- Garcia-Mazcorro, J.F.; Barcenas-Walls, J.R.; Suchodolski, J.S.; Steiner, J.M. Molecular assessment of the fecal microbiota in healthy cats and dogs before and during supplementation with fructo-oligosaccharides (FOS) and inulin using high-throughput 454-pyrosequencing. PeerJ 2017, 5, e3184. [Google Scholar] [CrossRef] [PubMed]
- Guard, B.C.; Mila, H.; Steiner, J.M.; Mariani, C.; Suchodolski, J.S.; Chastant-Maillard, S. Characterization of the fecal microbiome during neonatal and early pediatric development in puppies. PLoS ONE 2017, 12, e0175718. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; An, J.-U.; Kim, W.; Lee, S.; Cho, S. Differences in the gut microbiota of dogs (Canis lupus familiaris) fed a natural diet or a commercial feed revealed by the Illumina MiSeq platform. Gut Pathog. 2017, 9, 68. [Google Scholar] [CrossRef] [PubMed]
- Sandri, M.; Dal Monego, S.; Conte, G.; Sgorlon, S.; Stefanon, B. Raw meat based diet influences faecal microbiome and end products of fermentation in healthy dogs. BMC Vet. Res. 2017, 13, 65. [Google Scholar] [CrossRef]
- Herstad, K.M.V.; Moen, A.E.F.; Gaby, J.C.; Moe, L.; Skancke, E. Characterization of the fecal and mucosa-associated microbiota in dogs with colorectal epithelial tumors. PLoS ONE 2018, 13, e0198342. [Google Scholar] [CrossRef]
- Omatsu, T.; Omura, M.; Katayama, Y.; Kimura, T.; Okumura, M.; Okumura, A.; Murata, Y.; Mizutani, T. Molecular diversity of the faecal microbiota of Toy Poodles in Japan. J. Vet. Med. Sci. 2018, 80, 749–754. [Google Scholar] [CrossRef]
- Salas-Mani, A.; Jeusette, I.; Castillo, I.; Manuelian, C.L.; Lionnet, C.; Iraculis, N.; Sanchez, N.; Fernández, S.; Vilaseca, L.; Torre, C. Fecal microbiota composition changes after a BW loss diet in Beagle dogs. J. Anim. Sci. 2018, 96, 3102–3111. [Google Scholar] [CrossRef]
- Schmid, S.M.; Suchodolski, J.S.; Price, J.M.; Tolbert, M.K. Omeprazole minimally alters the fecal microbial community in six cats: A pilot study. Front. Vet. Sci. 2018, 5, 79. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.; Unterer, S.; Suchodolski, J.S.; Honneffer, J.B.; Guard, B.C.; Lidbury, J.A.; Steiner, J.M.; Fritz, J.; Kölle, P. The fecal microbiome and metabolome differs between dogs fed Bones and Raw Food (BARF) diets and dogs fed commercial diets. PLoS ONE 2018, 13, e0201279. [Google Scholar] [CrossRef]
- Ziese, A.-L.; Suchodolski, J.S.; Hartmann, K.; Busch, K.; Anderson, A.; Sarwar, F.; Sindern, N.; Unterer, S. Effect of probiotic treatment on the clinical course, intestinal microbiome, and toxigenic Clostridium perfringens in dogs with acute hemorrhagic diarrhea. PLoS ONE 2018, 13, e0204691. [Google Scholar] [CrossRef]
- Berry, A.S.F.; Kelly, B.J.; Barnhart, D.; Kelly, D.J.; Beiting, D.P.; Baldassano, R.N.; Redding, L.E. Gut microbiota features associated with Clostridioides difficile colonization in puppies. PLoS ONE 2019, 14, e0215497. [Google Scholar] [CrossRef] [PubMed]
- Blake, A.B.; Guard, B.C.; Honneffer, J.B.; Lidbury, J.A.; Steiner, J.M.; Suchodolski, J.S. Altered microbiota, fecal lactate, and fecal bile acids in dogs with gastrointestinal disease. PLoS ONE 2019, 14, e0224454. [Google Scholar] [CrossRef] [PubMed]
- Bresciani, F.; Minamoto, Y.; Suchodolski, J.S.; Galiazzo, G.; Vecchiato, C.G.; Pinna, C.; Biagi, G.; Pietra, M. Effect of an extruded animal protein-free diet on fecal microbiota of dogs with food-responsive enteropathy. J. Vet. Intern. Med. 2019, 32, 1903–1910. [Google Scholar] [CrossRef] [PubMed]
- Giaretta, P.R.; Suchodolski, J.S.; Jergens, A.E.; Steiner, J.M.; Lidbury, J.A.; Cook, A.K.; Hanifeh, M.; Spillmann, T.; Kilpinen, S.; Syrjä, P.; et al. Bacterial biogeography of the colon in dogs with chronic inflammatory enteropathy. Vet. Pathol. 2020, in press. [Google Scholar] [CrossRef] [PubMed]
- Jergens, A.E.; Guard, B.C.; Redfern, A.; Rossi, G.; Mochel, J.P.; Pilla, R.; Chandra, L.; Seo, Y.-J.; Steiner, J.M.; Lidbury, J.; et al. Microbiota-related changes in unconjugated fecal bile acids are associated with naturally occurring, insulin-dependent diabetes mellitus in dogs. Front. Vet. Sci. 2019, 6, 199. [Google Scholar] [CrossRef]
- Jha, A.R.; Shmalberg, J.; Tanprasertsuk, J.; Perry, L.; Massey, D.; Honaker, R.W. Characterization of gut microbiomes of household pets in the United States using a direct-to-consumer approach. PLoS ONE 2020, 15, e0227289. [Google Scholar] [CrossRef]
- Manchester, A.C.; Webb, C.B.; Blake, A.B.; Sarwar, F.; Lidbury, J.A.; Steiner, J.M.; Suchodolski, J.S. Long-term impact of tylosin on fecal microbiota and fecal bile acids of healthy dogs. J. Vet. Intern. Med. 2019, 33, 2605–2617. [Google Scholar] [CrossRef]
- Minamoto, Y.; Minamoto, T.; Isaiah, A.; Sattasathuchana, P.; Buono, A.; Rangachari, V.R.; McNeely, I.H.; Lidbury, J.; Steiner, J.M.; Suchodolski, J.S. Fecal short-chain fatty acid concentrations and dysbiosis in dogs with chronic enteropathy. J. Vet. Intern. Med. 2019, 33, 1608–1618. [Google Scholar] [CrossRef]
- Nogueira, J.P.S.; He, F.; Mangian, H.F.; Oba, P.M.; De Godoy, M.R.C. Dietary supplementation of a fiber-prebiotic and saccharin-eugenol blend in extruded diets fed to dogs. J. Anim. Sci. 2019, 97, 4519–4531. [Google Scholar] [CrossRef]
- Park, J.S.; Guevarra, R.B.; Kim, B.R.; Lee, J.H.; Lee, S.H.; Cho, J.H.; Kim, H.; Cho, J.H.; Song, M.; Lee, J.H.; et al. Intestinal microbial dysbiosis in Beagles naturally infected with canine parvovirus. J. Microbiol. Biotechnol. 2019, 29, 1391–1400. [Google Scholar] [CrossRef]
- Pilla, R.; Guard, B.C.; Steiner, J.M.; Gaschen, F.P.; Olson, E.; Werling, D.; Allenspach, K.; Salavati Schmitz, S.; Suchodolski, J.S. Administration of a synbiotic containing Enterococcus faecium does not significantly alter fecal microbiota richness or diversity in dogs with and without food-responsive chronic enteropathy. Vet. Sci. 2019, 6, 277. [Google Scholar] [CrossRef] [PubMed]
- Stone, N.E.; Nunnally, A.E.; Jimenez, V., Jr.; Cope, E.K.; Sahl, J.W.; Sheridan, K.; Hornstra, H.M.; Vinocur, J.; Settles, E.W.; Headley, K.C.; et al. Domestic canines do not display evidence of gut microbial dysbiosis in the presence of Clostridioides (Clostridium) difficile, despite cellular susceptibility to its toxins. Anaerobe 2019, 58, 53–72. [Google Scholar] [CrossRef] [PubMed]
- Whittemore, J.C.; Stokes, J.E.; Price, J.M.; Suchodolski, J.S. Effects of a synbiotic on the fecal microbiome and metabolomics profiles of healthy research cats administered clindamycin: A randomized, controlled trial. Gut Microbes 2019, 10, 521–539. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Huang, W.; Hou, Q.; Kwok, L.-Y.; Laga, W.; Wang, Y.; Ma, H.; Sun, Z.; Zhang, H. Oral administration of compound probiotics improved canine feed intake, weight gain, immunity and intestinal microbiota. Front. Immunol. 2019, 10, 666. [Google Scholar] [CrossRef]
- Sturgeon, A.; Stull, J.W.; Costa, M.C.; Weese, J.S. Metagenomic analysis of the canine oral cavity as revealed by high-throughput pyrosequencing of the 16S rRNA gene. Vet. Microbiol. 2013, 162, 891–898. [Google Scholar] [CrossRef]
- McDonald, J.E.; Larsen, N.; Pennington, A.; Connolly, J.; Wallis, C.; Rooks, D.J.; Hall, N.; McCarthy, A.J.; Allison, H.E. Characterising the canine oral microbiome by direct sequencing of reverse-transcribed rRNA molecules. PLoS ONE 2016, 11, e0157046. [Google Scholar] [CrossRef]
- Keijser, B.J.F.; Zaura, E.; Huse, S.M.; van der Vossen, J.M.B.M.; Schuren, F.H.J.; Montijn, R.C.; ten Cate, J.M.; Crielaard, W. Pyrosequencing analysis of the oral microflora of healthy adults. J. Dent. Res. 2008, 87, 1016–1020. [Google Scholar] [CrossRef]
- Lazarevic, V.; Whiteson, K.; Huse, S.; Hernandez, D.; Farinelli, L.; Østerås, M.; Schrenzel, J.; François, P. Metagenomic study of the oral microbiota by Illumina high-throughput sequencing. J. Microbiol. Met. 2009, 79, 266–271. [Google Scholar] [CrossRef]
- Bik, E.M.; Long, C.D.; Armitage, G.C.; Loomer, P.; Emerson, J.; Mongodin, E.F.; Nelson, K.E.; Gill, S.R.; Fraser-Liggett, C.M.; Relman, D.A. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010, 4, 962–974. [Google Scholar] [CrossRef]
- Shaw, L.; Ribeiro, A.L.R.; Levine, A.P.; Pontikos, N.; Balloux, F.; Segal, A.W.; Roberts, A.P.; Smith, A.M. The human salivary microbiome is shaped by shared environment rather than genetics: Evidence from a large family of closely related individuals. mBio 2017, 8, e01237-17. [Google Scholar] [CrossRef]
- Ye, F.; Shen, H.; Li, Z.; Meng, F.; Li, L.; Yang, J.; Chen, Y.; Bo, X.; Zhang, X.; Ni, M. Influence of the biliary system on biliary bacteria revealed by bacterial communities of the human biliary and upper digestive tracts. PLoS ONE 2016, 11, e0150519. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Yu, W.H.; Izard, J.; Baranova, O.V.; Lakshmanan, A.; Dewhirst, F.E. The human oral microbiome database: A web accessible resource for investigating oral microbe taxonomic and genomic information. Database 2010, 2010, baq013. [Google Scholar] [CrossRef] [PubMed]
- Dewhirst, F.E.; Chen, T.; Izard, J.; Paster, B.J.; Tanner, A.C.R.; Yu, W.H.; Lakshmanan, A.; Wade, W.G. The human oral microbiome. J. Bacteriol. 2010, 192, 5002–5017. [Google Scholar] [CrossRef]
- Ouwerkerk, J.P.; van der Ark, K.C.; Davids, M.; Claassens, N.J.; Robert Finestra, T.; de Vos, W.; Belzer, C. Adaptation of Akkermansia muciniphila to the oxic-anoxic interface of the mucus layer. Appl. Environ. Microbiol. 2016, 82, 6983–6993. [Google Scholar] [CrossRef] [PubMed]
- Mark Welch, J.L.; Rossetti, B.J.; Rieken, C.W.; Dewhirst, F.E.; Borisy, G.G. Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. USA 2016, 113, E791–E800. [Google Scholar] [CrossRef] [PubMed]
- Huck, O.; Mulhall, H.; Rubin, G.; Kizelnik, Z.; Iyer, R.; Perpich, J.D.; Haque, N.; Cani, P.D.; de Vos, W.M.; Amar, S. Akkermansia muciniphila reduces Porphyromonas gingivalis-induced inflammation and periodontal bone destruction. J. Clin. Periodontol. 2020. [CrossRef] [PubMed]
- Priestnall, S.L.; Wiinberg, B.; Spohr, A.; Neuhaus, B.; Kuffer, M.; Wiedmann, M.; Simpson, K.W. Evaluation of “Helicobacter heilmannii” subtypes in the gastric mucosas of cats and dogs. J. Clin. Microbiol. 2004, 42, 2144–2151. [Google Scholar] [CrossRef]
- Bik, E.M.; Eckburg, P.B.; Gill, S.R.; Nelson, K.E.; Purdom, E.A.; Francois, F.; Perez-Perez, G.; Blaser, M.J.; Relman, D.A. Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl. Acad. Sci. USA 2006, 103, 732–737. [Google Scholar] [CrossRef]
- Gantuya, B.; El-Serag, H.B.; Matsumoto, T.; Ajami, N.J.; Oyuntsetseg, K.; Azzaya, D.; Uchida, T.; Yamaoka, Y. Gastric microbiota in Helicobacter pylori-negative and –positive gastritis among high incidence of gastric cancer area. Cancers 2019, 11, 504. [Google Scholar] [CrossRef]
- Cassmann, E.; White, R.; Atherly, T.; Wang, C.; Sun, Y.; Khoda, S.; Mosher, C.; Ackermann, M.; Jergens, A. Alterations of the ileal and colonic mucosal microbiota in canine chronic enteropathies. PLoS ONE 2016, 11, e0147321. [Google Scholar] [CrossRef]
- Xiao, L.; Feng, Q.; Liang, S.; Brask Sonne, S.; Xia, Z.; Qiu, X.; Li, X.; Long, H.; Zhang, J.; Zhang, D.; et al. A catalog of the mouse gut metagenome. Nat. Biotechnol. 2015, 33, 1103–1108. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; DiBaise, J.K.; Zuccolo, A.; Kudrna, D.; Braidotti, M.; Yu, Y.; Parameswaran, P.; Crowell, M.D.; Wing, R.; Rittmann, B.E.; et al. Human gut microbiota in obesity and after gastric bypass. Proc. Natl. Acad. Sci. USA 2009, 106, 2365–2370. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Ahrné, S.; Jeppsson, B.; Molin, G. Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol. Ecol. 2005, 54, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Yildirim, S.; Yeoman, C.J.; Sipos, M.; Torralba, M.; Wilson, B.A.; Goldberg, T.L.; Stumpf, R.M.; Leigh, S.R.; White, B.A.; Nelson, K.E. Characterization of the fecal microbiome from non-human wild primates reveals species specific microbial communities. PLoS ONE 2010, 5, e13963. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.R.; Cornell, K.K.; Parnell, N.K.; Salisbury, S.K. Pancreatic abscess in 36 dogs: A retrospective analysis of prognostic indicators. J. Am. Anim. Hosp. Assoc. 2008, 44, 171–179. [Google Scholar] [CrossRef]
- Burton, E.N.; Cohn, L.A.; Reinero, C.N.; Rindt, H.; Moore, S.G.; Ericsson, A.C. Characterization of the urinary microbiome in healthy dogs. PLoS ONE 2017, 12, e0177783. [Google Scholar] [CrossRef]
- Vientós-Plotts, A.I.; Ericsson, A.C.; Rindt, H.; Grobman, M.E.; Graham, A.; Bishop, K.; Cohn, L.A.; Reinero, C.R. Dynamic changes of the respiratory microbiota and its relationship to fecal and blood microbiota in healthy young cats. PLoS ONE 2017, 12, e0173818. [Google Scholar] [CrossRef]
- Ericsson, A.C.; Personett, A.R.; Grobman, M.E.; Rindt, H.; Reinero, C.R. Composition and predicted metabolic capacity of upper and lower airway microbiota of healthy dogs in relation to the fecal microbiota. PLoS ONE 2016, 11, e0154646. [Google Scholar] [CrossRef]
- Rodrigues Hoffmann, A.; Patterson, A.P.; Diesel, A.; Lawhon, S.D.; Jaclyn Ly, H.; Elkins Stephenson, C.; Mansell, J.; Steiner, J.M.; Dowd, S.E.; Olivry, T.; et al. The skin microbiome in healthy and allergic dogs. PLoS ONE 2013, 9, e83197. [Google Scholar] [CrossRef]
- Cuscó, A.; Belanger, J.M.; Gershony, L.; Islas-Trejo, A.; Levy, K.; Medrano, J.F.; Sánchez, A.; Oberbauer, A.M.; Francino, O. Individual signatures and environmental factors shape skin microbiota in healthy dogs. Microbiome 2017, 5, 139. [Google Scholar] [CrossRef]
- Older, C.E.; Diesel, A.; Patterson, A.P.; Meason-Smith, C.; Johnson, T.J.; Mansell, J.; Suchodolski, J.S.; Rodrigues Hoffmann, A. The feline skin microbiota: The bacteria inhabiting the skin of healthy and allergic cats. PLoS ONE 2017, 12, e0178555. [Google Scholar] [CrossRef] [PubMed]
- Goddard, M.R.; Greig, D. Saccharomyces cerevisiae: A nomadic yeast with no niche? FEMS Yeast Res. 2015, 15, fov009. [Google Scholar] [CrossRef] [PubMed]
- Pereira, F.C.; Berry, D. Microbial nutrient niches in the gut. Environ. Microbiol. 2017, 19, 1366–1378. [Google Scholar] [CrossRef] [PubMed]
- Costea, P.I.; Hildebrand, F.; Arumugam, M.; Bäckhed, F.; Blaser, M.J.; Bushman, F.D.; de Vos, W.M.; Ehrlich, S.D.; Fraser, C.M.; Hattori, M.; et al. Enterotypes in the landscape of gut microbial community composition. Nat. Microbiol. 2018, 3, 8–16. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Gordon, J.I. The core gut microbiome, energy balance and obesity. J. Physiol. 2009, 587, 4153–4158. [Google Scholar] [CrossRef]
- Arumugan, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.-M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef]
- Claussen, J.C.; Skieceviĉienė, J.; Wang, J.; Rausch, P.; Karlsen, T.H.; Lieb, W.; Baines, J.F.; Franke, A.; Hütt, M.T. Boolean analysis reveals systematic interactions among low-abundance species in the human gut microbiome. PLoS Comput. Biol. 2017, 13, e1005361. [Google Scholar] [CrossRef]
- Navas-Molina, J.A.; Peralta-Sánchez, J.M.; González, A.; McMurdie, P.J.; Vázquez-Baeza, Y.; Xu, Z.; Ursell, L.K.; Lauber, C.; Zhou, H.; Jin Song, S.; et al. Advancing our understanding of the human microbiome using QIIME. Methods Enzymol. 2013, 531, 371–444. [Google Scholar]
- Dawson, W.; Hör, J.; Egert, M.; van Kleunen, M.; Pester, M. A small number of low-abundance bacteria dominate plant species-specific responses during rhizosphere colonization. Front. Microbiol. 2017, 8, 975. [Google Scholar] [CrossRef]
- Jousset, A.; Bienhold, C.; Chatzinotas, A.; Gallien, L.; Gobet, A.; Kurm, V.; Küsel, K.; Rilling, M.C.; Rivett, D.W.; Salles, J.F.; et al. Where less may be more: How the rare biosphere pulls ecosystems strings. ISME J. 2017, 11, 853–862. [Google Scholar] [CrossRef]
- Garcia-Mazcorro, J.F.; Lage, N.N.; Mertens-Talcott, S.; Talcott, S.; Chew, B.; Dowd, S.E.; Kawas, J.R.; Noratto, G.D. Effect of dark sweet cherry powder consumption on the gut microbiota, short-chain fatty acids, and biomarkers of gut health in obese db/db mice. PeerJ 2018, 6, e4195. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, M.L.; Swecker, W.S., Jr.; Jensen, R.V.; Ponder, M.A. Characterization of the fecal bacteria communities of forage-fed horses by pyrosequencing of 16S rRNA V4 gene amplicons. FEMS Microbiol. Lett. 2012, 326, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Berry, D.; Stecher, B.; Schintlmeister, A.; Reichert, J.; Brugiroux, S.; Wild, B.; Wanek, W.; Richter, A.; Rauch, I.; Decker, T.; et al. Host-compound foraging by intestinal microbiota revealed by single-cell stable isotope probing. Proc. Natl. Acad. Sci. USA 2013, 110, 4720–4725. [Google Scholar] [CrossRef] [PubMed]
- McGuckin, M.A.; Lindén, S.K.; Sutton, P.; Florin, T.H. Mucin dynamics and enteric pathogens. Nat. Rev. Microbiol. 2011, 9, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Limenitakis, J.P.; Fuhrer, T.; Geuking, M.B.; Lawson, M.A.; Wyss, M.; Brugiroux, S.; Keller, I.; Macpherson, J.A.; Rupp, S.; et al. The outer mucus layer hosts a distinct intestinal microbial niche. Nat. Commun. 2015, 6, 8292. [Google Scholar] [CrossRef] [PubMed]
- Berry, D.; Reinisch, W. Intestinal microbiota: A source of novel biomarkers in inflammatory bowel diseases? Best Pract. Res. Clin. Gastroenterol. 2013, 27, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Lee, S.E.; Kim, H.B.; Isaacson, R.E.; Seo, K.W.; Song, K.H. Association of obesity with serum leptin, adiponectin, and serotonin and gut microflora in beagle dogs. J. Vet. Intern. Med. 2015, 29, 43–50. [Google Scholar] [CrossRef]
- Kieler, I.N.; Mølbak, L.; Hansen, L.L.; Hermann-Bank, M.L.; Bjornvad, C.R. Overweight and the feline gut microbiome—A pilot study. J. Anim. Physiol. Anim. Nutr. 2016, 100, 478–484. [Google Scholar] [CrossRef]
- Fischer, M.M.; Kessler, A.M.; Kieffer, D.A.; Knotts, T.A.; Kim, K.; Wei, A.; Ramsey, J.J.; Fascetti, A.J. Effects of obesity, energy restriction and neutering on the faecal microbiota of cats. Br. J. Nutr. 2017, 118, 513–524. [Google Scholar] [CrossRef]
- Kieler, I.N.; Shamzir Kamal, S.; Vitger, A.D.; Nielsen, D.S.; Lauridsen, C.; Bjornvad, C.R. Gut microbiota composition may relate to weight loss rate in obese pet dogs. Vet. Med. Sci. 2017, 3, 252–262. [Google Scholar] [CrossRef]
- Xu, J.; Verbrugghe, A.; Lourenço, M.; Cools, A.; Liu, D.J.X.; Van de Wiele, T.; Marzorati, M.; Eeckhaut, V.; Van Immerseel, F.; Vanhaecke, L.; et al. The response of canine faecal microbiota to increased dietary protein is influenced by body condition. BMC Vet. Res. 2017, 13, 374. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; de Vos, W.M. Next-generation beneficial microbes: The case of Akkermansia muciniphila. Front Microbiol. 2017, 8, 1765. [Google Scholar] [CrossRef] [PubMed]
- Plovier, H.; Everard, A.; Druart, C.; Depommier, C.; Van Hul, M.; Geurts, L.; Chilloux, J.; Ottman, N.; Duparc, T.; Lichtenstein, L.; et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 2017, 23, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Seedorf, H.; Griffin, N.W.; Ridaura, V.K.; Reyes, A.; Cheng, J.; Rey, F.E.; Smith, M.I.; Simon, G.M.; Scheffrahn, R.H.; Woebken, D.; et al. Bacteria from diverse habitats colonize and compete in the mouse gut. Cell 2014, 159, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Abreu, M.T. Toll-like receptor signaling in the intestinal epithelium: How bacterial recognition shapes intestinal function. Nat. Rev. Immunol. 2010, 10, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Jergens, A.E.; Simpson, K.W. Inflammatory bowel disease in veterinary medicine. Front. Biosci. 2012, 4, 1404–1419. [Google Scholar] [CrossRef]
- Zhou, K. Strategies to promote abundance of Akkermansia muciniphila, an emerging probiotics in the gut, evidence from dietary intervention studies. J. Funct. Foods 2017, 33, 194–201. [Google Scholar] [CrossRef]
- Anhê, F.F.; Pilon, G.; Roy, D.; Desjardins, Y.; Levy, E.; Marette, A. Triggering Akkermansia with dietary polyphenols: A new weapon to combat the metabolic syndrome? Gut Microbes 2016, 7, 146–153. [Google Scholar] [CrossRef]
- Shin, J.; Noh, J.-R.; Chang, D.-H.; Kim, Y.-H.; Kim, M.H.; Lee, E.S.; Cho, S.; Ku, B.J.; Rhee, M.-S.; Kim, B.-C.; et al. Elucidation of Akkermansia muciniphila probiotic traits driven by mucin depletion. Front. Microbiol. 2019, 10, 1137. [Google Scholar] [CrossRef]
- Buntin, N.; de Vos, W.M.; Hongpattarakere, T. Variation of mucin adhesion, cell Surface characteristics, and molecular mechanisms among Lactobacillus plantarum isolated from different habitats. Appl. Microbiol. Biotechnol. 2017, 101, 7663–7674. [Google Scholar] [CrossRef]
- Święch, E.; Tuśnio, A.; Barszcz, M.; Taciak, M.; Siwiak, E. Goblet cells and mucus layer in the gut of young pigs: Response to dietary contents of threonine and non-essential amino acids. J. Anim. Physiol. Anim. Nutr. (Berl.) 2019, 103, 894–905. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Zhou, G.; Liu, X.; Song, S.; Xu, X.; Hooiveld, G.; Müller, M.; Liu, L.; Kristiansen, K.; Li, C. Dietary protein sources differentially affect the growth of Akkermansia muciniphila and maintenance of the gut mucus barrier in mice. Mol. Nutr. Food Res. 2019, 2019, e1900589. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, B.O. Fight them or feed them: How the intestinal mucus layer manages the gut microbiota. Gastroenterol. Rep. 2019, 7, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Derrien, M.; van Hylckama Vlieg, J.E.T. Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol. 2015, 23, 354–366. [Google Scholar] [CrossRef]
- Schmidt, T.S.B.; Hayward, M.R.; Coelho, L.P.; Li, S.S.; Costea, P.I.; Voigt, A.Y.; Wirbel, J.; Maistrenko, O.M.; Alves, R.J.C.; Bergsten, E.; et al. Extensive transmission of microbes along the gastrointestinal tract. eLife 2019, 8, e42693. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).