Characterization of Ethanol Extracted Cell Wall Components of Mycobacterium avium Subsp. paratuberculosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Antigen Preparation
2.2. Antibodies
2.3. Folch Lipid Extraction, Thin Layer Chromatography, and Lectin Binding Assays
2.4. Flow Cytometry Analysis
2.5. ELISA to Measure Antibody Binding
2.6. Construction and Screening of a Map Genomic Expression Library
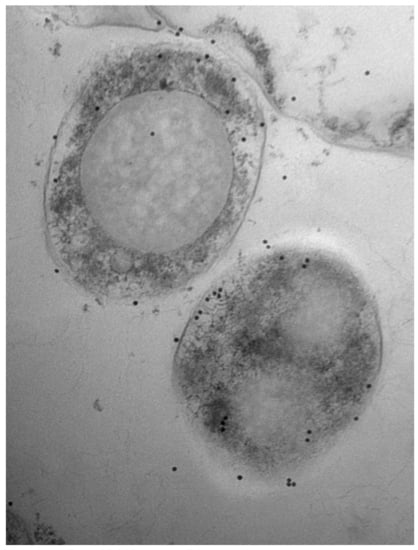
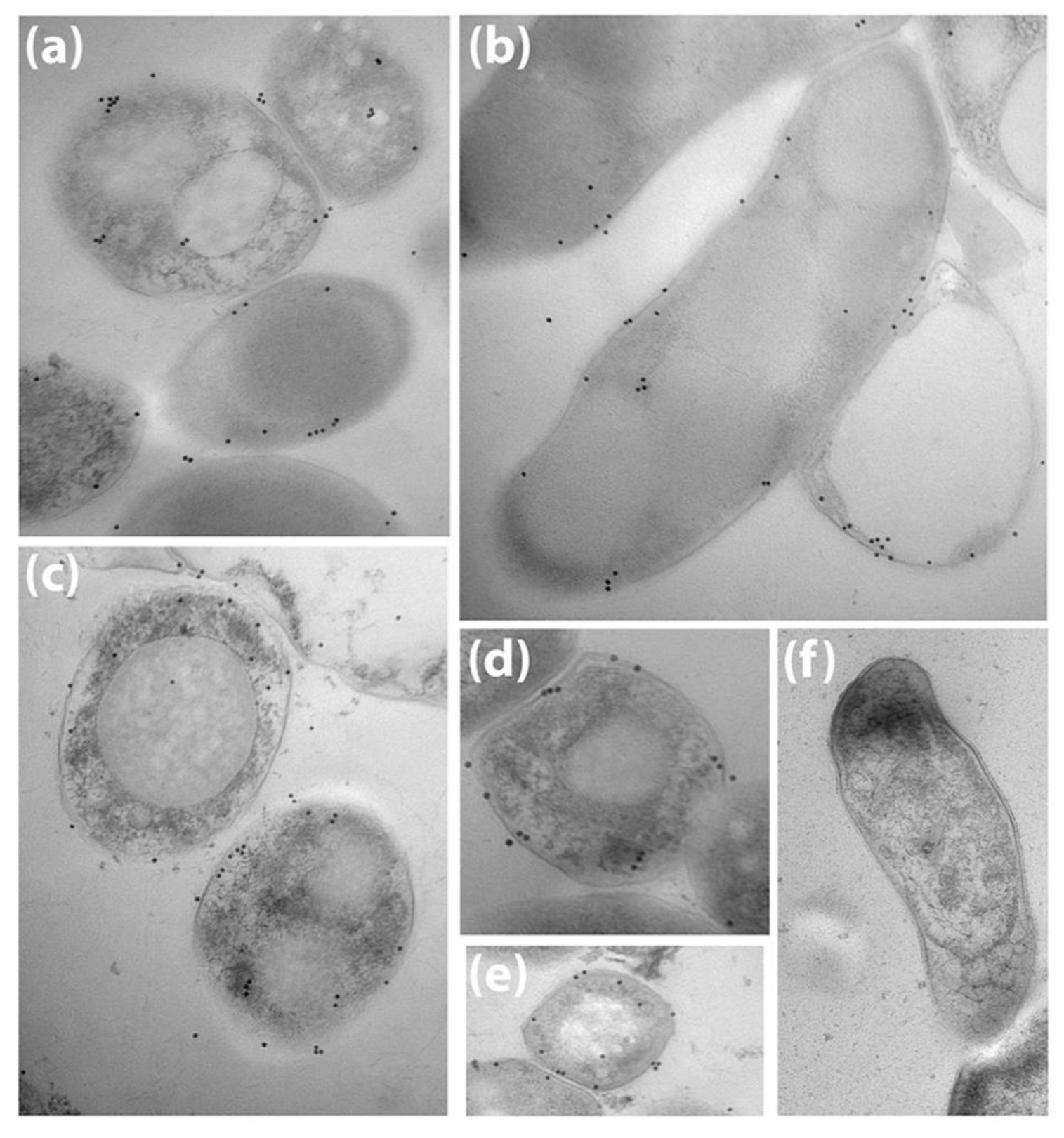
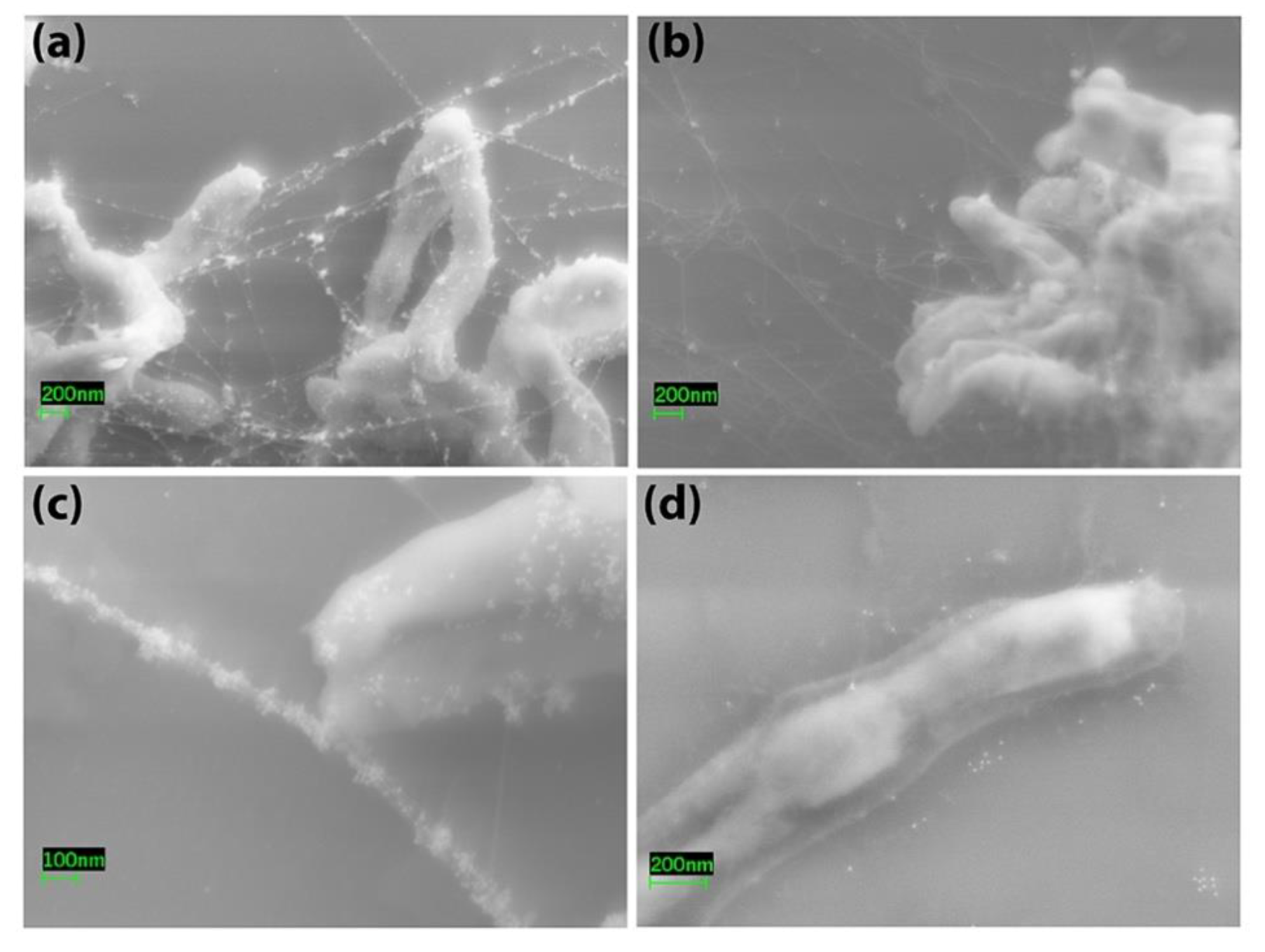
2.7. Electron Microscopy
2.8. Statistical Analysis
3. Results
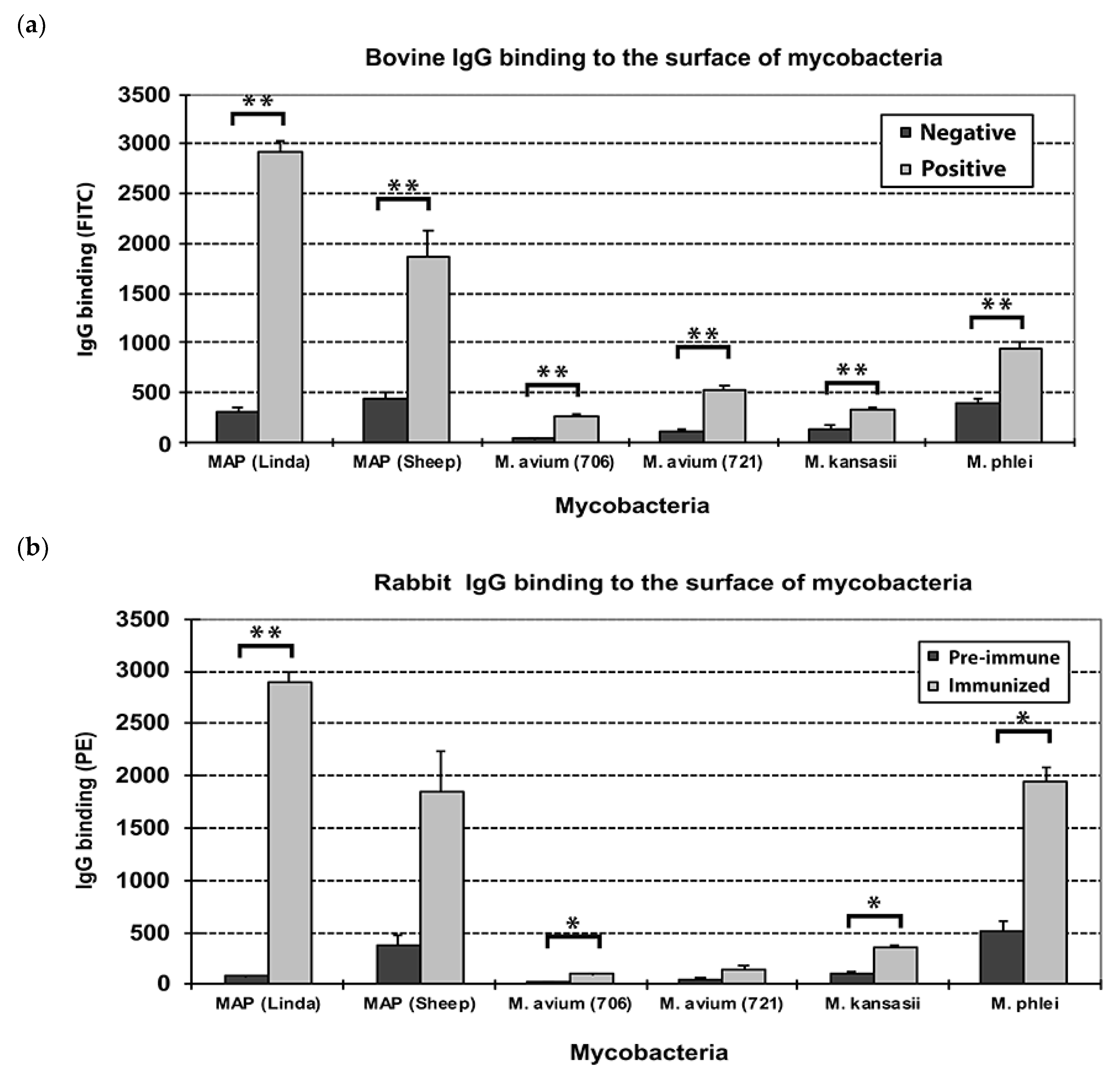
3.1. Bovine and Rabbit Antibodies React to the Surface of Map Bacteria
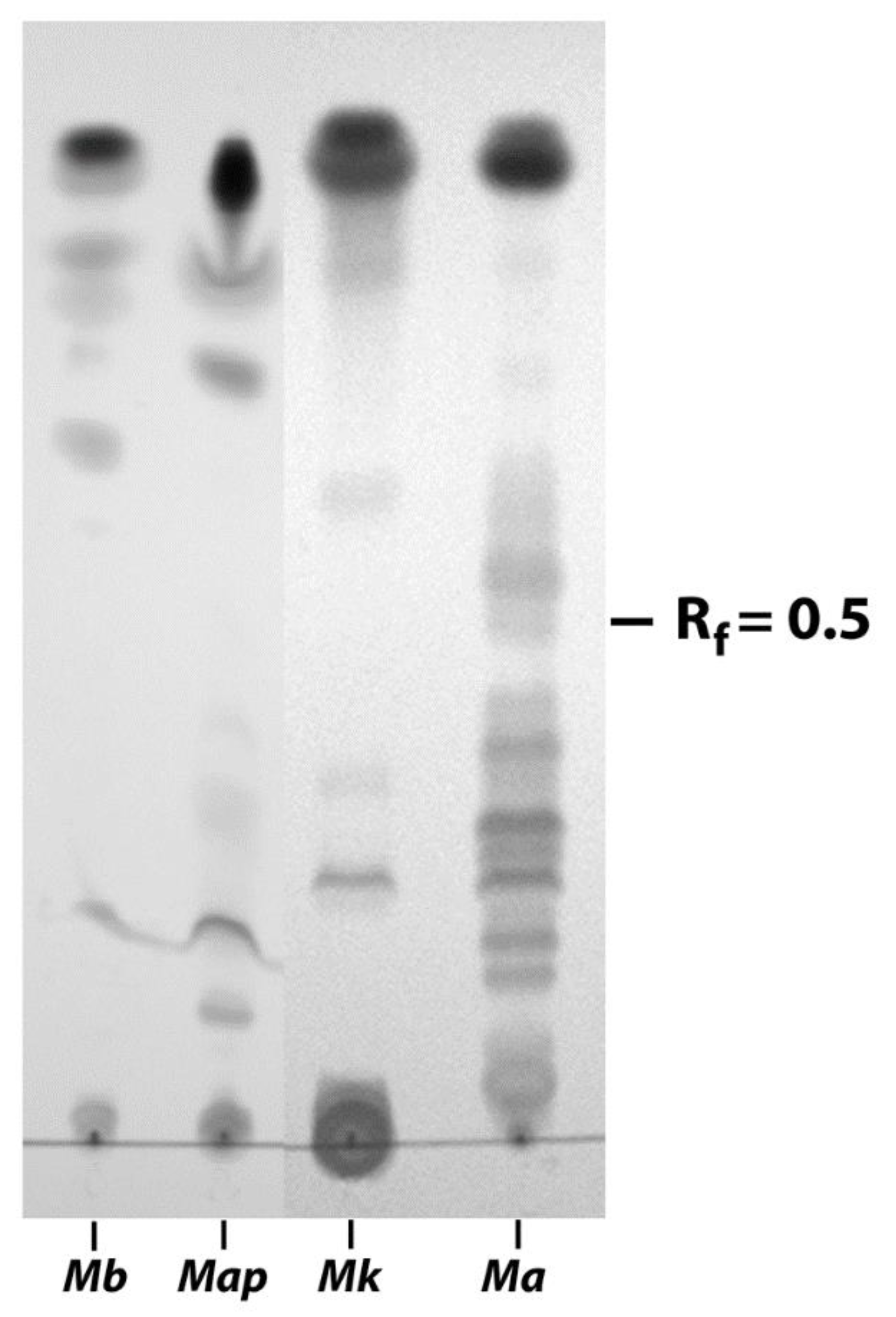
3.2. EtOH Extracts of Mycobacterial Species Show Markedly Different TLC Profiles
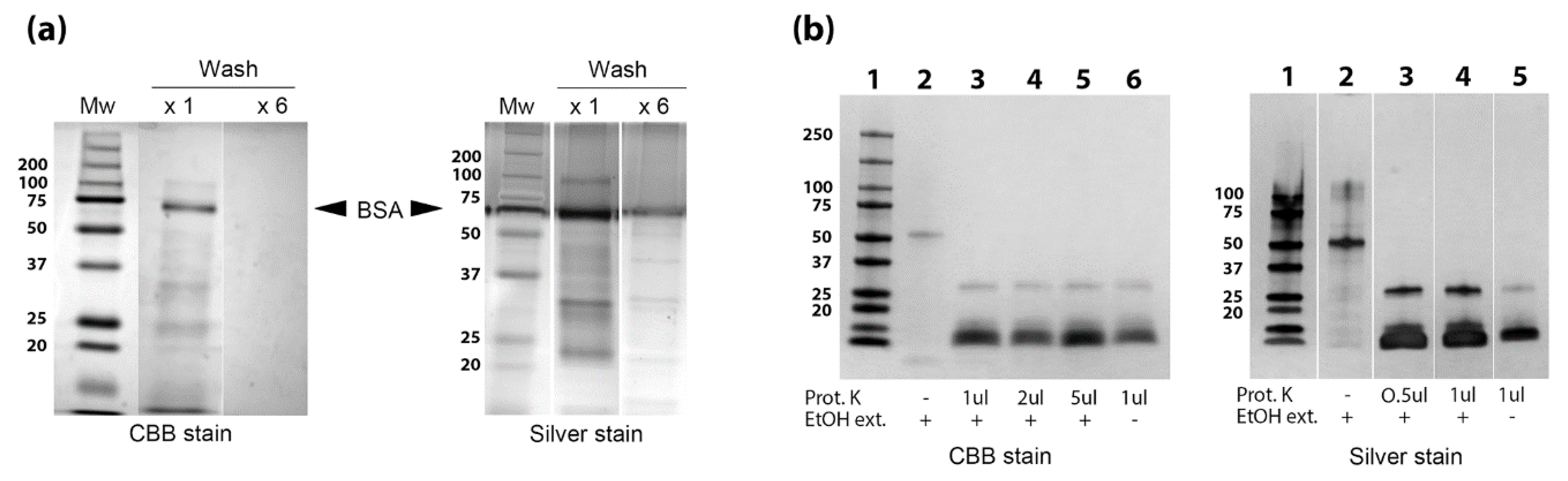
3.3. Identification of Proteins, Lipids and Carbohydrates in the EtOH Extract
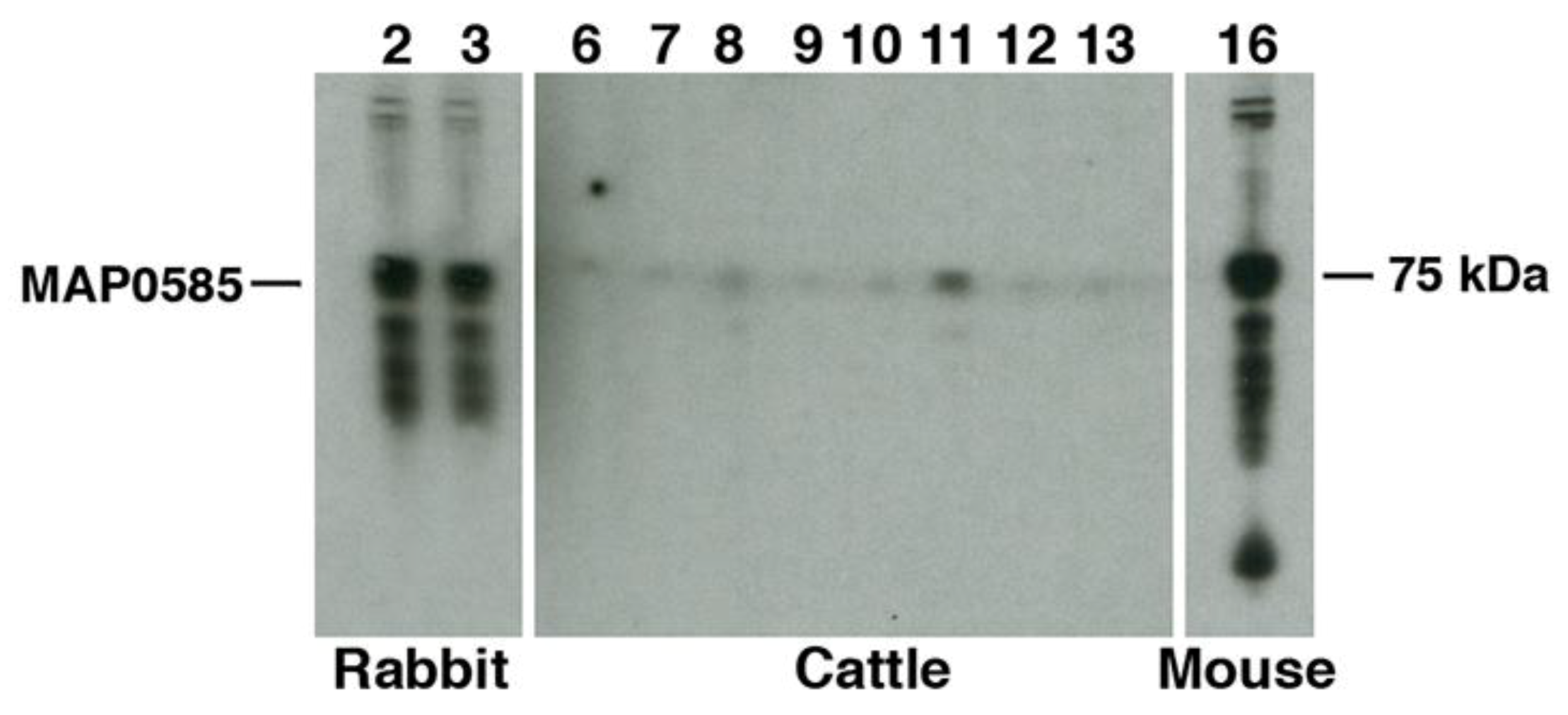
3.4. Identification of MAP_0585 in a Map Expression Library
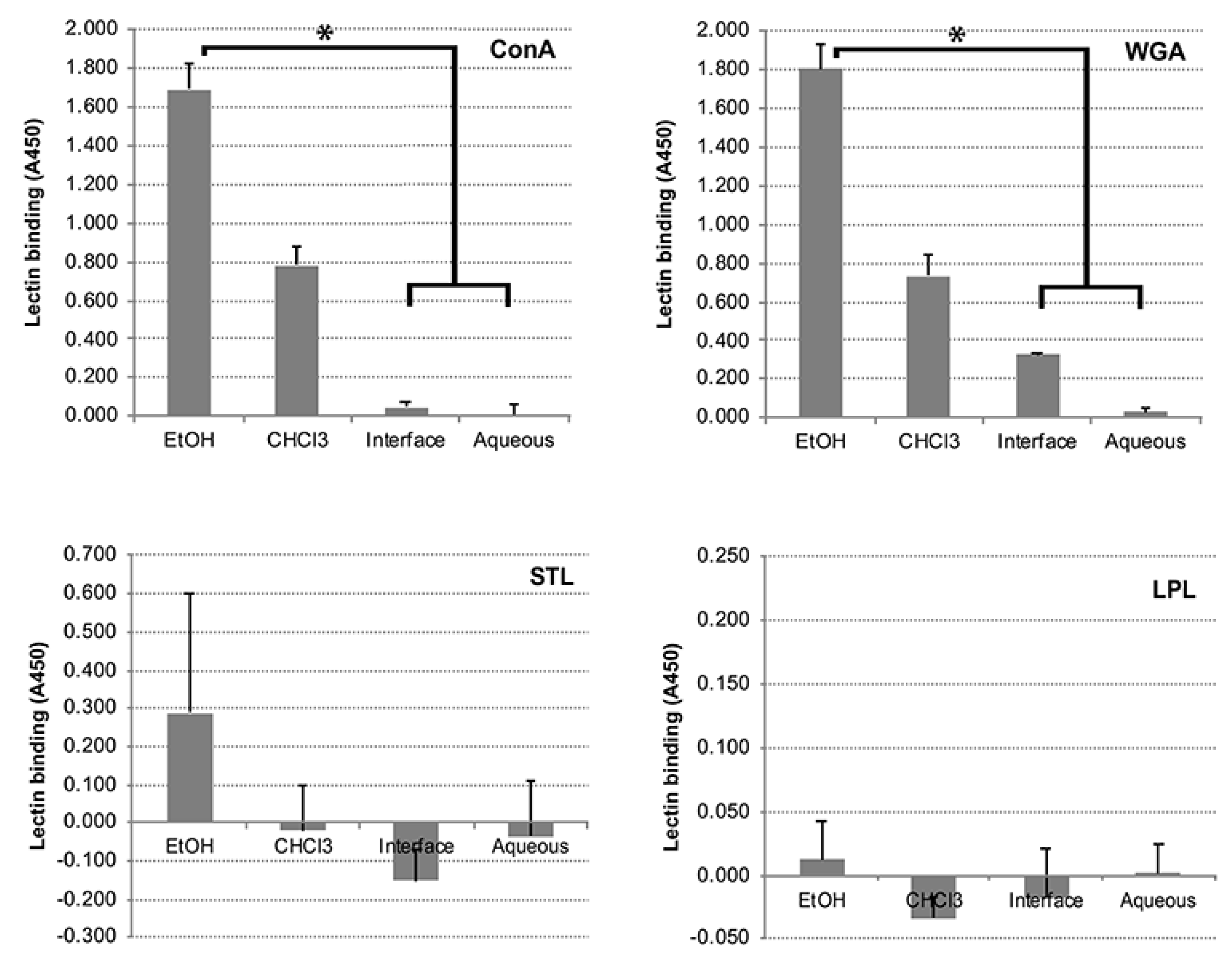
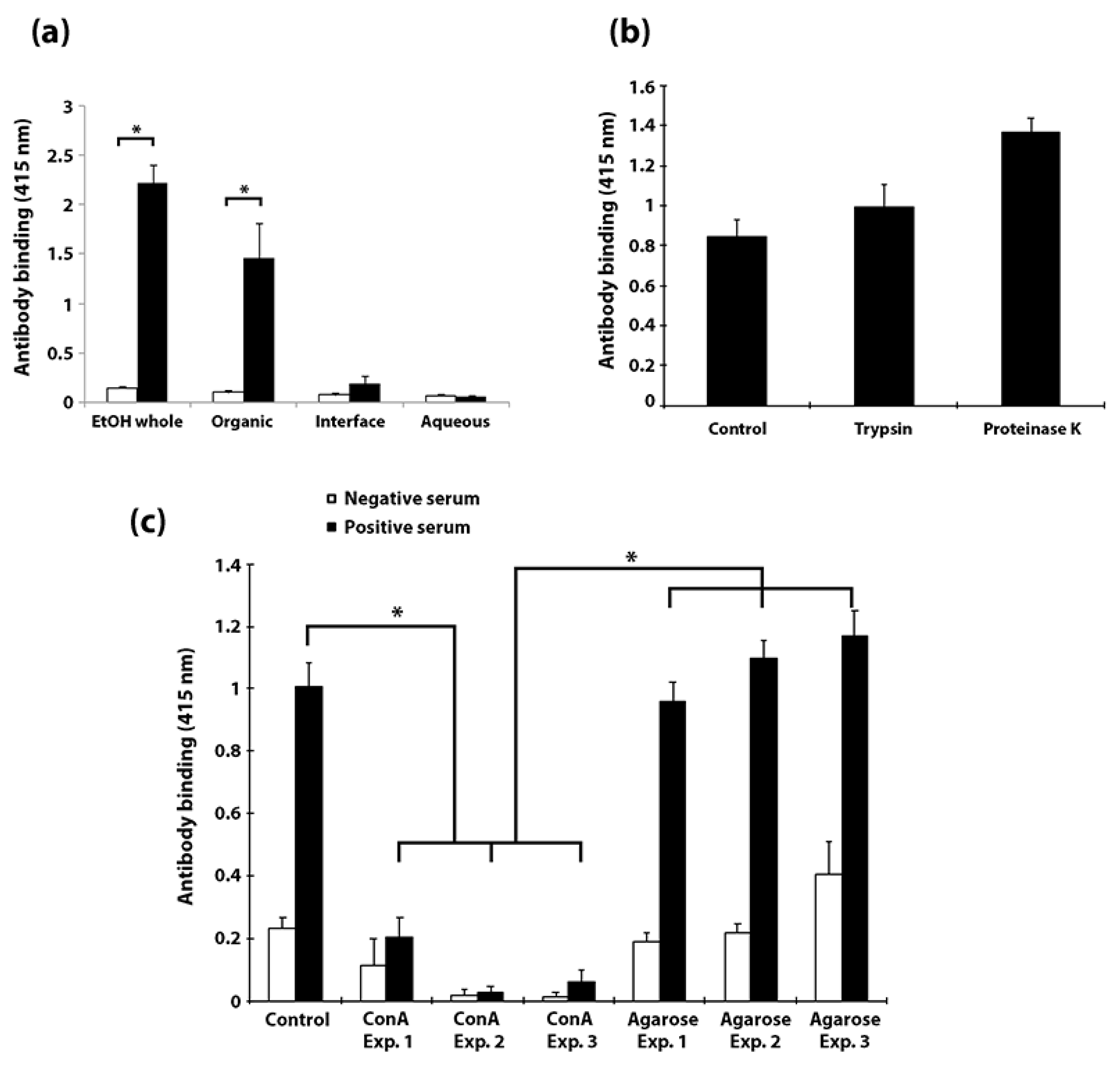
3.5. Carbohydrates, but Not Proteins in the EtOH Extract Are Immunogenic in Cattle
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Eda, S.; Elliott, B.; Scott, M.C.; Waters, W.R.; Bannantine, J.P.; Whitlock, R.H.; Speer, C.A. New method of serological testing for mycobacterium avium subsp. Paratuberculosis (johne’s disease) by flow cytometry. Foodborne Pathog. Dis. 2005, 2, 250–262. [Google Scholar] [CrossRef] [PubMed]
- Speer, C.A.; Scott, M.C.; Bannantine, J.P.; Waters, W.R.; Mori, Y.; Whitlock, R.H.; Eda, S. A novel enzyme-linked immunosorbent assay for diagnosis of mycobacterium avium subsp. Paratuberculosis infections (johne’s disease) in cattle. Clin. Vaccine Immunol. 2006, 13, 535–540. [Google Scholar] [CrossRef] [PubMed]
- Eda, S.; Bannantine, J.P.; Waters, W.R.; Mori, Y.; Whitlock, R.H.; Scott, M.C.; Speer, C.A. A highly sensitive and subspecies-specific surface antigen enzyme- linked immunosorbent assay for diagnosis of johne’s disease. Clin. Vaccine Immunol. 2006, 13, 837–844. [Google Scholar] [CrossRef] [PubMed]
- Wadhwa, A.; Johonson, R.E.; Eda, K.; Waters, W.R.; Palmer, M.V.; Bannantine, J.P.; Eda, S. Evaluation of ethanol vortex elisa for detection of bovine tuberculosis in cattle and deer. BMC Vet. Res. 2014, 10, 147. [Google Scholar] [CrossRef] [PubMed]
- Wadhwa, A.; Johnson, R.E.; Mackintosh, C.G.; Griffin, J.F.; Waters, W.R.; Bannantine, J.P.; Eda, S. Use of ethanol extract of mycobacterium bovis for detection of specific antibodies in sera of farmed red deer (cervus elaphus) with bovine tuberculosis. BMC Vet. Res. 2013, 9, 256. [Google Scholar] [CrossRef]
- Wadhwa, A.; Bannantine, J.P.; Byrem, T.M.; Stein, T.L.; Saxton, A.M.; Speer, C.A.; Eda, S. Optimization of serum evelisa for milk testing of johne’s disease. Foodborne Pathog. Dis 2012, 9, 749–754. [Google Scholar] [CrossRef]
- Eckstein, T.M.; Chandrasekaran, S.; Mahapatra, S.; McNeil, M.R.; Chatterjee, D.; Rithner, C.D.; Ryan, P.W.; Belisle, J.T.; Inamine, J.M. A major cell wall lipopeptide of mycobacterium avium subspecies paratuberculosis. J. Biol. Chem. 2006, 281, 5209–5215. [Google Scholar] [CrossRef]
- Bannantine, J.P.; Etienne, G.; Laval, F.; Stabel, J.R.; Lemassu, A.; Daffe, M.; Bayles, D.O.; Ganneau, C.; Bonhomme, F.; Branger, M.; et al. Cell wall peptidolipids of mycobacterium avium: From genetic prediction to exact structure of a nonribosomal peptide. Mol. Microbiol. 2017, 105, 525–539. [Google Scholar] [CrossRef]
- Mitachi, K.; Sharma Gautam, L.N.; Rice, J.H.; Eda, K.; Wadhwa, A.; Momotani, E.; Hlopak, J.P.; Eda, S.; Kurosu, M. Structure determination of lipopeptides from mycobacterium avium subspecies paratuberculosis and identification of antigenic lipopeptide probes. Anal. Biochem. 2016, 505, 29–35. [Google Scholar] [CrossRef]
- Thirunavukkarasu, S.; Plain, K.M.; Eckstein, T.M.; de Silva, K.; Whittington, R.J. Cellular and humoral immunogenicity of mycobacterium avium subsp. Paratuberculosis specific lipopentapeptide antigens. Res. Vet. Sci. 2013, 95, 123–129. [Google Scholar] [CrossRef]
- Bannantine, J.P.; Stabel, J.R.; Lippolis, J.D.; Reinhardt, T.A. Membrane and cytoplasmic proteins of mycobacterium avium subspecies paratuberculosis that bind to novel monoclonal antibodies. Microorganisms 2018, 6, 127. [Google Scholar] [CrossRef] [PubMed]
- Bannantine, J.P.; Radosevich, T.J.; Stabel, J.R.; Sreevatsan, S.; Kapur, V.; Paustian, M.L. Development and characterization of monoclonal antibodies and aptamers against major antigens of mycobacterium avium subsp. Paratuberculosis. Clin. Vaccine Immunol. 2007, 14, 518–526. [Google Scholar] [CrossRef] [PubMed]
- McNair, J.; Corbett, D.M.; Girvin, R.M.; Mackie, D.P.; Pollock, J.M. Characterization of the early antibody response in bovine tuberculosis: Mpb83 is an early target with diagnostic potential. Scand. J. Immunol. 2001, 53, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Bannantine, J.P.; Stabel, J.R. Identification of two mycobacterium avium subspecies paratuberculosis gene products differentially recognised by sera from rabbits immunised with live mycobacteria but not heat-killed mycobacteria. J. Med. Microbiol. 2001, 50, 795–804. [Google Scholar] [CrossRef] [PubMed]
- Friedman, M. Applications of the ninhydrin reaction for analysis of amino acids, peptides, and proteins to agricultural and biomedical sciences. J. Agric. Food Chem. 2004, 52, 385–406. [Google Scholar] [CrossRef] [PubMed]
- Bannantine, J.P.; Stabel, J.R. Killing of mycobacterium avium subspecies paratuberculosis within macrophages. BMC Microbiol. 2002, 2, 2. [Google Scholar] [CrossRef]
- Dahl, J.L. Scanning electron microscopy analysis of aged mycobacterium tuberculosis cells. Can. J. Microbiol. 2005, 51, 277–281. [Google Scholar] [CrossRef]
- Bech-Nielsen, S.; Jorgensen, J.B.; Ahrens, P.; Feld, N.C. Diagnostic accuracy of a mycobacterium phlei-absorbed serum enzyme-linked immunosorbent assay for diagnosis of bovine paratuberculosis in dairy cows. J. Clin. Microbiol. 1992, 30, 613–618. [Google Scholar]
- Bannantine, J.P.; Zhang, Q.; Li, L.L.; Kapur, V. Genomic homogeneity between mycobacterium avium subsp. Avium and mycobacterium avium subsp. Paratuberculosis belies their divergent growth rates. BMC Microbiol. 2003, 3, 10. [Google Scholar] [CrossRef]
- Cummings, R.D.; Darvill, A.G.; Etzler, M.E.; Hahn, M.G. Glycan-recognizing probes as tools. In Essentials of Glycobiology, 3rd ed.; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor (NY): New York, NY, USA, 2015; pp. 611–625. [Google Scholar]
- Aub, J.C.; Sanford, B.H.; Cote, M.N. Studies on reactivity of tumor and normal cells to a wheat germ agglutinin. Proc. Natl. Acad. Sci. USA 1965, 54, 396–399. [Google Scholar] [CrossRef]
- Gurung, R.B.; Purdie, A.C.; Whittington, R.J.; Begg, D.J. Cellular and humoral immune responses in sheep vaccinated with candidate antigens MAP2698c and MAP3567 from Mycobacterium avium subspecies paratuberculosis. Front. Cell. Infect. Microbiol. 2014, 4, 93. [Google Scholar] [CrossRef] [PubMed]
- Bannantine, J.P.; Radosevich, T.J.; Stabel, J.R.; Berger, S.; Griffin, J.F.; Paustian, M.L. Production and characterization of monoclonal antibodies against a major membrane protein of mycobacterium avium subsp. Paratuberculosis. Clin. Vaccine Immunol. 2007, 14, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Vosloo, W.; Tippoo, P.; Hughes, J.E.; Harriman, N.; Emms, M.; Beatty, D.W.; Zappe, H.; Steyn, L.M. Characterisation of a lipoprotein in mycobacterium bovis (bcg) with sequence similarity to the secreted protein mpb70. Gene 1997, 188, 123–128. [Google Scholar] [CrossRef]
- Harboe, M.; Wiker, H.G.; Ulvund, G.; Lund-Pedersen, B.; Andersen, A.B.; Hewinson, R.G.; Nagai, S. MPB70 and MPB83 as indicators of protein localization in mycobacterial cells. Infect. Immun. 1998, 66, 289–296. [Google Scholar] [PubMed]
- Bannantine, J.P.; Stabel, J.R.; Bayles, D.O.; Geisbrecht, B.V. Characteristics of an extensive mycobacterium avium subspecies paratuberculosis recombinant protein set. Protein Expr. Purif. 2010, 72, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Bannantine, J.P.; Campo, J.J.; Randall, A.; Grohn, Y.T.; Katani, R.; Schilling, M.; Radzio-Basu, J.; Kapur, V. Identification of sero-reactive antigens for the early diagnosis of johne’s disease in cattle. PLoS ONE 2017, 12, e0184373. [Google Scholar] [CrossRef] [PubMed]
- Stokes, R.W.; Norris-Jones, R.; Brooks, D.E.; Beveridge, T.J.; Doxsee, D.; Thorson, L.M. The glycan-rich outer layer of the cell wall of mycobacterium tuberculosis acts as an antiphagocytic capsule limiting the association of the bacterium with macrophages. Infect. Immun. 2004, 72, 5676–5686. [Google Scholar] [CrossRef]
- Bannantine, J.P.; Huntley, J.F.; Miltner, E.; Stabel, J.R.; Bermudez, L.E. The mycobacterium avium subsp. Paratuberculosis 35 kda protein plays a role in invasion of bovine epithelial cells. Microbiology 2003, 149, 2061–2069. [Google Scholar] [CrossRef]
- Mishra, A.K.; Driessen, N.N.; Appelmelk, B.J.; Besra, G.S. Lipoarabinomannan and related glycoconjugates: Structure, biogenesis and role in mycobacterium tuberculosis physiology and host-pathogen interaction. FEMS Microbiol. Rev. 2011, 35, 1126–1157. [Google Scholar] [CrossRef]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J.; et al. Psortb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef]
- Reichel, M.P.; Kittelberger, R.; Penrose, M.E.; Meynell, R.M.; Cousins, D.; Ellis, T.; Mutharia, L.M.; Sugden, E.A.; Johns, A.H.; de Lisle, G.W. Comparison of serological tests and faecal culture for the detection of mycobacterium avium subsp. Paratuberculosis infection in cattle and analysis of the antigens involved. Vet. Microbiol. 1999, 66, 135–150. [Google Scholar] [CrossRef]
- Sugden, E.A.; Stilwell, K.; Michaelides, A. A comparison of lipoarabinomannan with other antigens used in absorbed enzyme immunoassays for the serological detection of cattle infected with mycobacterium paratuberculosis. J. Vet. Diagn. Investig. 1997, 9, 413–417. [Google Scholar] [CrossRef] [PubMed]
- McKenna, S.L.; Sockett, D.C.; Keefe, G.P.; McClure, J.; VanLeeuwen, J.A.; Barkema, H.W. Comparison of two enzyme-linked immunosorbent assays for diagnosis of mycobacterium avium subsp. Paratuberculosis. J. Vet. Diagn. Investig. 2005, 17, 463–466. [Google Scholar] [CrossRef] [PubMed]
- Coulibaly, F.S.; Youan, B.B. Concanavalin a-polysaccharides binding affinity analysis using a quartz crystal microbalance. Biosens. Bioelectron. 2014, 59, 404–411. [Google Scholar] [CrossRef]
- Herrmann, J.L.; Delahay, R.; Gallagher, A.; Robertson, B.; Young, D. Analysis of post-translational modification of mycobacterial proteins using a cassette expression system. FEBS Lett. 2000, 473, 358–362. [Google Scholar] [CrossRef]

| Name | Type | Antigen 1 | Description |
|---|---|---|---|
| 17A12 | Murine mAb IgG1 | MAP_1025 | Proline rich antigen; RDD family protein |
| 14C5 | Murine mAb IgG2a | MAP_1272c | NlpC/P60; peptidoglycan hydrolase |
| 8G6 | Murine mAb IgG1 | MAP_1272c | NlpC/P60; peptidoglycan hydrolase |
| 9G10 | Murine mAb IgG2a | MAP_1643 | Isocitrate lyase, AceAb |
| 8G2 | Murine mAb IgG1 | MAP_2121c | Major membrane protein (MMP) |
| 14G3 | Murine mAb IgG2a | unknown | |
| 14D4 | Murine mAb IgG3 | MAP_2698c | Fatty acid desaturase |
| 6C9 | Murine mAb IgG1 | MAP_3060c | Electron transfer protein, α-subunit |
| 9H3 | Murine mAb IgG2b | MAP_3404 | Acetyl-CoA, biotin carboxylase subunit |
| 11G4 | Murine mAb IgG1 | MAP_3840 | DnaK chaperone; Heat shock protein |
| 14G11 | Murine mAb IgG1 | MAP_3976 | Lipoprotein anchoring transpeptidase |
| 7A6 | Murine mAb IgG2a | MAP_3936 | Molecular chaperone, GroEL2 |
| 12C9 | Murine mAb IgG1 | MAP_4145 | Memb. protein, short C-terminal domain |
| 13E1 | Murine mAb IgG1 | MAP_2121c | Major membrane protein (MMP) |
| 7C8 | Murine mAb IgG2a | MAP_3404 | Acetyl-CoA, biotin carboxylase subunit |
| p9270 | Human mAb IgG1 | LAM 2 | Cell wall lipopolysaccharide |
| p9045 | Human mAb IgG1 | LAM 2 | Cell wall lipopolysaccharide |
| 3993 | Rabbit polyclonal ab | EtOH 3 extract | |
| 3995 | Rabbit polyclonal ab | EtOH extract |
| Protein1 | Description | mAb | Map K-10 | M. Bovis |
|---|---|---|---|---|
| MAP_1025 | Proline rich antigen | 17A12 | - | - |
| MAP_1272c | NlpC/P60 protein | 14C5 | - | - |
| MAP_1272c | NlpC/P60 protein | 8G6 | - | - |
| MAP_1643 | Isocitrate lyase, AceAb | 11F6 | - | - |
| MAP_2121c | Membrane protein | 8G2 | - | - |
| MAP_2121c | Membrane protein | 13E1 | - | - |
| MAP_2698c | Fatty acid desaturase | 14D4 | + | - |
| MAP_3060c | Electron transfer protein | 6C9 | - | - |
| MAP_3404 | Acetyl-CoA carboxylase | 9H3 | - | - |
| MAP_3404 | Acetyl-CoA carboxylase | 11B8 | - | - |
| MAP_3840 | DnaK chaperone | 11G4 | - | - |
| MAP_3976 | Lipoprotein anchoring transpeptidase | 14G11 | - | - |
| MAP_3936 | Molecular chaperone GroEL2 | 7A6 | - | - |
| MAP_4145 | Membrane protein | 12C9 | - | - |
| LAM | Lipoarabinomannan | p9045 | + | + |
| LAM | Lipoarabinomannan | p9270 | + | + |
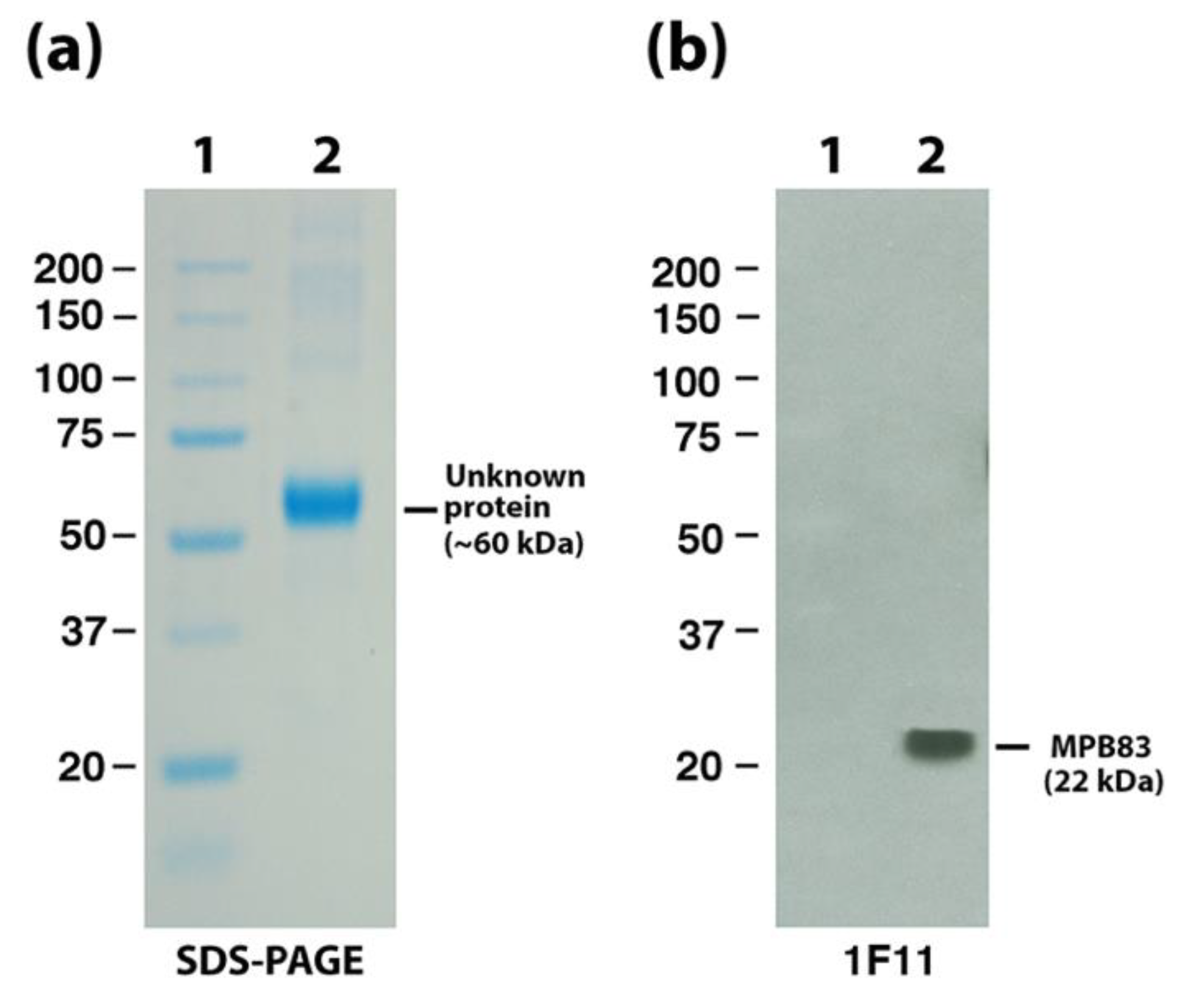
| Mb2898 | Surface lipoprotein MPB83 | 11F1 | - | + |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bannantine, J.P.; Wadhwa, A.; Stabel, J.R.; Eda, S. Characterization of Ethanol Extracted Cell Wall Components of Mycobacterium avium Subsp. paratuberculosis. Vet. Sci. 2019, 6, 88. https://doi.org/10.3390/vetsci6040088
Bannantine JP, Wadhwa A, Stabel JR, Eda S. Characterization of Ethanol Extracted Cell Wall Components of Mycobacterium avium Subsp. paratuberculosis. Veterinary Sciences. 2019; 6(4):88. https://doi.org/10.3390/vetsci6040088
Chicago/Turabian StyleBannantine, John P., Ashutosh Wadhwa, Judith R. Stabel, and Shigetoshi Eda. 2019. "Characterization of Ethanol Extracted Cell Wall Components of Mycobacterium avium Subsp. paratuberculosis" Veterinary Sciences 6, no. 4: 88. https://doi.org/10.3390/vetsci6040088
APA StyleBannantine, J. P., Wadhwa, A., Stabel, J. R., & Eda, S. (2019). Characterization of Ethanol Extracted Cell Wall Components of Mycobacterium avium Subsp. paratuberculosis. Veterinary Sciences, 6(4), 88. https://doi.org/10.3390/vetsci6040088