Detection of Bovine Leukemia Virus RNA in Blood Samples of Naturally Infected Dairy Cattle
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples under Study
2.2. Nucleic Acid Purification
2.3. Nucleic Acid Amplification
2.4. BLV Serology
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gutiérrez, G.; Carignano, H.; Alvarez, I.; Martínez, C.; Porta, N.; Politzki, R.; Gammella, M.; Lomonaco, M.; Fondevila, N.; Poli, M.; et al. Bovine leukemia virus p24 antibodies reflect blood proviral load. BMC Vet. Res. 2012, 8, 187. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, P.C.; Sordillo, L.M.; Byrem, T.M.; Norby, B.; Grooms, D.L.; Swenson, C.L.; Zalucha, J.; Erskine, R.J. Options for the control of bovine leukemia virus in dairy cattle. J. Am. Vet. Med. Assoc. 2014, 244, 914–922. [Google Scholar] [CrossRef] [PubMed]
- Walsh, R.B.; Kelton, D.F.; Hietala, S.K.; Duffield, T.F. Evaluation of enzyme-linked immunosorbent assays performed on milk and serum samples for detection of neosporosis and leukosis in lactating dairy cows. Can. Vet. J. La Rev. Vet. Can. 2013, 54, 347–352. [Google Scholar]
- Valor agregado institucional: captación de valor económico de innovaciones tecnológicas. Available online: https://inta.gob.ar/documentos/201cvalor-agregado-institucional-captacion-de-valor-economico-de-innovaciones-tecnologicas201d (accessed on 6 August 2019).
- Gillet, N.; Florins, A.; Boxus, M.; Burteau, C.; Nigro, A.; Vandermeers, F.; Balon, H.; Bouzar, A.B.; Defoiche, J.; Burny, A.; et al. Mechanisms of leukemogenesis induced by bovine leukemia virus: Prospects for novel anti-retroviral therapies in human. Retrovirology 2007, 4, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Rovnak, J.; Casey, J.W. Assessment of Bovine Leukemia Virus Transcripts In Vivo Reverse transcriptase PCR (RT-PCR) consistently detected bovine leukemia virus transcripts in fresh cells, and competitive RT-PCR enumerated these transcripts. The detection of transcripts in limited. J. Virol. 1999, 73, 8890–8897. [Google Scholar] [PubMed]
- Jensen, W.A.; Rovnak, J.; Cockerell, G.L. In Vivo Transcription of the Bovine Leukemia Virus taxlrex Region in Normal and Neoplastic Lymphocytes of Cattle and Sheep. J. Virol. 1991, 65, 2484–2490. [Google Scholar]
- Wu, D.; Murakami, K.; Morooka, A.; Jin, H.; Inoshima, Y.; Sentsui, H. In vivo transcription of bovine leukemia virus and bovine immunodeficiency-like virus. Virus Res. 2003, 97, 81–87. [Google Scholar] [CrossRef]
- Kuckleburg, C.J.; Chase, C.C.; Nelson, E.A.; Marras, S.A.; Dammen, M.A.; Christopher-Hennings, J. Detection of bovine leukemia virus in blood and milk by nested and real-time polymerase chain reactions. J. Vet. Diagn. Investig. 2003, 15, 72–76. [Google Scholar] [CrossRef]
- Jaworski, J.P.; Porta, N.G.; Gutierrez, G.; Politzki, R.P.; Álvarez, I.; Galarza, R.; Abdala, A.; Calvinho, L.; Trono, K.G. Relationship between the level of bovine leukemia virus antibody and provirus in blood and milk of cows from a naturally infected herd. J. Dairy Sci. 2016, 99, 5629–5634. [Google Scholar] [CrossRef]
- Petersen, M.I.; Alvarez, I.; Trono, K.G.; Jaworski, J.P. Quantification of bovine leukemia virus proviral DNA using a low-cost real-time polymerase chain reaction. J. Dairy Sci. 2018, 101, 6366–6374. [Google Scholar] [CrossRef]
- Jaworski, J.P.; Pluta, A.; Rola-Łuszczak, M.; McGowan, S.L.; Finnegan, C.; Heenemann, K.; Carignano, H.A.; Alvarez, I.; Murakami, K.; Willems, L.; et al. Interlaboratory Comparison of Six Real-Time PCR Assays for Detection of Bovine Leukemia Virus Proviral DNA. J. Clin. Microbiol. 2018, 56, e00304–e00318. [Google Scholar] [CrossRef] [PubMed]
- Trono, K.G.; Pérez-Filgueira, D.M.; Duffy, S.; Borca, M.V.; Carrillo, C. Seroprevalence of bovine leukemia virus in dairy cattle in Argentina: Comparison of sensitivity and specificity of different detection methods. Vet. Microbiol. 2001, 83, 235–248. [Google Scholar] [CrossRef]
- Alexandersen, S.; Carpenter, S.; Christensen, J.; Storgaard, T.; Viuff, B.; Wannemuehler, Y.; Belousov, J.; Roth, J.A. Identification of Alternatively Spliced mRNAs Encoding Potential New Regulatory Proteins in Cattle Infected with Bovine Leukemia Virus. J. Virol. 1993, 67, 39–52. [Google Scholar] [PubMed]
- Haas, L.; Divers, T.; Casey, J.W. Bovine Leukemia Virus Gene Expression In Vivo. J. Virol. 1992, 66, 6223–6225. [Google Scholar] [PubMed]
- Frie, M.C.; Droscha, C.J.; Greenlick, A.E.; Coussens, P.M. MicroRNAs Encoded by Bovine Leukemia Virus (BLV) Are Associated with Reduced Expression of B Cell Transcriptional Regulators in Dairy Cattle Naturally Infected with BLV. Front. Vet. Sci. 2017, 4, 245. [Google Scholar] [CrossRef] [PubMed]
- Jensen, W.A.; Sheehy, S.E.; Fox, M.H.; Davis, W.C.; Cockerell, G.L. In vitro expression of bovine leukemia virus in isolated B-lymphocytes of cattle and sheep. Vet. Immunol. Immunopathol. 1990, 26, 333–342. [Google Scholar] [CrossRef]
- Sherman, M.P.; Ehrlich, G.D.; Ferrer, J.F.; Sninsky, J.J.; Zandomeni, R.; Dock, N.L.; Poiesz1, B.J. Amplification and Analysis of Specific DNA and RNA Sequences of Bovine Leukemia Virus from Infected Cows by Polymerase Chain Reaction. J. Clin. Microbiol. 1992, 30, 185–191. [Google Scholar] [PubMed]
- Driscoll, D.M.; Olson, C. Bovine leukemia virus-associated antigens in lymphocyte cultures. Am. J. Vet. Res. 1977, 38, 1897–1898. [Google Scholar]
- Kerkhofs, P.; Adam, E.; Droogmans, L.; Portetelle, D.; Mammerickx, M.; Burny, A.; Kettmann, R.; Willems, L. Cellular Pathways Involved in the Ex Vivo Expression of Bovine Leukemia Virus. J. Virol. 1996, 70, 2170–2177. [Google Scholar]
- Mammerickx, M.; Portetelle, D.; de Clercq, K.; Burny, A. Experimental transmission of enzootic bovine leukosis to cattle, sheep and goats: infectious doses of blood and incubation period of the disease. Leuk. Res. 1987, 11, 353–358. [Google Scholar] [CrossRef]
- Mirsky, M.L.; Olmstead, C.A.; Da, Y.; Lewin, H.A. The prevalence of proviral bovine leukemia virus in peripheral blood mononuclear cells at two subclinical stages of infection. J. Virol. 1996, 70, 2178–2183. [Google Scholar] [PubMed]
- Carpentier, A.; Barez, P.-Y.; Hamaidia, M.; Gazon, H.; de Brogniez, A.; Perike, S.; Gillet, N.; Willems, L. Modes of Human T Cell Leukemia Virus Type 1 Transmission, Replication and Persistence. Viruses 2015, 7, 3603–3624. [Google Scholar] [CrossRef] [PubMed]
- Mellors, J.W.; Munoz, A.; Giorgi, J.V.; Margolick, J.B.; Tassoni, C.J.; Gupta, P.; Kingsley, L.A.; Todd, J.A.; Saah, A.J.; Detels, R.; et al. Plasma Viral Load and CD4+ Lymphocytes as Prognostic Markers of HIV-1 Infection. Ann. Intern. Med. 1997, 126, 946–954. [Google Scholar] [CrossRef] [PubMed]
- Cabral, F.; Barbara Arruda, L.; Ladeira de Araújo, M.; Montanheiro, P.; Smid, J.; César Penalva de Oliveira, A.; Duarte, A.J.; Casseb, J. Detection of human T-cell lymphotropic virus type 1 in plasma samples. Virus Res. 2012, 163, 87–90. [Google Scholar] [CrossRef] [PubMed]
- Demontis, M.A.; Sadiq, M.T.; Golz, S.; Taylor, G.P. HTLV-1 viral RNA is detected rarely in plasma of HTLV-1 infected subjects. J. Med. Virol. 2015, 87, 2130–2134. [Google Scholar] [CrossRef] [PubMed]
- Jaworski, J.P.; Petersen, M.I.; Carignano, H.A.; Trono, K.G. Spontaneous virus reactivation in cattle chronically infected with bovine leukemia virus. BMC Vet. Res. 2019, 15, 150. [Google Scholar]
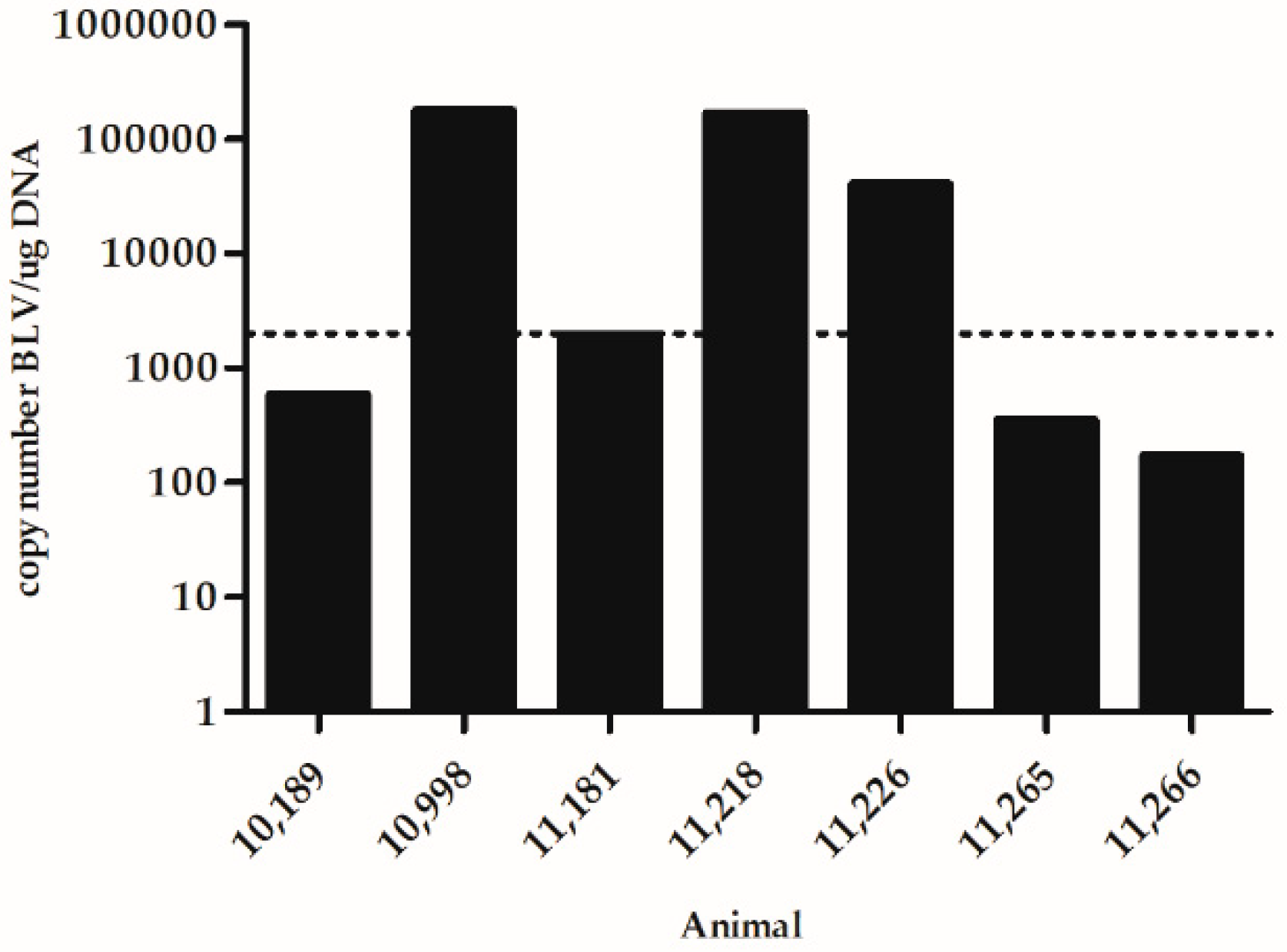
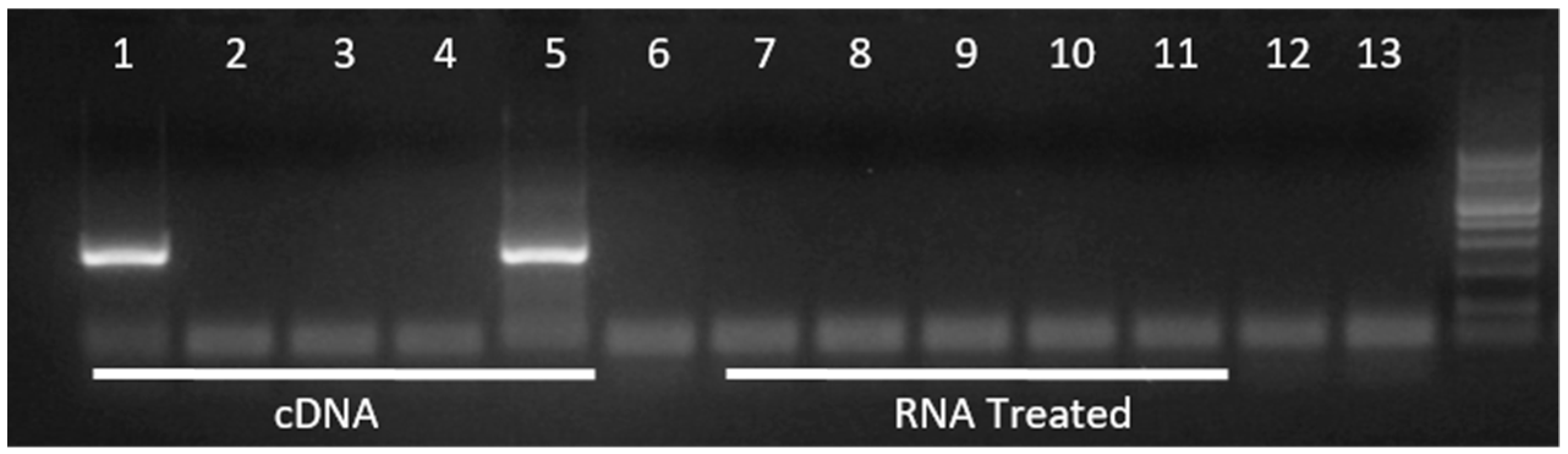
| High PVL 2 | Low PVL 2 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time Point 1 | 10,998 | 11,218 | 11,226 | 10,189 | 11,181 | 11,265 | 11,266 | |||||||
| Tax | Pol | Tax | Pol | Tax | Pol | Tax | Pol | Tax | Pol | Tax | Pol | Tax | Pol | |
| 5 BD | + | − | − | − | − | − | − | − | − | − | − | − | − | − |
| D1 | − | − | + | + | − | − | − | − | + | − | NE | NE | − | − |
| 5 AD | NE | NE | + | − | − | − | − | − | − | − | − | − | − | − |
| 2 M | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 4 M | + | − | − | + | − | − | − | − | − | − | − | − | − | − |
| 6 M | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 8 M | − | − | + | + | − | − | NE | NE | − | − | − | − | − | − |
| 10 M | − | − | − | − | − | − | NE | NE | − | − | − | − | − | − |
| 5 BD2 | − | − | − | − | NE | NE | NE | NE | − | − | − | − | NE | NE |
| D2 | + | − | + | − | + | + | NE | NE | − | − | − | − | NE | NE |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alvarez, I.; Porta, N.G.; Trono, K. Detection of Bovine Leukemia Virus RNA in Blood Samples of Naturally Infected Dairy Cattle. Vet. Sci. 2019, 6, 66. https://doi.org/10.3390/vetsci6030066
Alvarez I, Porta NG, Trono K. Detection of Bovine Leukemia Virus RNA in Blood Samples of Naturally Infected Dairy Cattle. Veterinary Sciences. 2019; 6(3):66. https://doi.org/10.3390/vetsci6030066
Chicago/Turabian StyleAlvarez, Irene, Natalia Gabriela Porta, and Karina Trono. 2019. "Detection of Bovine Leukemia Virus RNA in Blood Samples of Naturally Infected Dairy Cattle" Veterinary Sciences 6, no. 3: 66. https://doi.org/10.3390/vetsci6030066
APA StyleAlvarez, I., Porta, N. G., & Trono, K. (2019). Detection of Bovine Leukemia Virus RNA in Blood Samples of Naturally Infected Dairy Cattle. Veterinary Sciences, 6(3), 66. https://doi.org/10.3390/vetsci6030066





