Quantitative Single-Cell Transcript Assessment of Biomarkers Supports Cellular Heterogeneity in the Bovine IVD
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Collection and IVD Isolation
2.2. Scanning Electron Microscopy (SEM)
2.3. Fluorescent RNA in situ Hybridization (FL-RISH) and Immunohistochemistry (IHC)
2.4. Selection of Investigated Biomarkers
2.5. Data Collection and Analysis
3. Results
3.1. Morphology of Outer AF and NP Cells in their ECM Environment
3.2. Subcellular Transcript Localization of Proposed Biomarkers in Outer AF and NP Cells
3.3. Confirmation of Biomarkers Using Transcript Quantification Through Population Averaging
3.4. Transcriptional Heterogeneity in IVD Cells is Not a Sole Consequence of Cell Cycle Arrest
3.5. Single Cell Transcript Analysis Indicates Further Heterogeneity in Transcriptionally Active Cells.
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Humzah, M.D.; Soames, R.W. Human intervertebral disc: structure and function. Anat Rec. 1988, 220, 337–356. [Google Scholar] [CrossRef] [PubMed]
- Kraus, P.; Li, K.; Sipes, D.; Varden, L.; Yerden, R.; Henderson, A.; Sur, S.; Lufkin, T. Single-Cell Phenotyping of Complex Heterogeneous Tissue. In Handbook of Single Cell Technologies; Santra, T.S., Tseng, F.-G., Eds.; Springer: Singapore, 2019; pp. 1–17. [Google Scholar]
- Li, K.; Kapper, D.; Youngs, B.; Kocsis, V.; Mondal, S.; Kraus, P.; Lufkin, T. Potential biomarkers of the mature intervertebral disc identified at the single cell level. J. Anat. 2019, 234, 16–32. [Google Scholar] [CrossRef] [PubMed]
- Stergar, J.; Gradisnik, L.; Velnar, T.; Maver, U. Intervertebral disc tissue engineering: A brief review. Bosn. J. Basic. Med. Sci. 2019. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.J.; Silverman, L.; Sakai, D.; Le Maitre, C.L.; Mauck, R.L.; Malhotra, N.R.; Lotz, J.C.; Buckley, C.T. Advancing cell therapies for intervertebral disc regeneration from the lab to the clinic: Recommendations of the ORS spine section. JOR Spine 2018, 1, e1036. [Google Scholar] [CrossRef] [PubMed]
- McCann, M.R.; Bacher, C.A.; Seguin, C.A. Exploiting notochord cells for stem cell-based regeneration of the intervertebral disc. J. Cell Commun. Signal 2011, 5, 39–43. [Google Scholar] [CrossRef] [PubMed]
- Eyre, D.R.; Muir, H. Types I and II collagens in intervertebral disc. Interchanging radial distributions in annulus fibrosus. Biochem. J. 1976, 157, 267–270. [Google Scholar] [CrossRef] [PubMed]
- Bron, J.L.; Helder, M.N.; Meisel, H.J.; Van Royen, B.J.; Smit, T.H. Repair, regenerative and supportive therapies of the annulus fibrosus: achievements and challenges. Eur. Spine J. 2009, 18, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Sivakamasundari, V.; Lufkin, T. Stemming the Degeneration: IVD Stem Cells and Stem Cell Regenerative Therapy for Degenerative Disc Disease. Adv. Stem. Cells 2013, 2013. [Google Scholar] [CrossRef]
- Pennicooke, B.; Moriguchi, Y.; Hussain, I.; Bonssar, L.; Hartl, R. Biological Treatment Approaches for Degenerative Disc Disease: A Review of Clinical Trials and Future Directions. Cureus 2016, 8, e892. [Google Scholar] [CrossRef] [PubMed]
- Oehme, D.; Goldschlager, T.; Ghosh, P.; Rosenfeld, J.V.; Jenkin, G. Cell-Based Therapies Used to Treat Lumbar Degenerative Disc Disease: A Systematic Review of Animal Studies and Human Clinical Trials. Stem Cells Int. 2015, 2015, 946031. [Google Scholar] [CrossRef] [PubMed]
- Brown, C. Stem cell tourism poses risks. CMAJ 2012, 184, E121–122. [Google Scholar] [CrossRef]
- Rodrigues-Pinto, R.; Richardson, S.M.; Hoyland, J.A. Identification of novel nucleus pulposus markers: Interspecies variations and implications for cell-based therapiesfor intervertebral disc degeneration. Bone Joint Res. 2013, 2, 169–178. [Google Scholar] [CrossRef]
- Schubert, A.K.; Smink, J.J.; Arp, M.; Ringe, J.; Hegewald, A.A.; Sittinger, M. Quality Assessment of Surgical Disc Samples Discriminates Human Annulus Fibrosus and Nucleus Pulposus on Tissue and Molecular Level. Int. J. Mol. Sci. 2018, 19, 1761. [Google Scholar] [CrossRef]
- Rutges, J.; Creemers, L.B.; Dhert, W.; Milz, S.; Sakai, D.; Mochida, J.; Alini, M.; Grad, S. Variations in gene and protein expression in human nucleus pulposus in comparison with annulus fibrosus and cartilage cells: potential associations with aging and degeneration. Osteoarthritis Cartilage 2010, 18, 416–423. [Google Scholar] [CrossRef]
- Kraus, P.; Sivakamasundari, V.; Xing, X.; Lufkin, T. Generating mouse lines for lineage tracing and knockout studies. Methods Mol. Biol. 2014, 1194, 37–62. [Google Scholar] [CrossRef]
- Choi, K.S.; Harfe, B.D. Hedgehog signaling is required for formation of the notochord sheath and patterning of nuclei pulposi within the intervertebral discs. PNAS 2011, 108, 9484–9489. [Google Scholar] [CrossRef]
- Choi, K.S.; Lee, C.; Harfe, B.D. Sonic hedgehog in the notochord is sufficient for patterning of the intervertebral discs. Mech. Dev. 2012, 129, 255–262. [Google Scholar] [CrossRef]
- Smits, P.; Lefebvre, V. Sox5 and Sox6 are required for notochord extracellular matrix sheath formation, notochord cell survival and development of the nucleus pulposus of intervertebral discs. Development 2003, 130, 1135–1148. [Google Scholar] [CrossRef]
- Kraus, P.; Yerden, R.; Kocsis, V.; Lufkin, T. RNA in situ hybridization characterization of non-enzymatic derived bovine intervertebral disc cell lineages suggests progenitor cell potential. Acta Histochem. 2017. [Google Scholar] [CrossRef]
- Oshima, H.; Ishihara, H.; Urban, J.P.; Tsuji, H. The use of coccygeal discs to study intervertebral disc metabolism. J. Orthop. Res. 1993, 11, 332–338. [Google Scholar] [CrossRef]
- Demers, C.N.; Antoniou, J.; Mwale, F. Value and limitations of using the bovine tail as a model for the human lumbar spine. Spine (Phila Pa 1976) 2004, 29, 2793–2799. [Google Scholar] [CrossRef]
- Beckstein, J.C.; Sen, S.; Schaer, T.P.; Vresilovic, E.J.; Elliott, D.M. Comparison of animal discs used in disc research to human lumbar disc: axial compression mechanics and glycosaminoglycan content. Spine (Phila Pa 1976) 2008, 33, E166–173. [Google Scholar] [CrossRef]
- Michalek, A.J. A growth-based model for the prediction of fiber angle distribution in the intervertebral disc annulus fibrosus. Biomech. Model Mechanobiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Ekblom, M.; Falk, M.; Salmivirta, K.; Durbeej, M.; Ekblom, P. Laminin isoforms and epithelial development. Ann. N. Y. Acad. Sci. 1998, 857, 194–211. [Google Scholar] [CrossRef]
- Ahn, S.; Joyner, A.L. In vivo analysis of quiescent adult neural stem cells responding to Sonic hedgehog. Nature 2005, 437, 894–897. [Google Scholar] [CrossRef] [PubMed]
- Buttitta, L.; Mo, R.; Hui, C.C.; Fan, C.M. Interplays of Gli2 and Gli3 and their requirement in mediating Shh-dependent sclerotome induction. Development 2003, 130, 6233–6243. [Google Scholar] [CrossRef]
- Shin, S.H.; Kogerman, P.; Lindstrom, E.; Toftgard, R.; Biesecker, L.G. GLI3 mutations in human disorders mimic Drosophila cubitus interruptus protein functions and localization. PNAS 1999, 96, 2880–2884. [Google Scholar] [CrossRef]
- Abdelkhalek, H.B.; Beckers, A.; Schuster-Gossler, K.; Pavlova, M.N.; Burkhardt, H.; Lickert, H.; Rossant, J.; Reinhardt, R.; Schalkwyk, L.C.; Muller, I.; et al. The mouse homeobox gene Not is required for caudal notochord development and affected by the truncate mutation. Genes Dev. 2004, 18, 1725–1736. [Google Scholar] [CrossRef] [PubMed]
- McCann, M.R.; Tamplin, O.J.; Rossant, J.; Seguin, C.A. Tracing notochord-derived cells using a Noto-cre mouse: Implications for intervertebral disc development. Dis. Model Mech. 2012, 5, 73–82. [Google Scholar] [CrossRef]
- Cserjesi, P.; Brown, D.; Ligon, K.L.; Lyons, G.E.; Copeland, N.G.; Gilbert, D.J.; Jenkins, N.A.; Olson, E.N. Scleraxis: A basic helix-loop-helix protein that prefigures skeletal formation during mouse embryogenesis. Development 1995, 121, 1099–1110. [Google Scholar]
- Shukunami, C.; Takimoto, A.; Oro, M.; Hiraki, Y. Scleraxis positively regulates the expression of tenomodulin, a differentiation marker of tenocytes. Dev. Biol. 2006, 298, 234–247. [Google Scholar] [CrossRef]
- Niwa, H.; Ogawa, K.; Shimosato, D.; Adachi, K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature 2009, 460, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Kraus, P.; Sivakamasundari, V.; Yu, H.B.; Xing, X.; Lim, S.L.; Adler, T.; Pimentel, J.A.; Becker, L.; Bohla, A.; Garrett, L.; et al. Pleiotropic functions for transcription factor zscan10. PLoS ONE 2014, 9, e104568. [Google Scholar] [CrossRef]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Lyu, F.J.; Cheung, K.M.; Zheng, Z.; Wang, H.; Sakai, D.; Leung, V.Y. IVD progenitor cells: a new horizon for understanding disc homeostasis and repair. Nat. Rev. Rheumatol. 2019, 15, 102–112. [Google Scholar] [CrossRef]
- Wang, W.; Lo, P.; Frasch, M.; Lufkin, T. Hmx: an evolutionary conserved homeobox gene family expressed in the developing nervous system in mice and Drosophila. Mech. Dev. 2000, 99, 123–137. [Google Scholar] [CrossRef]
- Lohnes, D.; Dierich, A.; Ghyselinck, N.; Kastner, P.; Lampron, C.; LeMeur, M.; Lufkin, T.; Mendelsohn, C.; Nakshatri, H.; Chambon, P. Retinoid receptors and binding proteins. J. Cell Sci. Suppl. 1992, 16, 69–76. [Google Scholar] [CrossRef]
- Kraus, P.; Lufkin, T. Mammalian Dlx homeobox gene control of craniofacial and inner ear morphogenesis. J. Cell Biochem. 1999, Suppl 32–33, 133–140. [Google Scholar] [CrossRef]
- Chen, X.; Li, X.; Wang, W.; Lufkin, T. Dlx5 and Dlx6: an evolutionary conserved pair of murine homeobox genes expressed in the embryonic skeleton. Ann. N. Y. Acad. Sci. 1996, 785, 38–47. [Google Scholar] [CrossRef]
- Minogue, B.M.; Richardson, S.M.; Zeef, L.A.; Freemont, A.J.; Hoyland, J.A. Transcriptional profiling of bovine intervertebral disc cells: implications for identification of normal and degenerate human intervertebral disc cell phenotypes. Arthritis Res. Ther. 2010, 12, R22. [Google Scholar] [CrossRef]
- Minogue, B.M.; Richardson, S.M.; Zeef, L.A.; Freemont, A.J.; Hoyland, J.A. Characterization of the human nucleus pulposus cell phenotype and evaluation of novel marker gene expression to define adult stem cell differentiation. Arthritis Rheum. 2010, 62, 3695–3705. [Google Scholar] [CrossRef]
- Van den Akker, G.G.H.; Koenders, M.I.; van de Loo, F.A.J.; van Lent, P.; Blaney Davidson, E.; van der Kraan, P.M. Transcriptional profiling distinguishes inner and outer annulus fibrosus from nucleus pulposus in the bovine intervertebral disc. Eur. Spine J. 2017, 26, 2053–2062. [Google Scholar] [CrossRef]
- Chatterjee, S.; Kraus, P.; Sivakamasundari, V.; Xing, X.; Yap, S.P.; Jie, S.; Lufkin, T. A conditional mouse line for lineage tracing of Sox9 loss-of-function cells using enhanced green fluorescent protein. Biotechnol. Lett. 2013, 35, 1991–1996. [Google Scholar] [CrossRef] [PubMed]
- Kraus, P.; Sivakamasundari, V.; Olsen, V.; Villeneuve, V.; Hinds, A.; Lufkin, T. Klhl14 antisense RNA is a target of key skeletogenic transcription factors in the developing intervertebral disc. Spine (Phila Pa 1976) 2019, 44, E260–E268. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Sivakamasundari, V.; Yap, S.P.; Kraus, P.; Kumar, V.; Xing, X.; Lim, S.L.; Sng, J.; Prabhakar, S.; Lufkin, T. In vivo genome-wide analysis of multiple tissues identifies gene regulatory networks, novel functions and downstream regulatory genes for Bapx1 and its co-regulation with Sox9 in the mammalian vertebral column. BMC Genomics 2014, 15, 1072. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.J.; Chatterjee, S.; Yap, S.P.; Lim, S.L.; Xing, X.; Kraus, P.; Sun, W.; Hu, X.; Sivakamasundari, V.; Chan, H.Y.; et al. An Integrative Developmental Genomics and Systems Biology Approach to Identify an In Vivo Sox Trio-Mediated Gene Regulatory Network in Murine Embryos. Biomed. Res. Int. 2017, 2017, 8932583. [Google Scholar] [CrossRef] [PubMed]
- Sivakamasundari, V.; Kraus, P.; Jie, S.; Lufkin, T. Pax1(EGFP): New wildtype and mutant EGFP mouse lines for molecular and fate mapping studies. Genesis 2013, 51, 420–429. [Google Scholar] [CrossRef]
- Sivakamasundari, V.; Kraus, P.; Sun, W.; Hu, X.; Lim, S.L.; Prabhakar, S.; Lufkin, T. A developmental transcriptomic analysis of Pax1 and Pax9 in embryonic intervertebral disc development. Biol. Open 2017, 6, 187–199. [Google Scholar] [CrossRef]
- Murray, C.J.; Atkinson, C.; Bhalla, K.; Birbeck, G.; Burstein, R.; Chou, D.; Dellavalle, R.; Danaei, G.; Ezzati, M.; Fahimi, A.; et al. The state of US health, 1990-2010: burden of diseases, injuries, and risk factors. JAMA 2013, 310, 591–608. [Google Scholar] [CrossRef]
- Daly, C.; Ghosh, P.; Jenkin, G.; Oehme, D.; Goldschlager, T. A Review of Animal Models of Intervertebral Disc Degeneration: Pathophysiology, Regeneration, and Translation to the Clinic. Biomed. Res. Int. 2016, 2016, 5952165. [Google Scholar] [CrossRef]
- Kraus, P.; Lufkin, T. Implications for a Stem Cell Regenerative Medicine Based Approach to Human Intervertebral Disk Degeneration. Front. Cell Dev. Biol. 2017, 5, 17. [Google Scholar] [CrossRef]
- Lyons, G.; Eisenstein, S.M.; Sweet, M.B. Biochemical changes in intervertebral disc degeneration. Biochim. Biophys. Acta 1981, 673, 443–453. [Google Scholar] [CrossRef]
- Antoniou, J.; Steffen, T.; Nelson, F.; Winterbottom, N.; Hollander, A.P.; Poole, R.A.; Aebi, M.; Alini, M. The human lumbar intervertebral disc: evidence for changes in the biosynthesis and denaturation of the extracellular matrix with growth, maturation, ageing, and degeneration. J. Clin. Invest. 1996, 98, 996–1003. [Google Scholar] [CrossRef]
- Urban, J.P.; Roberts, S. Degeneration of the intervertebral disc. Arthritis Res. Ther. 2003, 5, 120–130. [Google Scholar] [CrossRef]
- Sakai, D.; Andersson, G.B. Stem cell therapy for intervertebral disc regeneration: obstacles and solutions. Nat. Rev. Rheumatol. 2015, 11, 243–256. [Google Scholar] [CrossRef] [PubMed]
- Lama, P.; Le Maitre, C.L.; Harding, I.J.; Dolan, P.; Adams, M.A. Nerves and blood vessels in degenerated intervertebral discs are confined to physically disrupted tissue. J. Anat. 2018, 233, 86–97. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, B.; Zhao, X.; Li, X.; Lou, Z.; Chen, X.; Zhang, F. Iron deficiency accelerates intervertebral disc degeneration through affecting the stability of DNA polymerase epsilon complex. Am. J. Transl. Res. 2018, 10, 3430–3442. [Google Scholar] [PubMed]
- Haidar, R.; Musallam, K.M.; Taher, A.T. Bone disease and skeletal complications in patients with beta thalassemia major. Bone 2011, 48, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Risbud, M.V.; Schoepflin, Z.R.; Mwale, F.; Kandel, R.A.; Grad, S.; Iatridis, J.C.; Sakai, D.; Hoyland, J.A. Defining the phenotype of young healthy nucleus pulposus cells: Recommendations of the Spine Research Interest Group at the 2014 annual ORS meeting. J. Orthop. Res. 2015, 33, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Risbud, M.V.; Shapiro, I.M. Notochordal cells in the adult intervertebral disc: new perspective on an old question. Crit. Rev. Eukaryot. Gene Expr. 2011, 21, 29–41. [Google Scholar] [CrossRef]
- Bushell, G.R.; Ghosh, P.; Taylor, T.F.; Akeson, W.H. Proteoglycan chemistry of the intervertebral disks. Clin. Orthop. Relat. Res. 1977, 115–123. [Google Scholar] [CrossRef]
- Sng, J.; Lufkin, T. Emerging stem cell therapies: treatment, safety, and biology. Stem Cells Int. 2012, 2012, 521343. [Google Scholar] [CrossRef]
- Urban, J.P.; Holm, S.; Maroudas, A.; Nachemson, A. Nutrition of the intervertebral disk. An in vivo study of solute transport. Clin. Orthop. Relat. Res. 1977, 101–114. [Google Scholar] [CrossRef]
- Wuertz, K.; Godburn, K.; Neidlinger-Wilke, C.; Urban, J.; Iatridis, J.C. Behavior of mesenchymal stem cells in the chemical microenvironment of the intervertebral disc. Spine (Phila Pa 1976) 2008, 33, 1843–1849. [Google Scholar] [CrossRef]
- Liang, C.Z.; Li, H.; Tao, Y.Q.; Zhou, X.P.; Yang, Z.R.; Li, F.C.; Chen, Q.X. The relationship between low pH in intervertebral discs and low back pain: a systematic review. Arch. Med. Sci. 2012, 8, 952–956. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Johnson, Z.I.; Risbud, M.V. Understanding nucleus pulposus cell phenotype: a prerequisite for stem cell based therapies to treat intervertebral disc degeneration. Curr. Stem Cell Res. Ther. 2015, 10, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Errington, R.J.; Puustjarvi, K.; White, I.R.; Roberts, S.; Urban, J.P. Characterisation of cytoplasm-filled processes in cells of the intervertebral disc. J. Anat. 1998, 192 (Pt 3), 369–378. [Google Scholar] [CrossRef]
- Huang, Y.C.; Urban, J.P.; Luk, K.D. Intervertebral disc regeneration: do nutrients lead the way? Nat. Rev. Rheumatol. 2014, 10, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Bedore, J.; Leask, A.; Seguin, C.A. Targeting the extracellular matrix: matricellular proteins regulate cell-extracellular matrix communication within distinct niches of the intervertebral disc. Matrix. Biol. 2014, 37, 124–130. [Google Scholar] [CrossRef]
- Walmsley, R. The development and growth of the intervertebral disc. Edinb. Med. J. 1953, 60, 341–364. [Google Scholar]
- Trout, J.J.; Buckwalter, J.A.; Moore, K.C. Ultrastructure of the human intervertebral disc: II. Cells of the nucleus pulposus. Anat. Rec. 1982, 204, 307–314. [Google Scholar] [CrossRef]
- Trout, J.J.; Buckwalter, J.A.; Moore, K.C.; Landas, S.K. Ultrastructure of the human intervertebral disc. I. Changes in notochordal cells with age. Tissue Cell 1982, 14, 359–369. [Google Scholar] [CrossRef]
- Chen, J.; Yan, W.; Setton, L.A. Molecular phenotypes of notochordal cells purified from immature nucleus pulposus. Eur. Spine J. 2006, 15 Suppl 3, S303–311. [Google Scholar] [CrossRef]
- Tanaka, M.; Sakai, D.; Hiyama, A.; Arai, F.; Nakajima, D.; Yokoyama, K.; Mochida, J. Evidence of Nonnotochordal Origin in Chondrocyte-like Cells of the Nucleus Pulposus Appearing in Early Stage Disk Degeneration in the Mouse Model. Global Spine J. 2012, 2. [Google Scholar] [CrossRef]
- Kraus, P.; Lufkin, T. Bovine annulus fibrosus cell lines isolated from intervertebral discs. Genom. Data 2016, 10, 83–84. [Google Scholar] [CrossRef]
- Kraus, P.; Kocsis, V.; Williams, C.; Youngs, B.; Lufkin, T. Plate in situ hybridization (PISH) as a time and cost effective RNA expression assay to study phenotypic heterogeneity in a population of cultured murine cells at single cell resolution. Biotechnol. Lett. 2015, 37, 1573–1577. [Google Scholar] [CrossRef]
- Kraus, P.; Yerden, R.; Sipes, D.; Sur, S.; Lufkin, T. A quantitative and qualitative RNA expression profiling assay for cell culture with single cell resolution. Cytotechnology 2018, 70, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Thorpe, A.A.; Binch, A.L.; Creemers, L.B.; Sammon, C.; Le Maitre, C.L. Nucleus pulposus phenotypic markers to determine stem cell differentiation: fact or fiction? Oncotarget 2016, 7, 2189–2200. [Google Scholar] [CrossRef]
- Akker, G.; Eijssen, L.M.T.; Richardson, S.M.; Rhijn, L.W.V.; Hoyland, J.A.; Welting, T.J.M.; Voncken, J.W. A Membranome-Centered Approach Defines Novel Biomarkers for Cellular Subtypes in the Intervertebral Disc. Cartilage 2018. [Google Scholar] [CrossRef] [PubMed]
- Scholzen, T.; Gerdes, J. The Ki-67 protein: from the known and the unknown. J. Cell Physiol. 2000, 182, 311–322. [Google Scholar] [CrossRef]
- Gerdes, J.; Schwab, U.; Lemke, H.; Stein, H. Production of a mouse monoclonal antibody reactive with a human nuclear antigen associated with cell proliferation. Int. J. Cancer 1983, 31, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues-Pinto, R.; Ward, L.; Humphreys, M.; Zeef, L.A.H.; Berry, A.; Hanley, K.P.; Hanley, N.; Richardson, S.M.; Hoyland, J.A. Human notochordal cell transcriptome unveils potential regulators of cell function in the developing intervertebral disc. Sci. Rep. 2018, 8, 12866. [Google Scholar] [CrossRef]
- Kraus, P.; Xing, X.; Lim, S.L.; Fun, M.E.; Sivakamasundari, V.; Yap, S.P.; Lee, H.; Karuturi, R.K.; Lufkin, T. Mouse strain specific gene expression differences for illumina microarray expression profiling in embryos. BMC Res. Notes 2012, 5, 232. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues-Pinto, R.; Berry, A.; Piper-Hanley, K.; Hanley, N.; Richardson, S.M.; Hoyland, J.A. Spatiotemporal analysis of putative notochordal cell markers reveals CD24 and keratins 8, 18, and 19 as notochord-specific markers during early human intervertebral disc development. J. Orthop. Res. 2016, 34, 1327–1340. [Google Scholar] [CrossRef] [PubMed]
- Lv, F.; Leung, V.Y.; Huang, S.; Huang, Y.; Sun, Y.; Cheung, K.M. In search of nucleus pulposus-specific molecular markers. Rheumatology (Oxford) 2014, 53, 600–610. [Google Scholar] [CrossRef]
- Power, K.A.; Grad, S.; Rutges, J.P.; Creemers, L.B.; van Rijen, M.H.; O’Gaora, P.; Wall, J.G.; Alini, M.; Pandit, A.; Gallagher, W.M. Identification of cell surface-specific markers to target human nucleus pulposus cells: expression of carbonic anhydrase XII varies with age and degeneration. Arthritis Rheum. 2011, 63, 3876–3886. [Google Scholar] [CrossRef] [PubMed]
- Sive, J.I.; Baird, P.; Jeziorsk, M.; Watkins, A.; Hoyland, J.A.; Freemont, A.J. Expression of chondrocyte markers by cells of normal and degenerate intervertebral discs. Mol. Pathol. 2002, 55, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Van den Akker, G.G.; Surtel, D.A.; Cremers, A.; Rodrigues-Pinto, R.; Richardson, S.M.; Hoyland, J.A.; van Rhijn, L.W.; Welting, T.J.; Voncken, J.W. Novel immortal human cell lines reveal subpopulations in the nucleus pulposus. Arthritis Res. Ther. 2014, 16, R135. [Google Scholar] [CrossRef]
- Risbud, M.V.; Guttapalli, A.; Tsai, T.T.; Lee, J.Y.; Danielson, K.G.; Vaccaro, A.R.; Albert, T.J.; Gazit, Z.; Gazit, D.; Shapiro, I.M. Evidence for skeletal progenitor cells in the degenerate human intervertebral disc. Spine (Phila Pa 1976) 2007, 32, 2537–2544. [Google Scholar] [CrossRef]
- Salviano-Silva, A.; Lobo-Alves, S.C.; Almeida, R.C.; Malheiros, D.; Petzl-Erler, M.L. Besides Pathology: Long Non-Coding RNA in Cell and Tissue Homeostasis. Noncoding RNA 2018, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Buxbaum, A.R.; Haimovich, G.; Singer, R.H. In the right place at the right time: visualizing and understanding mRNA localization. Nat. Rev. Mol. Cell. Biol. 2015, 16, 95–109. [Google Scholar] [CrossRef]
- Eliscovich, C.; Singer, R.H. RNP transport in cell biology: the long and winding road. Curr. Opin. Cell Biol. 2017, 45, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Mofatteh, M.; Bullock, S.L. SnapShot: Subcellular mRNA Localization. Cell 2017, 169, 178–178.e171. [Google Scholar] [CrossRef] [PubMed]
- Sirri, V.; Urcuqui-Inchima, S.; Roussel, P.; Hernandez-Verdun, D. Nucleolus: the fascinating nuclear body. Histochem. Cell Biol. 2008, 129, 13–31. [Google Scholar] [CrossRef] [PubMed]
- Spector, D.L.; Lamond, A.I. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef] [PubMed]

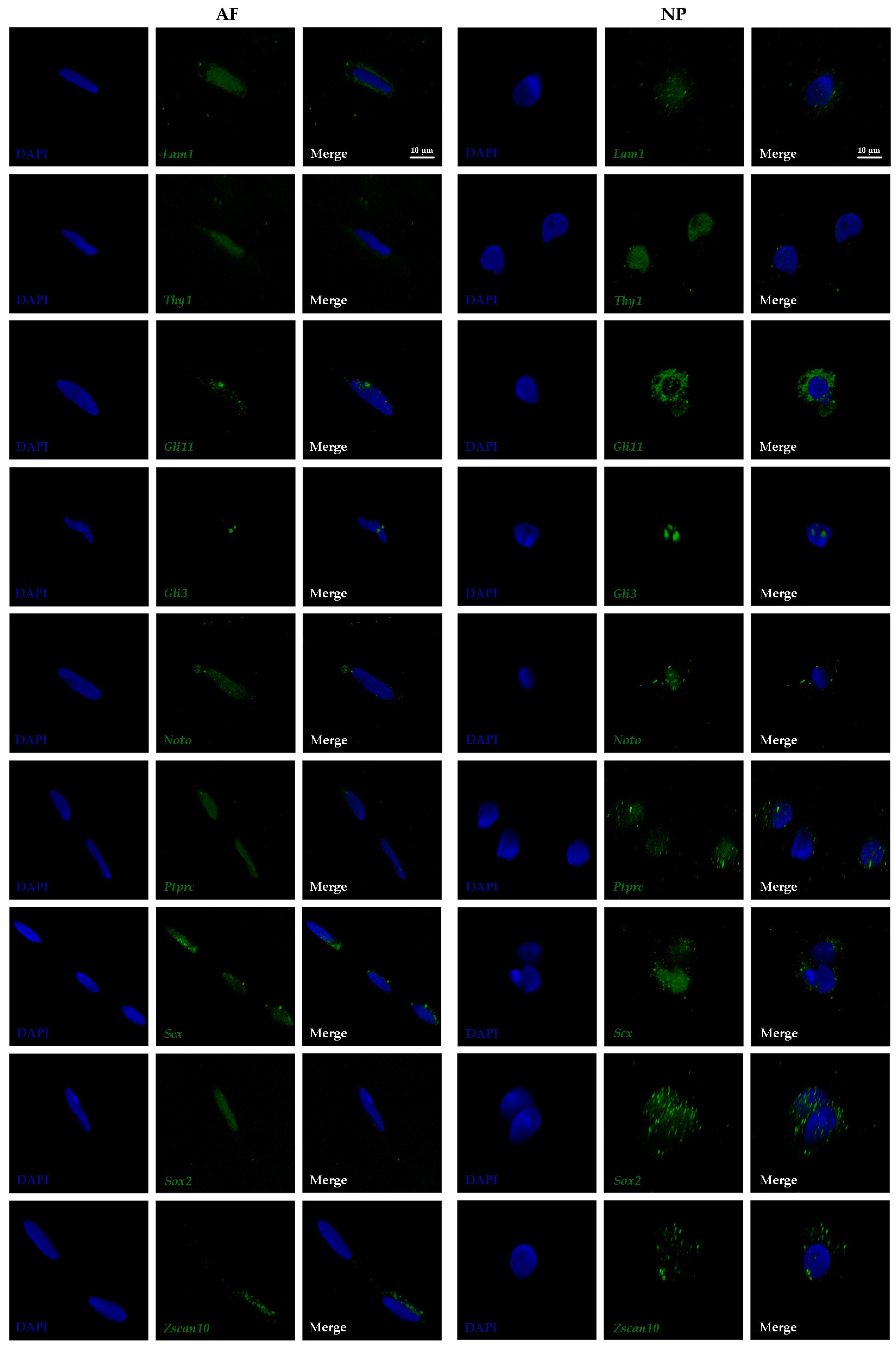
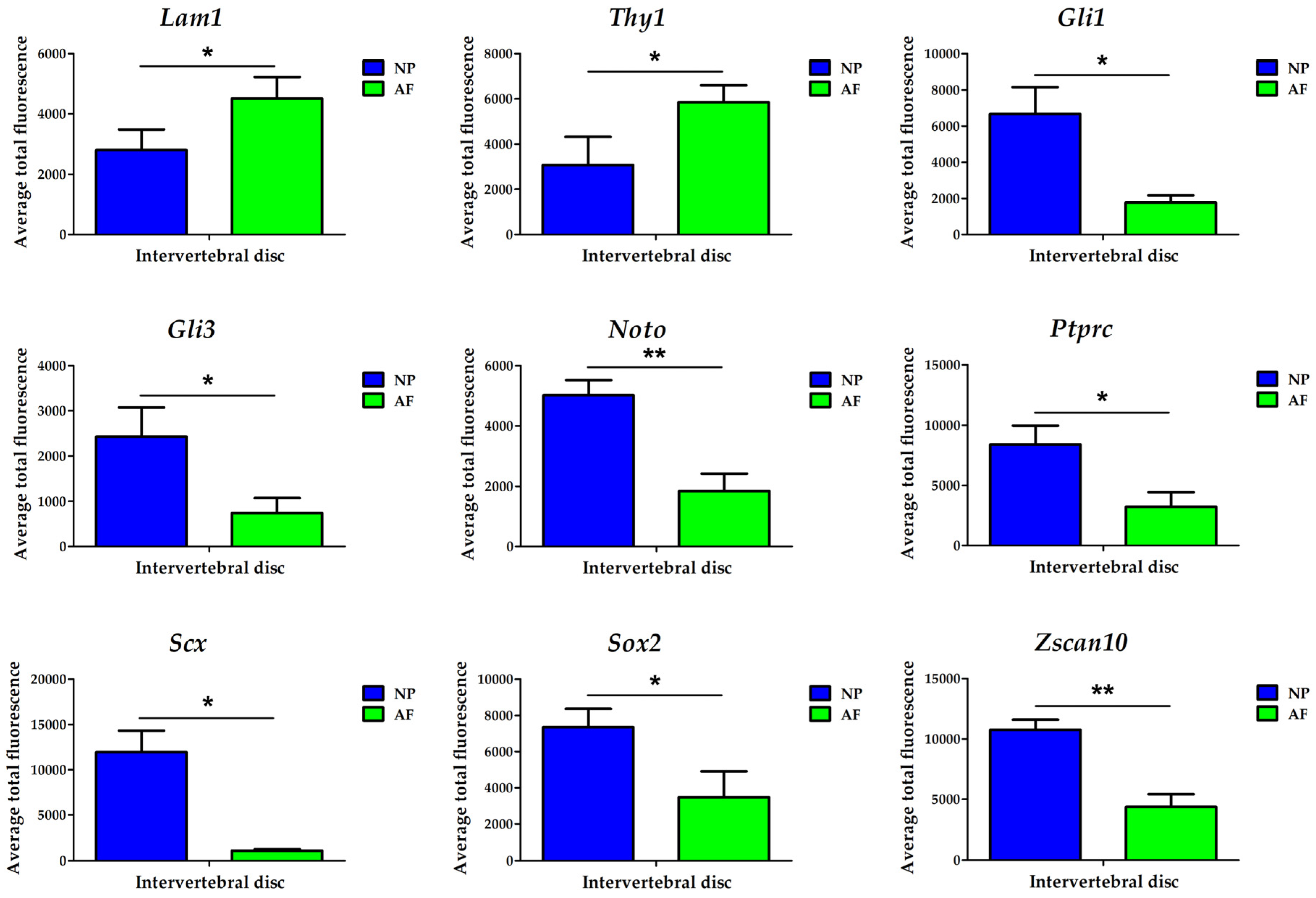
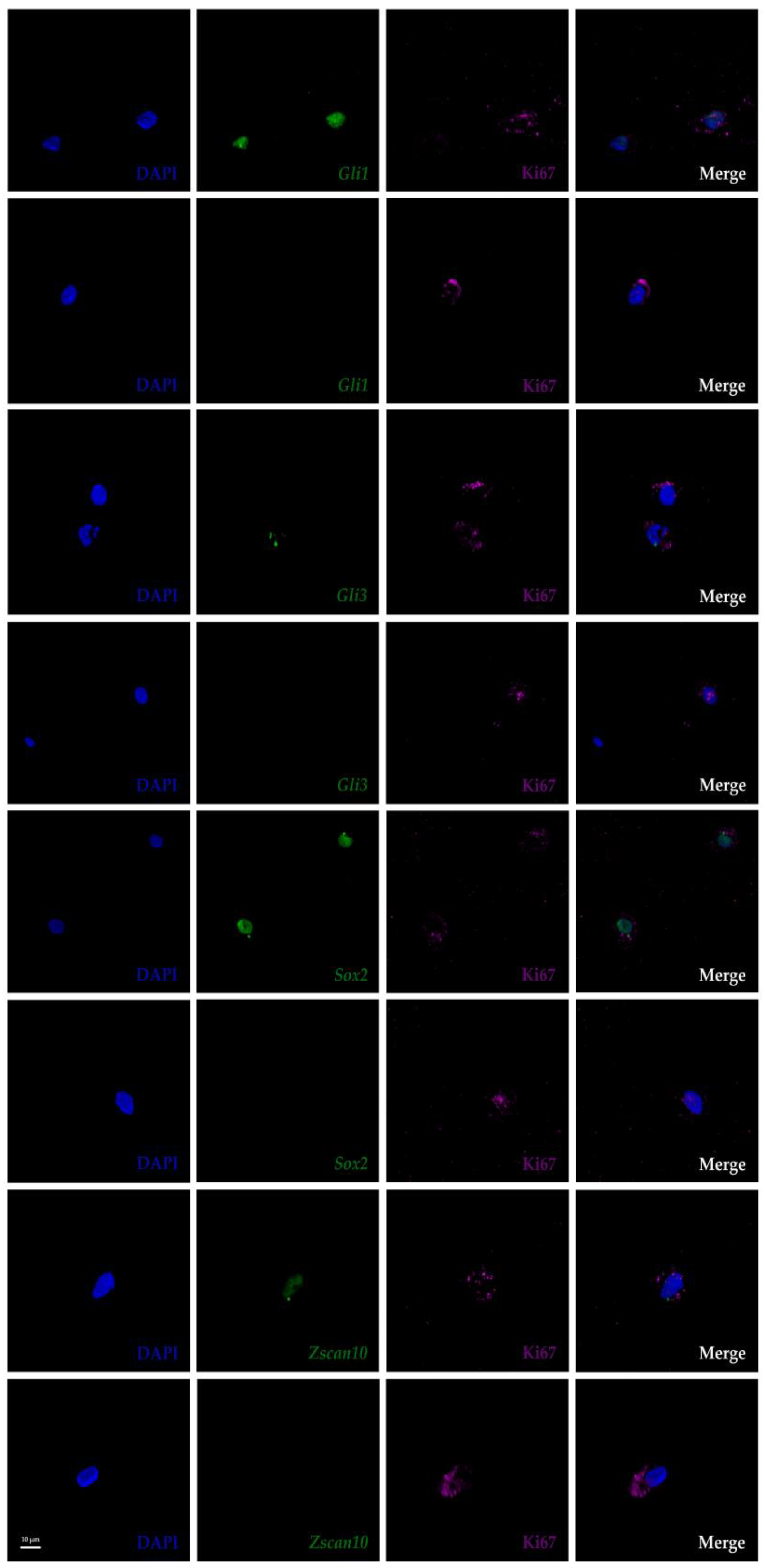

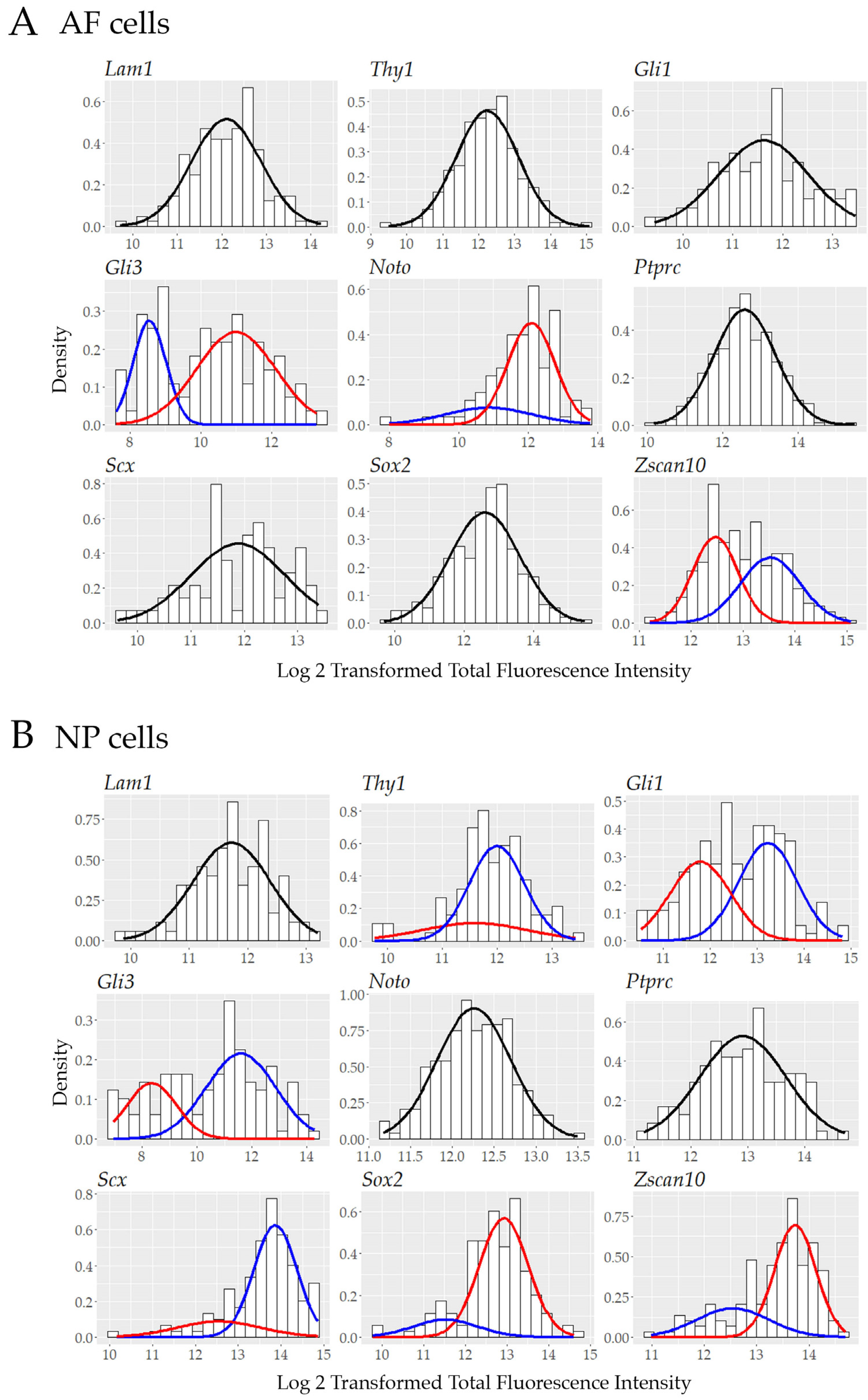
| Biomarker | AP-RISH and z-Proportion Test | FL-RISH and z-Proportion Test | FL-RISH and Population Averaging |
|---|---|---|---|
| NP/AF Ratio | NP/AF Ratio | FL in NP/FL in AF Ratio | |
| Lam1 | 0.3 | 0.8 | 0.6 |
| Thy1 | 0.2 | 0.7 | 0.5 |
| Gli1 | 2.2 | 2.0 | 3.8 |
| Gli3 | 1.5 | 2.0 | 3.3 |
| Noto | 4.3 | 2.4 | 2.7 |
| Ptprc | 2.3 | 2.2 | 2.6 |
| Scx | 2.2 | 3.4 | 10.8 |
| Sox2 | 3.2 | 2.2 | 2.1 |
| Zscan10 | 1.6 | 2.0 | 2.5 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, K.; Kapper, D.; Mondal, S.; Lufkin, T.; Kraus, P. Quantitative Single-Cell Transcript Assessment of Biomarkers Supports Cellular Heterogeneity in the Bovine IVD. Vet. Sci. 2019, 6, 42. https://doi.org/10.3390/vetsci6020042
Li K, Kapper D, Mondal S, Lufkin T, Kraus P. Quantitative Single-Cell Transcript Assessment of Biomarkers Supports Cellular Heterogeneity in the Bovine IVD. Veterinary Sciences. 2019; 6(2):42. https://doi.org/10.3390/vetsci6020042
Chicago/Turabian StyleLi, Kangning, Devin Kapper, Sumona Mondal, Thomas Lufkin, and Petra Kraus. 2019. "Quantitative Single-Cell Transcript Assessment of Biomarkers Supports Cellular Heterogeneity in the Bovine IVD" Veterinary Sciences 6, no. 2: 42. https://doi.org/10.3390/vetsci6020042
APA StyleLi, K., Kapper, D., Mondal, S., Lufkin, T., & Kraus, P. (2019). Quantitative Single-Cell Transcript Assessment of Biomarkers Supports Cellular Heterogeneity in the Bovine IVD. Veterinary Sciences, 6(2), 42. https://doi.org/10.3390/vetsci6020042





