Unraveling the Genetic Legacy: Comparative Analysis of Yucatán Black Hairless Pig and Worldwide Indigenous Breeds
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
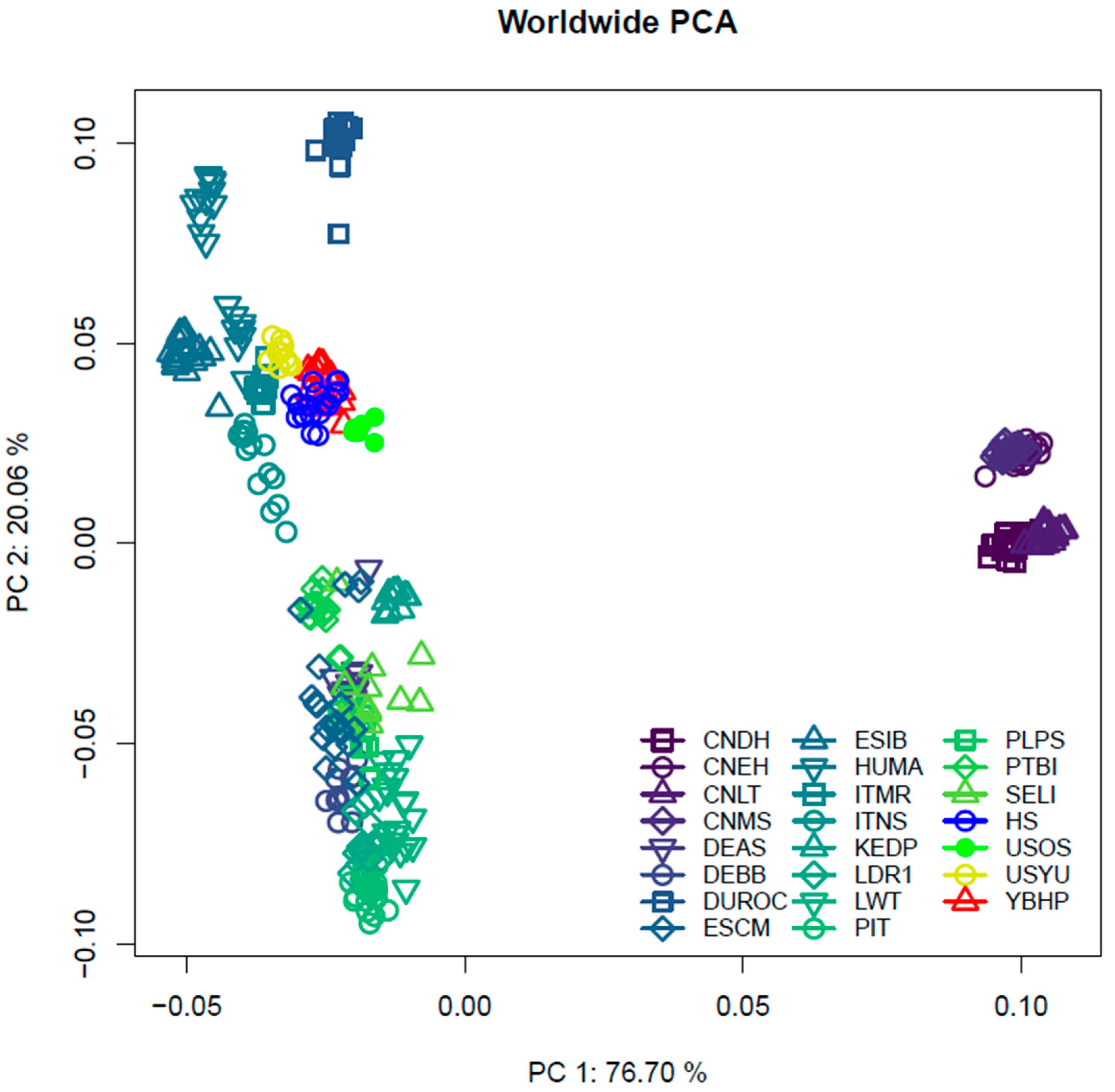
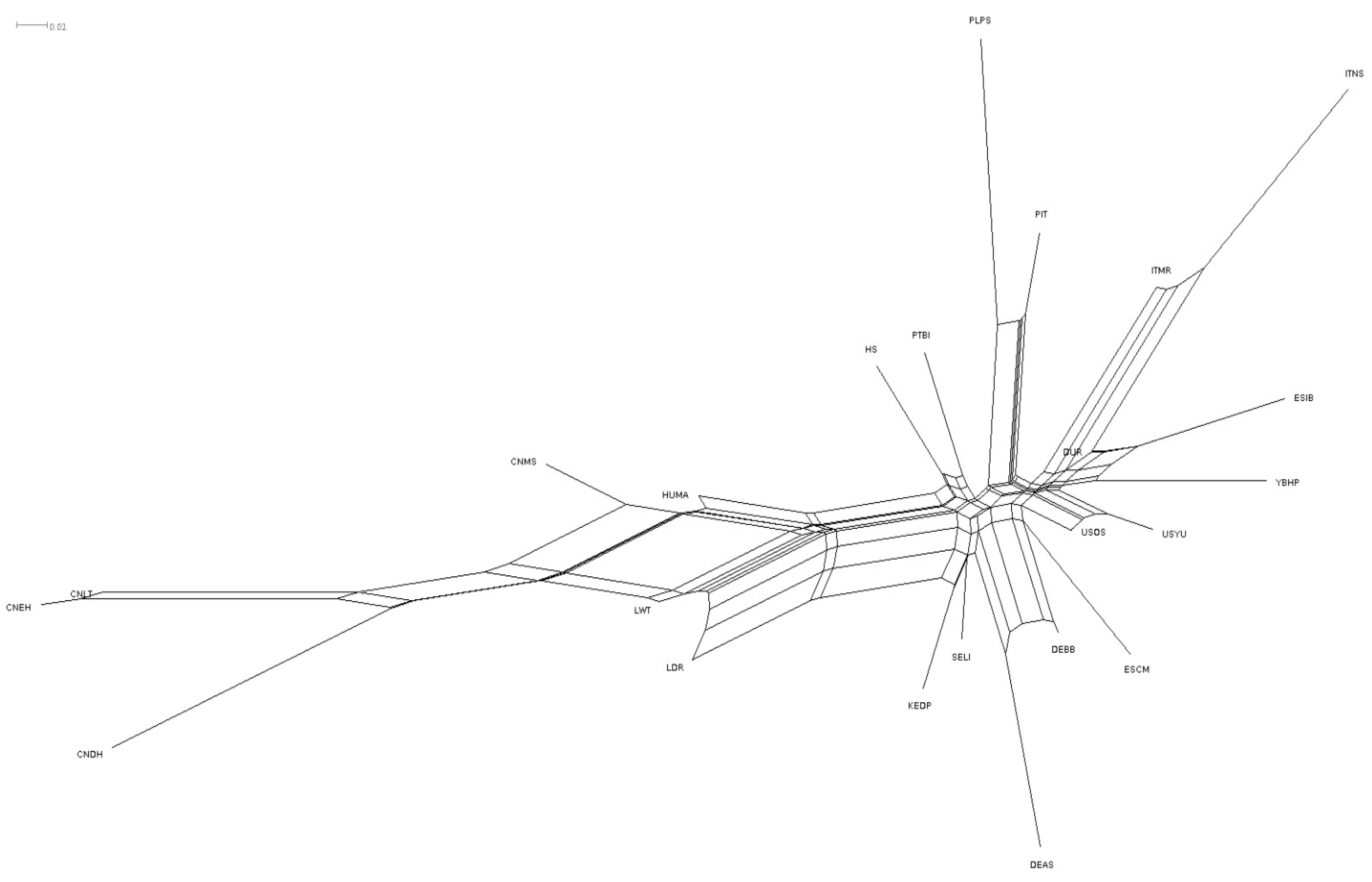
3. Results
4. Discussion
4.1. Genetic Diversity Indices
4.2. Population Structure
4.3. Admixture Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Candek-Potokar, M.; Nieto Linan, R.M. (Eds.) European Local Pig Breeds—Diversity and Performance. A Study of Project TREASURE; IntechOpen: Rijeka, Croatia, 2019; ISBN 978-1-78985-407-7. [Google Scholar]
- Groenen, M.A.M.; Archibald, A.L.; Uenishi, H.; Tuggle, C.K.; Takeuchi, Y.; Rothschild, M.F.; Rogel-Gaillard, C.; Park, C.; Milan, D.; Megens, H.-J.; et al. Analyses of Pig Genomes Provide Insight into Porcine Demography and Evolution. Nature 2012, 491, 393–398. [Google Scholar] [CrossRef]
- Larson, G.; Dobney, K.; Albarella, U.; Fang, M.; Matisoo-Smith, E.; Robins, J.; Lowden, S.; Finlayson, H.; Brand, T.; Willerslev, E.; et al. Worldwide Phylogeography of Wild Boar Reveals Multiple Centers of Pig Domestication. Science 2005, 307, 1618–1621. [Google Scholar] [CrossRef]
- Leroy, G.; Baumung, R.; Boettcher, P.; Besbes, B.; From, T.; Hoffmann, I. Animal Genetic Resources Diversity and Ecosystem Services. Glob. Food Sec. 2018, 17, 84–91. [Google Scholar] [CrossRef]
- Bordonaro, S.; Chessari, G.; Mastrangelo, S.; Senczuk, G.; Chessa, S.; Castiglioni, B.; Tumino, S.; Marletta, D.; Criscione, A. Genome-wide Population Structure, Homozygosity, and Heterozygosity Patterns of Nero Siciliano Pig in the Framework of Italian and Cosmopolitan Breeds. Anim. Genet. 2023, 54, 591–605. [Google Scholar] [CrossRef]
- Food and Agriculture Organization. FAO Domestic Animal Diversity Information System (DAD-IS); FAO: Rome, Italy, 2025. [Google Scholar]
- Alonso, I.; Ibáñez-Escriche, N.; Noguera, J.L.; Casellas, J.; Martín de Hijas-Villalba, M.; Gracia-Santana, M.J.; Varona, L. Genomic Differentiation among Varieties of Iberian Pig. Span. J. Agric. Res. 2020, 18, 1–20. [Google Scholar] [CrossRef]
- Yang, B.; Cui, L.; Perez-Enciso, M.; Traspov, A.; Crooijmans, R.P.M.A.; Zinovieva, N.; Schook, L.B.; Archibald, A.; Gatphayak, K.; Knorr, C.; et al. Genome-Wide SNP Data Unveils the Globalization of Domesticated Pigs. Genet. Sel. Evol. 2017, 49, 71. [Google Scholar] [CrossRef]
- Hernández, A.A.; García-Munguía, C.A.; García-Munguía, A.M.; Ortíz, J.R.; Sierra-Vásquez, A.C.; Morales-Flores, S. Sistema de Producción Del Cerdo Pelón Mexicano En La Península de Yucatán. Nova Sci. 2020, 12. [Google Scholar] [CrossRef]
- Ángel-Hernández, A.; García, M.C.A.; Valencia, P.M.; Gutiérrez, C.A.J.; García, M.A.M.; Gómez, S.J.A.; Morales-Flores, S. Estudio de Cerdos Criollos Mexicanos Para Instalación Del Centro de Conservación En La Universidad de Guanajuato, Mexico. Actas Iberoam. Conserv. Anim. 2019, 12, 77–84. [Google Scholar]
- Ramos-Canché, M.E.; Magaña-Magaña, M.A.; Aguilar-Urquizo, E.; Pech-Zapata, A.; Piñeiro-Vázquez, A.T.; Toledo-López, V.M.; Sangines-García, J.R. Óptimos Económicos En La Cría Del Cerdo Pelón Mexicano: Propuesta de Integración Para Cadena Productiva. Ecosistemas Recur. Agropecu. 2019, 7, 1. [Google Scholar] [CrossRef]
- Lemus-Flores, C.; Ulloa-Arvizu, R.; Ramos-Kuri, M.; Estrada, F.J.; Alonso, R.A. Genetic Analysis of Mexican Hairless Pig Populations. J. Anim. Sci. 2001, 79, 3021. [Google Scholar] [CrossRef]
- Lemus-Flores, C.; Bugarín Prado, J.O.; Valdivia Bernal, R.; Segura Correa, J.C.; Sansor-Nah, R. Genetic Relationships of the Yucatán Black Hairless Pig with Iberian Breeds Using Single Nucleotide Polymorfisms. Braz. J. Vet. Res. Anim. Sci. 2023, 60, e195697. [Google Scholar] [CrossRef]
- NOM-051-ZOO-1995; Trato Humanitario En La Movilización de Animales. Norma Oficial Mexicana; p. 23. Available online: http://publico.senasica.gob.mx/?doc=531 (accessed on 20 May 2025).
- NOM-062-ZOO-1999; Especificaciones Técnicas Para La Producción, Cuidado y Uso de Los Animales de Laboratorio. NORMA Oficial Mexicana; p. 58. Available online: http://publico.senasica.gob.mx/?doc=743 (accessed on 29 June 2025).
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-Generation PLINK: Rising to the Challenge of Larger and Richer Datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast Model-Based Estimation of Ancestry in Unrelated Individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; Version 4.3.1 R Core Team: Vienna, Austria, 2025. [Google Scholar]
- Huson, D.H.; Bryant, D. Application of Phylogenetic Networks in Evolutionary Studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Bovo, S.; Ribani, A.; Muñoz, M.; Alves, E.; Araujo, J.P.; Bozzi, R.; Charneca, R.; Di Palma, F.; Etherington, G.; Fernandez, A.I.; et al. Genome-wide Detection of Copy Number Variants in European Autochthonous and Commercial Pig Breeds by Whole-genome Sequencing of DNA Pools Identified Breed-characterising Copy Number States. Anim. Genet. 2020, 51, 541–556. [Google Scholar] [CrossRef]
- Dadousis, C.; Muñoz, M.; Óvilo, C.; Fabbri, M.C.; Araújo, J.P.; Bovo, S.; Potokar, M.Č.; Charneca, R.; Crovetti, A.; Gallo, M.; et al. Admixture and Breed Traceability in European Indigenous Pig Breeds and Wild Boar Using Genome-Wide SNP Data. Sci. Rep. 2022, 12, 7346. [Google Scholar] [CrossRef]
- Lemus-Flores, C.; Alonso-Morales, R.; Toledo-Alvarado, H.; Sansor-Nah, R.; Burgos-Paz, W.; Dzib-Cauich, D. Diversidad Genética y Estructura Poblacional Del Cerdo Negro Lampiño de Yucatán Usando Chip SNP50. Abanico Vet. 2020, 10. [Google Scholar] [CrossRef]
- Burgos-Paz, W.; Souza, C.A.; Megens, H.J.; Ramayo-Caldas, Y.; Melo, M.; Lemús-Flores, C.; Caal, E.; Soto, H.W.; Martínez, R.; Álvarez, L.A.; et al. Porcine Colonization of the Americas: A 60k SNP Story. Heredity 2013, 110, 321–330. [Google Scholar] [CrossRef]
- Babigumira, B.M.; Sölkner, J.; Mészáros, G.; Pfeiffer, C.; Lewis, C.R.G.; Ouma, E.; Wurzinger, M.; Marshall, K. A Mix of Old British and Modern European Breeds: Genomic Prediction of Breed Composition of Smallholder Pigs in Uganda. Front. Genet. 2021, 12, 676047. [Google Scholar] [CrossRef]
- Ai, H.; Huang, L.; Ren, J. Genetic Diversity, Linkage Disequilibrium and Selection Signatures in Chinese and Western Pigs Revealed by Genome-Wide SNP Markers. PLoS ONE 2013, 8, e56001. [Google Scholar] [CrossRef]
- Lee, S.H.; Seo, D.W.; Cho, E.S.; Choi, B.H.; Kim, Y.M.; Hong, J.K.; Han, H.D.; Jung, Y.B.; Kim, D.J.; Choi, T.J.; et al. Genetic Diversity and Ancestral Study for Korean Native Pigs Using 60K SNP Chip. Animals 2020, 10, 760. [Google Scholar] [CrossRef]
- Muñoz, M.; Bozzi, R.; García-Casco, J.; Núñez, Y.; Ribani, A.; Franci, O.; García, F.; Škrlep, M.; Schiavo, G.; Bovo, S.; et al. Genomic Diversity, Linkage Disequilibrium and Selection Signatures in European Local Pig Breeds Assessed with a High Density SNP Chip. Sci. Rep. 2019, 9, 13546. [Google Scholar] [CrossRef]
- Lemus, G.; Rodríguez, J.; Burgos, W.; Lemus, C.; Carmona, C. Exploring the Genetic of Three Hairless Pig Breed Populations in Mexico. Rev. Fac. Agron. Univ. Zulia 2024, 41, e244122. [Google Scholar] [CrossRef]
- Tang, H.; Ouyang, J.; Liu, S.; Xiong, Y.; Wu, Y.; Wang, L.; Wang, C.; Yan, X.; Shen, Y.; Chen, H. Population Structure of 3907 Worldwide Pigs and the Introgression of Chinese Indigenous Pigs by European Pigs. Anim. Genet. 2022, 53, 599–612. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Ye, X.; Zhang, Z.; Zhao, Q.; Xiang, Y.; Xu, N.; Wang, Q.; Pan, Y.; Guo, X.; Wang, Z. Genetic Diversity and Selection Signatures of Four Indigenous Pig Breeds from Eastern China. Anim. Genet. 2022, 53, 506–509. [Google Scholar] [CrossRef] [PubMed]
- Perezgrovas-Garza, R. Cría de Cerdos Autóctonos En Comunidades Indígenas; Perezgrovas Garza, R., Ed.; Instituto de Estudios Indígenas, Universidad Autónoma de Chiapas: San Cristóbal de Las Casas, Mexico, 2007. [Google Scholar]
- Alders, R.G.; Campbell, A.; Costa, R.; Guèye, E.F.; Ahasanul Hoque, M.; Perezgrovas-Garza, R.; Rota, A.; Wingett, K. Livestock across the World: Diverse Animal Species with Complex Roles in Human Societies and Ecosystem Services. Anim. Front. 2021, 11, 20–29. [Google Scholar] [CrossRef] [PubMed]

| Breed | Country | Continent | Code | Size | Type | References |
|---|---|---|---|---|---|---|
| YUCATÁN BLACK HAIRLESS PIG | MEXICO | AMERICA | YBHP | 20 | INDIGENOUS | This Study |
| OSSABAW | USA | AMERICA | OSSABAW | 6 | INDIGENOUS | [8] |
| US YUCATÁN MINI PIG | USA | AMERICA | USYU | 10 | INDIGENOUS | [8] |
| KENYA LOCAL | KENYA | AFRICA | KENYA/KEDP | 9 | INDIGENOUS | [8] |
| GUANGDONGDAHUABAI | CHINA | ASIA | CNDH | 20 | INDIGENOUS | [8] |
| ERHUALIAN | CHINA | ASIA | CNEH | 20 | INDIGENOUS | [8] |
| LANTANG | CHINA | ASIA | CNLT | 20 | INDIGENOUS | [8] |
| MEIXAN | CHINA | ASIA | CHMS | 20 | INDIGENOUS | [8] |
| ANGLER SATTLESCHWEIN | GERMANY | EUROPE | DEAS | 10 | INDIGENOUS | [8] |
| BUNTE BENTHEIMER | GERMANY | EUROPE | DEBB | 12 | INDIGENOUS | [8] |
| CHATO MURCIANO | SPAIN | EUROPE | ESCM | 20 | INDIGENOUS | [8] |
| IBERIAN | SPAIN | EUROPE | IBERIAN | 20 | INDIGENOUS | [8] |
| MANGALICA | HUNGARY | EUROPE | HUMA | 20 | INDIGENOUS | [8] |
| MORA ROMAGNOLA | ITALY | EUROPE | ITMR | 9 | INDIGENOUS | [8] |
| NERO SICILIANO | ITALY | EUROPE | ITNS | 15 | INDIGENOUS | [8] |
| PULAWSKA | POLAN | EUROPE | PLPS | 15 | INDIGENOUS | [8] |
| BISARO | PORTUGAL | EUROPE | PORTUGAL | 14 | INDIGENOUS | [8] |
| LINDEROTH | SWEDEN | EUROPE | SELI | 15 | INDIGENOUS | [8] |
| DUROC | USA | COSMOPOLITAN | DUROC | 20 | COSMOPOLITAN | [8] |
| LANDRACE | DENMARK | COSMOPOLITAN | LDR/LDR1 | 20 | COSMOPOLITAN | [8] |
| LARGEWHITE | DENMARK | COSMOPOLITAN | LWT | 20 | COSMOPOLITAN | [8] |
| PIETRAIN | NETHERLANDS | COSMOPOLITAN | PIT | 20 | COSMOPOLITAN | [8] |
| HAMPSHIRE | UK | COSMOPOLITAN | HS | 20 | COSMOPOLITAN | [8] |
| Population | N | MAF | HO | HE | FIS |
|---|---|---|---|---|---|
| YBHP | 141 | 0.3325 ± 0.1997 | 0.3602 ± 0.0323 | 0.4247 ± 0001 | 0.1517 ± 0.0762 |
| OSSABAW | 6 | 0.3375 ± 0.2588 | 0.3666 ± 0.0305 | 0.4246 ± 0001 | 0.1366 ± 0.0718 |
| USYU | 10 | 0.3423 ± 0.2056 | 0.3745 ± 0.0387 | 0.4247 ± 0001 | 0.1181 ± 0.0913 |
| KENYA | 9 | 0.3398 ± 0.3882 | 0.3244 ± 0.0256 | 0.4247 ± 0.0001 | 0.2363 ± 0.0605 |
| CNDH | 20 | 0.3990 ± 0.3882 | 0.2035 ± 0.0118 | 0.4247 ± 0.0001 | 0.22474 ± 0.0277 |
| CNEH | 20 | 0.3877 ± 0.3888 | 0.1796 ± 0.0111 | 0.4247 ± 0.0001 | 0.15742 ± 0.0263 |
| CNLT | 20 | 0.4004 ± 0.3992 | 0.1716 ± 0.0228 | 0.4247 ± 0.0001 | 0.1595 ± 0.0537 |
| CHMS | 20 | 0.2468 ± 0.1476 | 0.3774 ± 0.0363 | 0.3474 ± 0.0001 | -0.0862 ± 0.1043 |
| DEAS | 10 | 0.3277 ± 0.2628 | 0.3118 ± 0.0276 | 0.4248 ± 0.0001 | 0.2659 ± 0.0649 |
| DEBB | 12 | 0.3153 ± 0.2147 | 0.2775 ± 0.0449 | 0.4247 ± 0.0001 | 0.2465 ± 0.1056 |
| ESCM | 20 | 0.2872 ± 0.2530 | 0.2053 ± 0.0494 | 0.4247 ± 0.0001 | 0.1547 ± 0.016 |
| IBERIAN | 20 | 0.2939 ± 0.2555 | 0.1931 ± 0.0571 | 0.4247 ± 0.0001 | 0.077 ± 0.0135 |
| HUMA | 20 | 0.3098 ± 0.2141 | 0.2931 ± 0.0704 | 0.4247 ± 0.0001 | 0.3099 ± 0.1659 |
| ITMR | 9 | 0.3298 ± 0.2032 | 0.3779 ± 0.0102 | 0.4247 ± 0.0001 | 0.1101 ± 0.0241 |
| ITNS | 15 | 0.3393 ± 0.2254 | 0.3847 ± 0.0166 | 0.4247 ± 0.0001 | 0.0904 ± 0.0391 |
| POLAND | 15 | 0.3331 ± 0.2065 | 0.3621 ± 0.0530 | 0.4248 ± 0.0008 | 0.1476 ± 0.1249 |
| PORTUGAL | 14 | 0.3378 ± 0.2475 | 0.3374 ± 0.0481 | 0.4247 ± 0.0001 | 0.2055 ± 0.1134 |
| SWEDEN | 15 | 0.3111 ± 0.2444 | 0.2714 ± 0.0360 | 0.4247 ± 0.0001 | 0.1609 ± 0.0847 |
| DUROC | 20 | 0.3240 ± 0.2824 | 0.2854 ± 0.0208 | 0.4177 ± 0.0001 | 0.1866 ± 0.0500 |
| LANDRACE | 20 | 0.3308 ± 0.2472 | 0.3191 ± 0.0252 | 0.4177 ± 0.0001 | 0.2360 ± 0.0603 |
| LW | 20 | 0.3463 ± 0.2174 | 0.3383 ± 0.0302 | 0.4176 ± 0.0001 | 0.1900 ± 0.0723 |
| PIETRAIN | 20 | 0.3335 ± 0.2318 | 0.3469 ± 0.0165 | 0.4176 ± 0.0001 | 0.1693 ± 0.0395 |
| HS | 20 | 0.3078 ± 0.2363 | 0.2944 ± 0.0287 | 0.4176 ± 0.0001 | 0.2950 ± 0.0688 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lara-Castillo, J.B.; Lemus-Flores, C.; Bugarín-Prado, J.O.; Grageola-Núñez, F.; Burgos-Paz, W.O. Unraveling the Genetic Legacy: Comparative Analysis of Yucatán Black Hairless Pig and Worldwide Indigenous Breeds. Vet. Sci. 2025, 12, 755. https://doi.org/10.3390/vetsci12080755
Lara-Castillo JB, Lemus-Flores C, Bugarín-Prado JO, Grageola-Núñez F, Burgos-Paz WO. Unraveling the Genetic Legacy: Comparative Analysis of Yucatán Black Hairless Pig and Worldwide Indigenous Breeds. Veterinary Sciences. 2025; 12(8):755. https://doi.org/10.3390/vetsci12080755
Chicago/Turabian StyleLara-Castillo, Jorge Barzilai, Clemente Lemus-Flores, Job Oswaldo Bugarín-Prado, Fernando Grageola-Núñez, and William Orlando Burgos-Paz. 2025. "Unraveling the Genetic Legacy: Comparative Analysis of Yucatán Black Hairless Pig and Worldwide Indigenous Breeds" Veterinary Sciences 12, no. 8: 755. https://doi.org/10.3390/vetsci12080755
APA StyleLara-Castillo, J. B., Lemus-Flores, C., Bugarín-Prado, J. O., Grageola-Núñez, F., & Burgos-Paz, W. O. (2025). Unraveling the Genetic Legacy: Comparative Analysis of Yucatán Black Hairless Pig and Worldwide Indigenous Breeds. Veterinary Sciences, 12(8), 755. https://doi.org/10.3390/vetsci12080755








