1. Introduction
Mastitis is an economically significant disease affecting dairy cattle worldwide, impacting animal health, welfare, and productivity [
1,
2,
3]. Bovine mastitis is most commonly caused by pathogens, typically classified as contagious or environmental based on their primary reservoir and transmission route [
4]. Contagious mastitis pathogens reside in the cow’s udder and on teat skin, colonizing and growing in the teat canal. They are primarily transmitted among cows through contact with infected milk during milking [
4]. Among these pathogens,
Staphylococcus aureus,
Streptococcus agalactiae, and
Mycoplasma bovis are the most significant, with
S. aureus considered to be the most common in North America [
4,
5]. On the other hand, environmental pathogens live in the cow’s environment, such as in the bedding and housing, and cause an infection when given the opportunity [
4]. A wide range of bacterial species cause environmental mastitis, with the most common pathogens including coliforms (
Escherichia coli,
Klebsiella spp.,
Enterobacter spp.),
Streptococcus species such as
Streptococcus uberis and
Streptococcus dysgalactiae, and
Pseudomonas spp. [
4]. Non-aureus staphylococci (NAS) are opportunistic bacteria that can cause intramammary infections [
6]. Pathogens can also be categorized as major and minor based on their prevalence and the severity of the symptoms they cause [
4].
Understanding the prevalence and distribution of mastitis-causing bacteria is crucial for controlling and preventing bovine mastitis [
7]. Milk samples are often submitted for bacteriological examination to identify the causative agent as a part of mastitis control programs. However, limited information is available on the distribution of mastitis-causing organisms in milk from individual dairy cows in North America [
8]. In California, one of the largest dairy-producing states, mastitis management is critical for maintaining productivity and profitability. California is the leading milk-producing state in the nation, housing 1.69 million dairy cows and accounting for 18% of the total milk produced in the United States. Over 90% of the cows are housed in dairies in the San Joaquin Valley [
9]. Moreover, the California dairy industry has undergone significant structural transformations over the past two decades [
10], including changes in environmental regulations and water usage, leading to shifts in bedding management and housing systems. Technological innovations, such as advances in milking technology and hygiene practices, alongside the adoption of genomic testing and improved diagnostic tools, have enhanced the ability to detect and manage mastitis. These advancements have led to better control of both environmental and contagious pathogens, likely influencing the health and management of dairy cows, as well as the profile of udder pathogens affecting them.
Therefore, the objective of this retrospective study was to describe the microbiologic culture results of dairy cows’ milk samples submitted from San Joaquin Valley dairies for routine microbiological testing to the Milk Quality Laboratory at the Veterinary Medicine Teaching and Research Center, UC Davis, Tulare, and to identify the prevalence, seasonal distribution, and annual trends of the most common mastitis pathogens isolated between January 2009 and December 2023.
4. Discussion
This is the first comprehensive study describing microbiological culture results from milk samples collected from dairy cows housed in California’s Central Valley. Understanding the profile of udder pathogens and monitoring trends over time is crucial for implementing effective mastitis control strategies. Milk samples are often collected from individual quarters or as composite milk samples (all quarters) to identify the pathogen causing an intramammary infection, or are taken from bulk tanks and string samples for herd surveillance. The current study evaluated records of milk samples from mastitis cases submitted to the Milk Quality Laboratory at the Veterinary Medicine Teaching and Research Center—the University of California, Davis, in Tulare, California, for routine milk culture between 2009 and 2023. The selection of the study laboratory was based on access to dairy cattle milk sample submissions. The study aimed to uncover patterns in the results of microbiological tests on milk samples originating from individual cows housed in California dairies. Notably, the results may underrepresent the prevalence of mastitis pathogens in California, as many dairy farms submit their milk samples to veterinary clinics or perform their own on-farm culturing.
Dairy farms rely on milk microbiological culturing to evaluate udder health status, which can be conducted using an on-farm culture (OFC) system or submitted to a diagnostic laboratory [
12]. The popularity of performing an OFC has increased in recent years because it provides results within 24–48 h of sample collection. In contrast, laboratory-submitted samples may take several days from submission to the delivery of results to producers. The growing use of OFCs could partially explain the decline in the number of submissions from 2009 to 2023, which was similarly reported in a study evaluating trends of pathogen isolation in milk samples collected from cows in Canada [
7]. Another factor that may have affected the number of submissions is a decrease in the number of dairies and animals in California over the years [
9].
Conventional bacteriological culturing remains the gold standard for evaluating milk samples and identifying mastitis-causing pathogens. However, many samples yield no bacterial growth, complicating identification [
13]. In this study, 27.44% of samples were NG, a result that is similar to findings from routine milk culture studies in Wisconsin [
14] but lower than what was reported in a Canadian study [
7]. A scoping review [
15] analyzing MALDI-TOF results from 50,429 healthy cows and 43,924 clinical mastitis cases in Canada, the U.S., and Brazil found NG in 68.2% of healthy cow samples, but only 39.6% of clinical mastitis cases, with the latter aligning with our results. A recent California study on Gram-negative mastitis treatment in three large dairies reported NG rates ranging from 20.1% to 52.0% [
2], suggesting variability based on farm conditions and sampling methods.
Several factors contribute to NG results in routine cultures, including infection clearance before sampling, prior antimicrobial use, improper sample handling, low bacterial loads, post-milking sampling, or cows not shedding the pathogen at the time of collection [
13,
14]. In this study, 98.68% of milk samples tested negative for
Mycoplasma spp., consistent with a Cornell study [
16] that found 98.03% of mastitic quarter samples showed no Mycoplasma growth. The low recovery of
Mycoplasma spp. may be due to pathogen absence, intermittent shedding, or sample contamination, as Mycoplasma can be easily overgrown by other bacteria [
17]. Seasonal trends also influenced NG results. Our study found a higher NG occurrence in Summer, aligning with a Canadian study [
7], while a Wisconsin study [
14] reported more NG cases in Winter. Regional climate differences may explain these variations—California’s Central Valley experiences hot, dry Summers exceeding 38 °C, whereas Wisconsin’s Summers are warm and wet (21–27 °C), and Canada’s range from 10 to 30 °C with high humidity.
Contamination of milk samples can occur at any point between collection and laboratory culturing. Common sources include the barn environment (e.g., feces, feed, bedding, air), teat skin, and the teat canal [
18]. To minimize contamination, aseptic techniques should be followed during sample collection. Contaminated samples complicate pathogen identification, potentially delaying or misguiding treatment and masking the presence of major contagious pathogens. A recent study [
15] reported contamination rates in U.S. dairies ranging from 2.5% to 39.7% for healthy cows and 2.4% for clinical mastitis cases. In our study, 5.63% of samples were contaminated, which was lower than in a similar Wisconsin study (15.3%) [
14] but higher than findings from a Canadian study [
7]. Contamination was more frequent in Winter, likely due to California’s wetter conditions. Since recycled manure is a common bedding material, increased organic matter on teats may contribute to higher contamination if teats are not properly cleaned. Interestingly, contamination rates were notably higher in 2020 and 2023. The COVID-19 pandemic in 2020 caused labor shortages and high turnover in the dairy industry [
19,
20], potentially impacting sample quality due to reduced training and workforce availability. Post-pandemic challenges continue to affect the agriculture sector [
21], possibly influencing sample handling. A contaminated sample is typically identified when a culture yields three or more dissimilar colony types. Contamination can result from dirty teat ends, improper handling (e.g., milk touching hands before entering the tube), nonsterile equipment, contaminated media, excess alcohol on teat ends, and poorly sealed containers leading to alcohol evaporation. In this study, contamination remained below 10%, with most cases occurring in Winter, reinforcing the impact of seasonal conditions on sample quality.
Our findings showed that environmental pathogens were the most frequently isolated bacteria, which agrees with similar studies [
7,
14,
15]. The environment plays a vital role in the growth and survival of environmental bacteria. Environmental mastitis pathogens are inherently present in the cows’ environment and often cause intramammary infections. Common environmental pathogens include major pathogens such as coliforms (
Escherichia coli,
Klebsiella spp., and
Enterobacter spp.) and
Streptococcus spp., as well as minor pathogens like NAS and
Corynebacterium spp. The type of bedding, bedding management, and climate greatly influence the prevalence of specific bacterial populations. This significantly impacts udder health and mastitis incidence [
22]. Organic materials such as composted manure bedding promote the rapid growth of environmental pathogens. Conversely, inorganic bedding, particularly sand, does not facilitate bacterial growth [
23]. Composted manure bedding, also referred to as recycled manure bedding, is the most commonly used bedding in California dairies [
24] and, if improperly managed, can be a substantial source of coliforms and other environmental pathogens.
Several Gram-negative pathogens can cause mastitis, with
Escherichia coli and
Klebsiella spp. being the most common, as they belong to a group commonly referred to as coliforms [
4]. Many intramammary infections caused by Gram-negative bacteria develop into clinical mastitis [
2]. Coliforms are the most common major environmental pathogens isolated in a pure culture. They naturally inhabit the soil and intestinal tract of animals, accumulating and multiplying in manure, as well as in contaminated bedding and water.
E. coli is one of the leading causes of bovine mastitis and is found in the cow’s environment, including the bedding material, flies, alleys, and even the bovine gastrointestinal tract, which is a common reservoir for many environmental pathogens [
25]. A recent study [
4] evaluating the efficacy of intramammary therapy against Gram-negative bacteria reported that only 9.1% of the cases were attributed to Gram-negative bacteria, with over 90% of the isolates being
E. coli. Wood-based bedding products are considered the primary source of
Klebsiella spp. on dairy farms, although these bacteria can also be present in herds that use recycled manure or sand for bedding [
26]. Any bedding contaminated with manure may contain
Klebsiella spp., and the nutrients and moisture in bedding enhance the growth of coliforms. Research [
26] has shown that healthy adult cows can shed Klebsiella organisms in their feces. The prevalence of
Klebsiella spp. varies geographically due to differences in climate and management practices [
27].
Staphylococcus spp. are differentiated in the lab into
S. aureus and NAS for mastitis management. While
S. aureus is considered a major contagious pathogen that causes a significant increase in somatic cell count and production losses, NAS are recognized as minor environmental mastitis pathogens, opportunists, and common skin inhabitants. They can be easily found in milking liners, the milker’s hands, bedding, floors, and air samples [
6]. Although their importance in intramammary infections has not been clearly delineated, NAS have been associated with mild clinical and subclinical mastitis and a high elevation of quarter somatic cell count compared to uninfected quarters [
6,
28]. NAS were the most prevalent among all pathogens found in the present study, with an increase of 1.3-fold from 2009 to 2023. A Canadian study [
7] reported a 17-fold increase in the prevalence of NAS from Canadian dairies between 2008 and 2017. A recent study also showed NAS as the most common pathogens isolated from clinical and subclinical cases of mastitis in Germany [
29]. It appears that the prevalence of NAS has been increasing over the years; understanding the prevalence of this minor pathogen is therefore important for implementing prevention and control protocols.
Streptococcus spp. was the most frequently isolated Gram-positive species among major mastitis pathogens. It is commonly present on the mucosal surfaces and skin of animals and humans [
30,
31]. Specific tests are required to differentiate environmental
Streptococcus spp., such as
S. uberis,
S. dysgalactiae, and other
Streptococcus spp., from the contagious
S. agalactiae. Several
Streptococci species can cause bovine mastitis. However, there have been some instances where
S. uberis acted as a contagious pathogen [
30].
Streptococcus uberis and
S. agalactiae can induce chronic mastitis [
31].
The pathogens
Pseudomonas aeruginosa,
Trueperella pyogenes,
Nocardia spp.,
Mycobacterium,
Serratia spp.,
Bacillus spp., Fungi, Algae (
Prototheca spp.), and Yeast are considered uncommon causes of mastitis, typically leading to sporadic infections that affect only a few cows within a herd. These opportunistic pathogens often exploit compromised udder health, such as teat injuries or suboptimal milking practices, to establish infections. Our study identified these pathogens in less than 2% of samples, indicating their relatively low significance as mastitis agents in the dairies. This observation aligns with findings from a survey of mastitis pathogens in Australia [
32], which reported that the most prevalent isolates were
Streptococcus uberis,
Staphylococcus aureus, and
Escherichia coli, with other pathogens constituting a minor fraction of cases. Additionally,
Prototheca zopfii is recognized as an uncommon cause of bovine mastitis, typically leading to sporadic infections within herds. These findings underscore the importance of maintaining proper milking hygiene and udder health to prevent opportunistic infections by these less common mastitis pathogens.
The primary reservoir of contagious pathogens is the udders of infected cows, and transmission occurs during milking through the milkers’ hands, the liners of the milking unit, and cloths. This study found a low prevalence of contagious pathogens, which could be associated with effective mastitis control programs [
33].
Staphylococcus aureus, a major mastitis pathogen, can cause a substantial economic loss once introduced into a herd [
4,
6]. It can colonize the scabs and damaged skin of cows, other animals, and humans and has been isolated from flies and environmental sites [
34]. Moreover,
S. aureus can be transmitted to heifers before calving by horn flies, and these infections can serve as a source of re-infection for cows in the herd [
34]. Although it had low prevalence compared to environmental microorganisms,
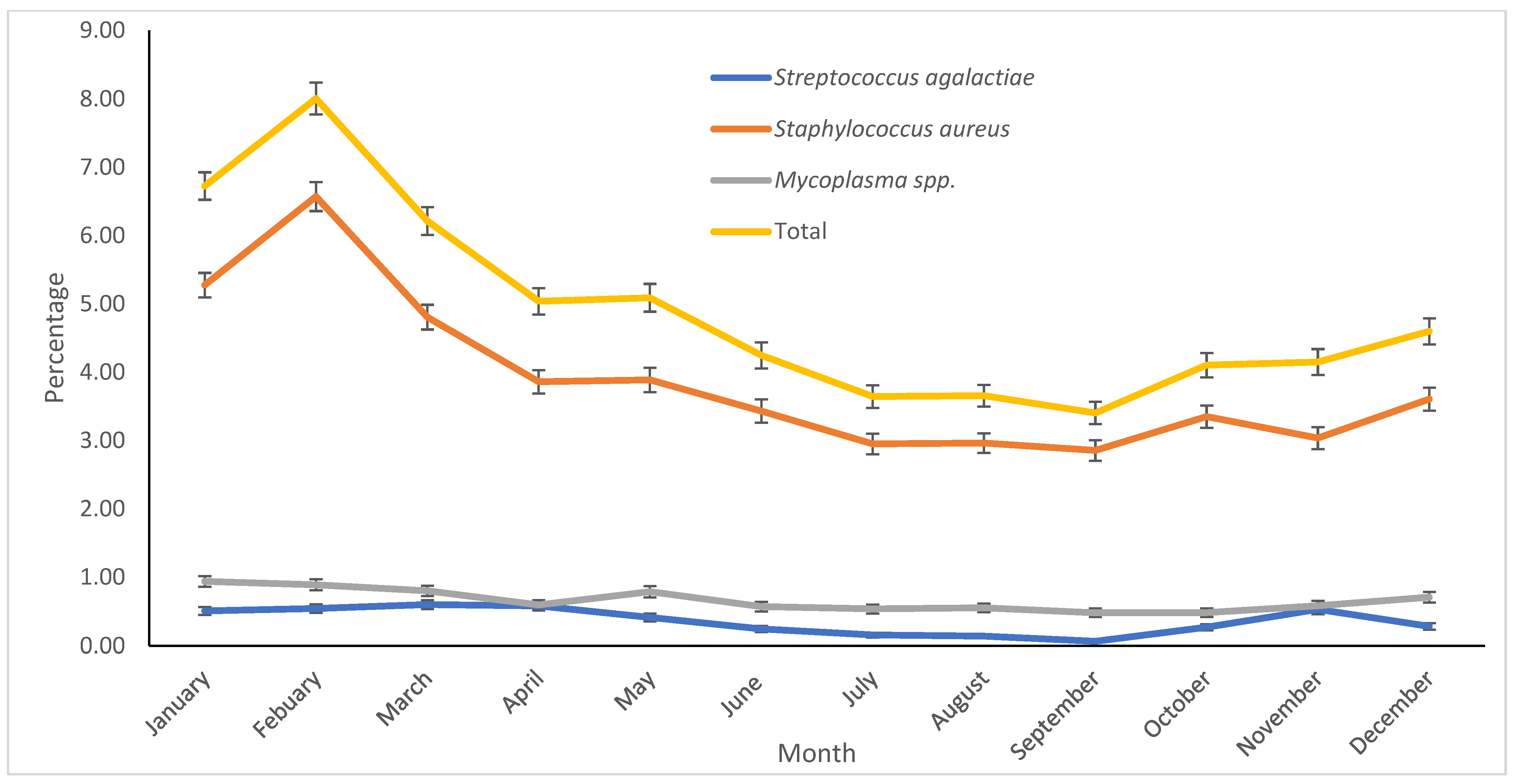
S. aureus was found to be the most common contagious mastitis pathogen isolated from cows in the study. We found a significant increase in the percentage of samples classified as positive for
S. aureus from 2013 to 2015, decreasing thereafter. We suspect that some herds had outbreaks of this pathogen during this period.
Streptococcus agalactiae has been considered a very contagious obligatory intramammary pathogen primarily transmitted from cow to cow during milking, infecting many cows in the herd. However, recent reports suggest that it can be found in extramammary sources [
30,
31]. In our study,
S. agalactiae was identified in a lower percentage (0.4%) of samples.
Streptococcus agalactiae is often shed in high numbers in milk, leading to elevated bacterial counts in bulk tank milk [
31].
Mycoplasma bovis and other
Mycoplasma species have been reported as important contagious mastitis pathogens, with
M. bovis being the most common species and likely causing the most severe mastitis problems [
35].
Mycoplasma spp. have been detected in California dairies since the 1970s [
35].
Mycoplasma spp. were also isolated in a small proportion of samples [1.02%], with the majority classified as
Mycoplasma bovis. The low recovery of mycoplasma from the samples could be due to the methodology used (mycoplasma culture), as this method is relatively slow, often taking one to two weeks, with potential non-growth of these bacteria due to their fastidious culture requirements. However, mycoplasma mastitis should be suspected when milk samples from cows with clinical mastitis routinely test negative for pathogens by standard routine culture methods and when multiple quarters, often all four, are affected in individual cows. Other signs of Mycoplasma mastitis include sudden onset, rapid spread within the herd, a marked reduction in milk production, and resistance to treatment [
15,
16].
The incidence of clinical mastitis is greatly influenced by weather factors, which affect the seasonal isolation of mastitis-causing pathogens [
36]. In our study, the season significantly impacted the proportion of pathogens. Most pathogens were isolated in Winter, except for NAS, which were primarily found in Summer. Reports suggest that the prevalence and distribution of pathogens vary greatly depending on the region [
7,
15,
27]. Differences in the distribution of seasonal pathogens are likely associated with climatological variations. The Midwest and East Coast regions typically experience increased humidity and temperature in Summer, possibly leading to higher bacterial counts in bedding material. Conversely, Winter typically brings freezing conditions, reducing the bacterial population in the environment. California’s Winter tends to be humid, with precipitation levels that are higher than in other seasons, while Summer is usually dry and hot. Furthermore, recycled manure bedding on most dairy farms, combined with increased humidity during Winter, may elevate the risk of coliform exposure. It has been noted that the proportion of contagious pathogens worldwide has been decreasing, likely due to effective contagious mastitis control programs [
33]. This suggests that the use of pre- and post-milking teat disinfectants, good milking hygiene, antimicrobial treatments, and dry cow therapy implemented in recent years could contribute to changes in the prevalence and distribution of contagious mastitis pathogens [
37]. In our study, contagious pathogens (
S. aureus,
S. agalactiae, and
Mycoplasma spp.), although isolated in low proportions, were more likely to be found in Winter. Infections acquired in Winter may persist into Spring, potentially increasing the likelihood of isolating contagious pathogens, as observed in the current study. The results of this study offer valuable insights into the prevalence and distribution of mastitis-causing pathogens in dairies within the Central Valley of California. The data indicate that both contagious and environmental pathogens are present, with significant variation in prevalence by season. These findings emphasize the importance of season-specific mastitis management strategies that address local pathogen profiles, particularly during Winter.