First Molecular Evidence and Phylogeny of Hepatozoon sp. and Theileria sp. in Saudi Rodents
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Host Species Molecular Characterization
2.3. Molecular Identification of Hepatozoon and Theileria
2.3.1. DNA Extraction and 18S rRNA Gene Amplification and Sequencing
2.3.2. Haplotype and Phylogenetic Analysis of Hepatozoon and Theileria
3. Results
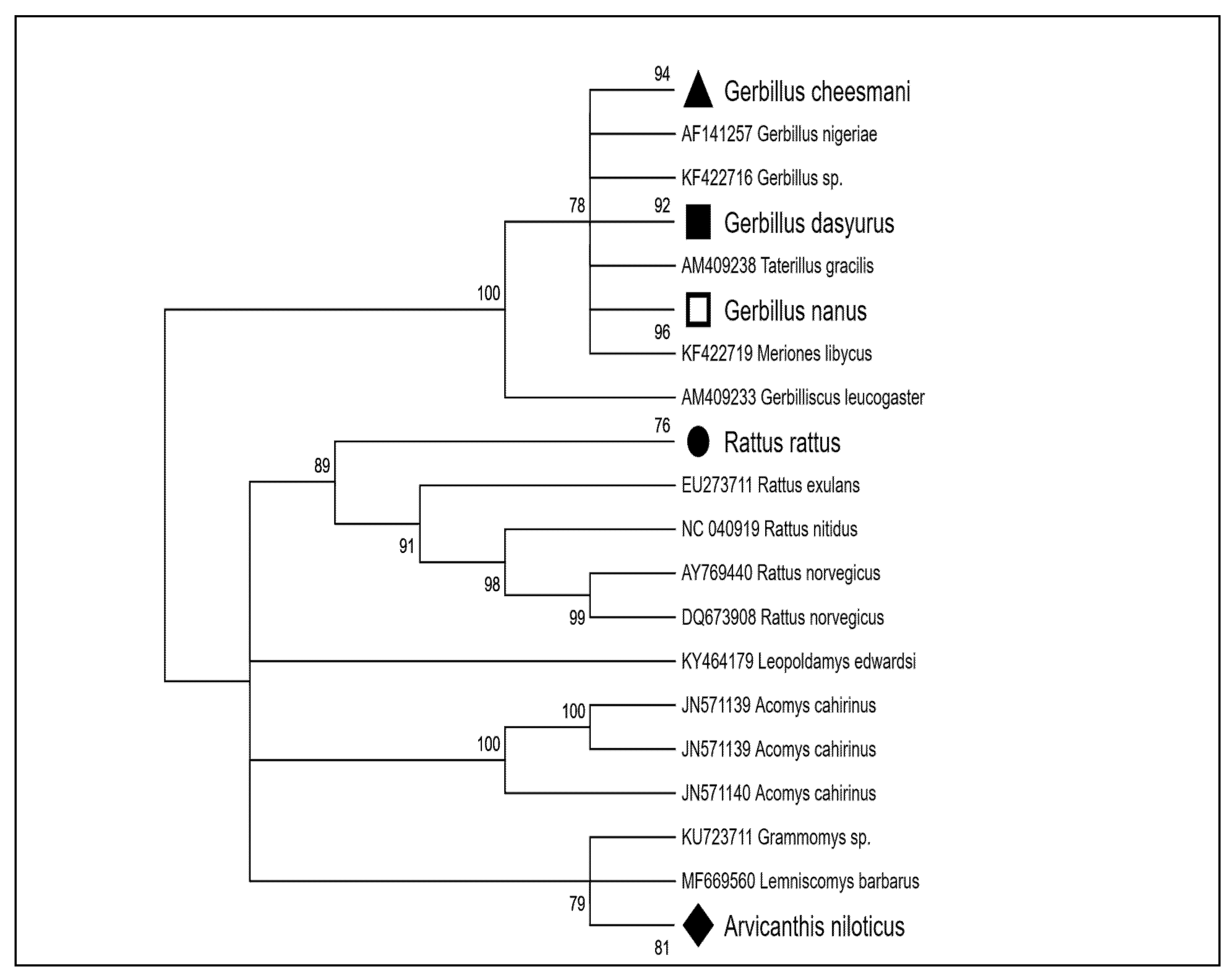
3.1. Characterization of Rodent Species
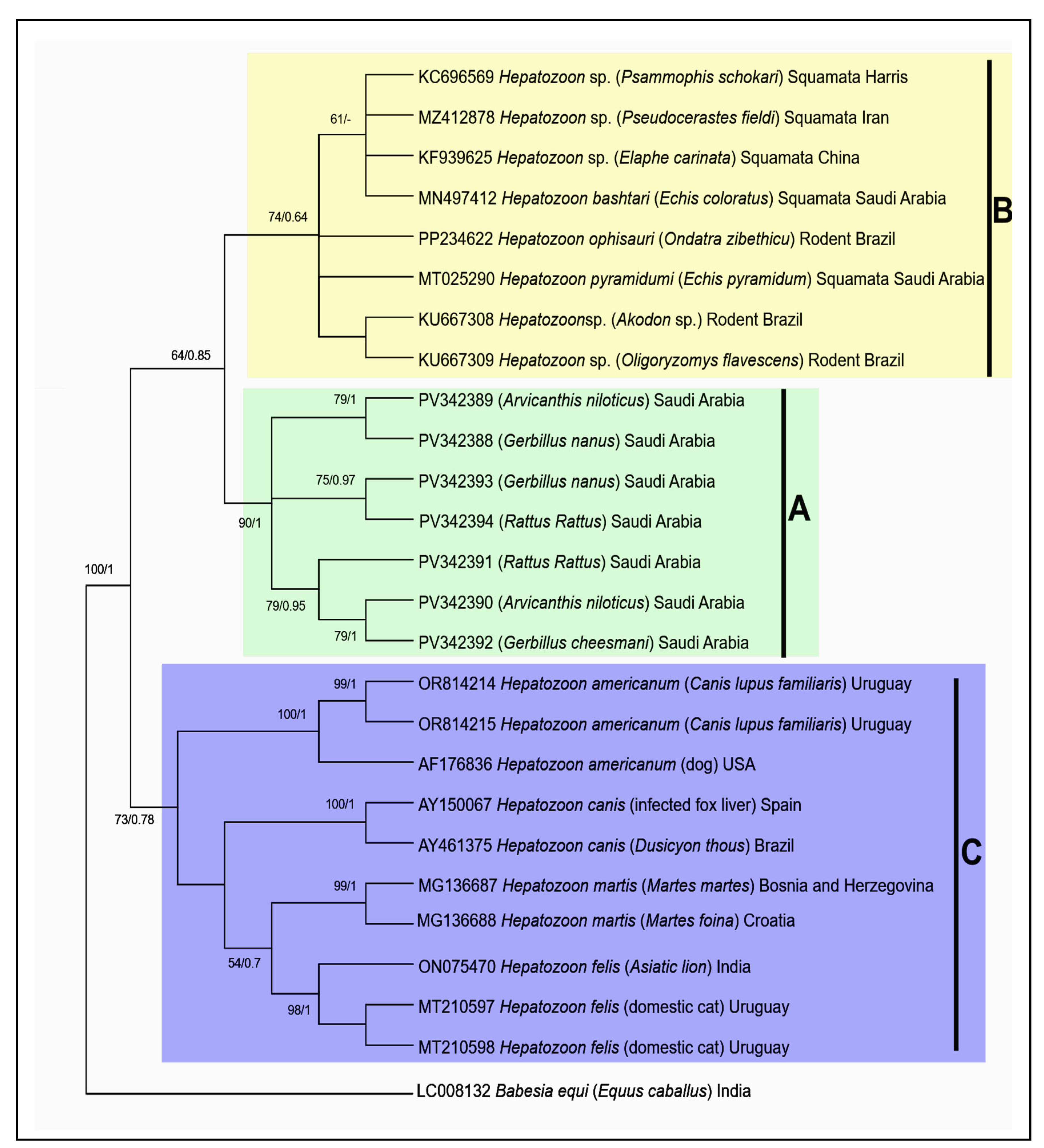
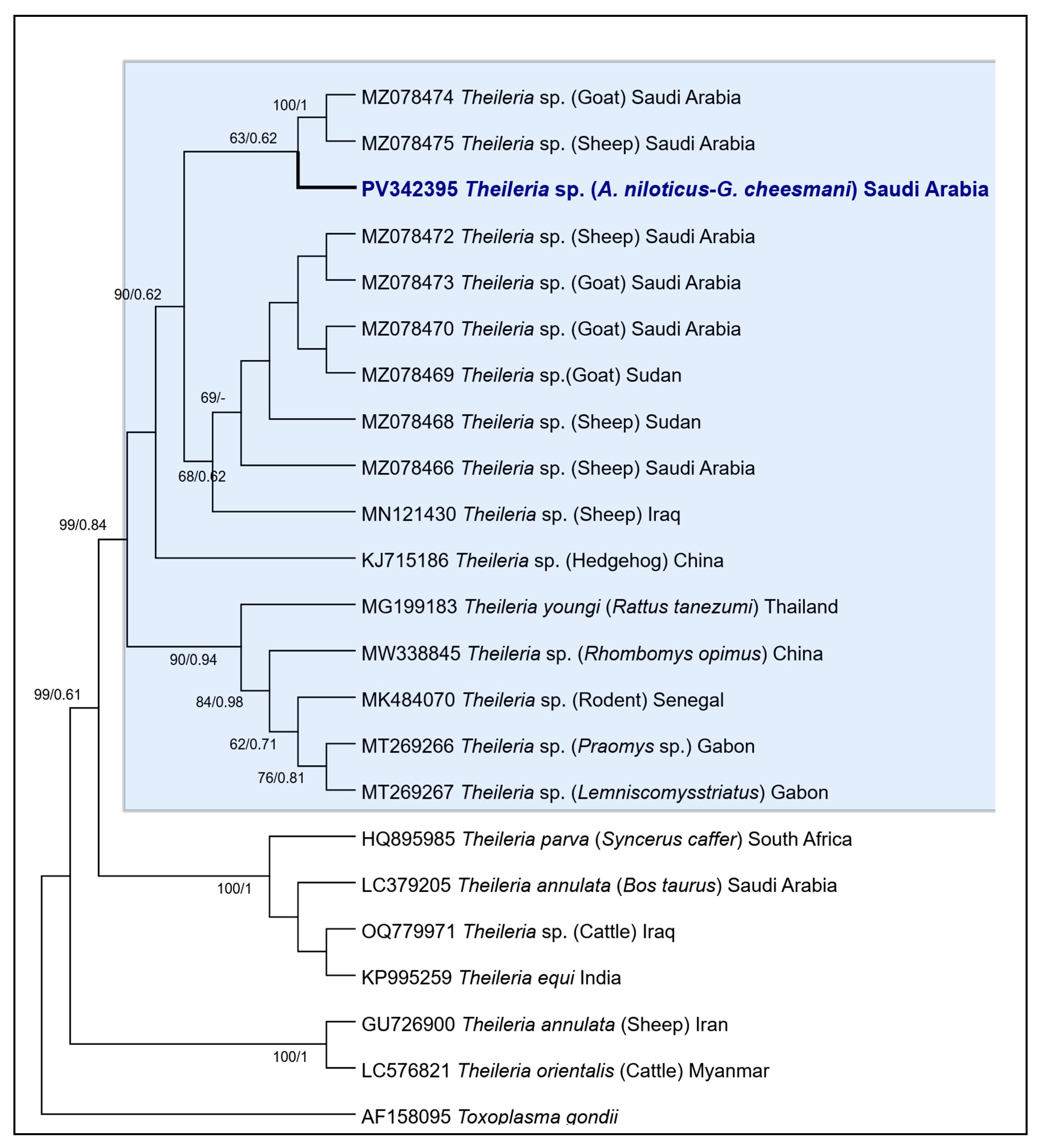
3.2. Molecular Characterization of Hepatozoon sp. and Theileria sp.
3.2.1. Prevalence of Hepatozoon and Theileria in Examined Rodents
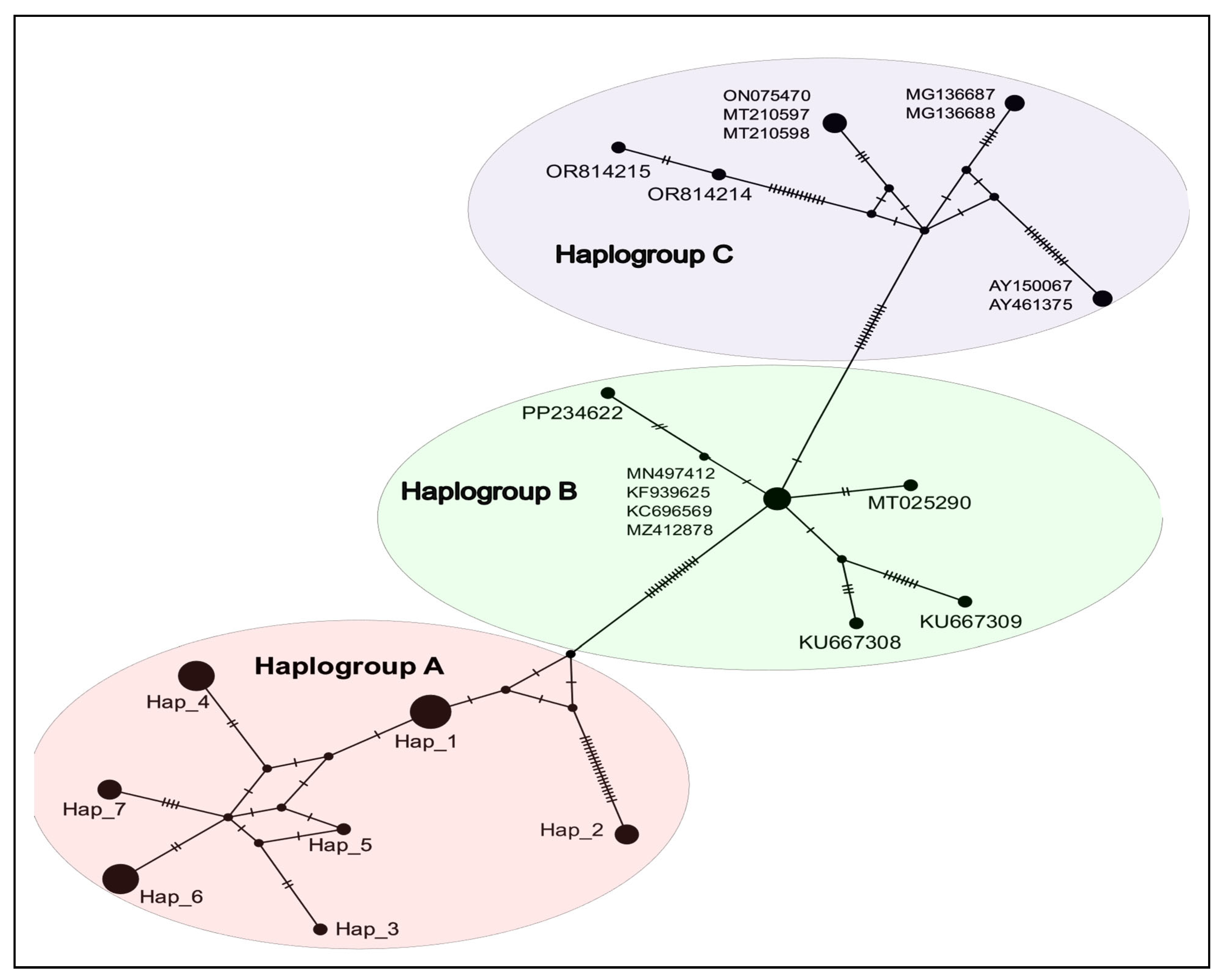
3.2.2. Population Genetic Analysis
3.2.3. Phylogenetic Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Criado-Fornelio, A.; Ruas, J.L.; Casado, N.; Farias, N.A.; Soares, M.P.; Müller, G. New molecular data on mammalian Hepatozoon species (Apicomplexa: Adeleorina) from Brazil and Spain. J. Parasitol. 2006, 92, 93–94. [Google Scholar] [CrossRef]
- Baneth, G. Perspectives on canine and feline hepatozoonosis. Vet. Par. 2011, 181, 3–11. [Google Scholar] [CrossRef]
- Smith, T.G. The genus Hepatozoon (Apicomplexa: Adeleina). J. Parasitol. 1996, 82, 565–585. [Google Scholar] [CrossRef]
- Johnson, E.M.; Allen, K.E.; Panciera, R.J.; Ewing, S.A.; Little, S.E.; Reichard, M.V. Field survey of rodents for Hepatozoon infections in an endemic focus of American canine hepatozoonosis. Vet. Parasitol. 2007, 150, 27–32. [Google Scholar] [CrossRef]
- Johnson, E.M.; Panciera, R.J.; Allen, K.E.; Sheets, M.E.; Beal, J.D.; Ewing, S.A. Alternate pathway of infection with Hepatozoon americanum and the epidemiologic importance of predation. J. Vet. Intern. Med. 2009, 23, 1315–1318. [Google Scholar] [CrossRef]
- D’Oliveira, C.; Van der Weide, M.; Habela, M.A.; Jacquiet, P.; Jongejan, F. Detection of Theileria annulata in blood samples of carrier cattle by PCR. J. Clin. Microbiol. 1995, 33, 2665–2669. [Google Scholar] [CrossRef]
- Ghafar, M.W.; Amer, S.A.M. A preliminary molecular survey of Babesia divergens and first evidence of Theileria annulata in cattle from Saudi Arabia. Vet. World 2019, 12, 266–270. [Google Scholar] [CrossRef]
- Metwally, D.M.; Alajmi, R.; Alsulami, M.N.; Al-Turaiki, I.M.; Abdel-Gaber, R.; Alkhuriji, A.F.; Albohiri, H.H.; Mohamed, K.; Baghdadi, H.B.; El-Khadragy, M.F.; et al. Identification of Theileria spp. in sheep and goats from Jeddah, Saudi Arabia, using molecular techniques. Peer J. 2021, 9, e12596. [Google Scholar] [CrossRef]
- Alanazi, A.D.; Nguyen, V.L.; Alyousif, M.S.; Manoj, R.R.S.; Alouffi, A.S.; Donato, R.; Sazmand, A.; Mendoza-Roldan, J.A.; Dantas-Torres, F.; Otranto, D. Ticks and associated pathogens in camels (Camelus dromedarius) from Riyadh Province, Saudi Arabia. Parasit. Vectors 2020, 13, 110. [Google Scholar] [CrossRef]
- Chandra, S.; Smith, K.; Alanazi, A.D.; Alyousif, M.S.; Emery, D.; Šlapeta, J. Rhipicephalus sanguineus sensu lato from dogs and dromedary camels in Riyadh, Saudi Arabia: Low prevalence of vector-borne pathogens in dogs detected using multiplexed tandem PCR panel. Folia Parasitol. 2019, 66, 007. [Google Scholar] [CrossRef]
- Dantas-Torres, F. Canine vector-borne diseases in Brazil. Parasit. Vectors 2008, 8, 25. [Google Scholar] [CrossRef]
- Mansour, L.; Abdel-Haleem, H.M.; Al-Malki, E.S.; Al-Quraishy, S.; Abdel-Baki, A.A.S. Hepatozoon pyramidumi sp. n. (Apicomplexa: Adeleorina) from the blood of Echis pyramidum: Morphology and SSU rDNA sequence. Braz. J. Vet. Parasitol. 2020, 29, e002420. [Google Scholar] [CrossRef]
- Abdel-Baki, A.S.; Mansour, L.; Al-Malki, E.S. Morphometric and molecular characterisation of Hepatozoon bashtari n. sp. in painted saw-scaled viper, Echis coloratus (Ophidia, Viperidae). Parasitol. Res. 2020, 119, 3793–3801. [Google Scholar] [CrossRef]
- Mohammed, O.B.; Amor, N.; Omer, S.A.; Alagaili, A.N. Molecular detection and characterization of Theileria sp. from hedgehogs (Paraechinus aethiopicus) in Saudi Arabia. Lett. Appl. Microbiol. 2021, 72, 476–483. [Google Scholar] [CrossRef]
- Dahmana, H.; Granjon, L.; Diagne, C.; Davoust, B.; Fenollar, F.; Mediannikov, O. Rodents as Hosts of Pathogens and Related Zoonotic Disease Risk. Pathogens 2020, 9, 202. [Google Scholar] [CrossRef] [PubMed]
- Perkins, S.L.; Schall, J. A molecular phylogeny of malarial parasites recovered from cytochrome b gene sequences. J. Parasitol. 2002, 88, 972–978. [Google Scholar] [CrossRef]
- Martinsen, E.S.; Paperna, I.; Schall, J.J. Morphological versus molecular identification of avian Haemosporidia: An exploration of three species concepts. Parasitology 2006, 133, 279–288. [Google Scholar] [CrossRef]
- Oyamada, M.; Davoust, B.; Boni, M.; Dereure, J.; Bucheton, B.; Hammad, A.; Itamoto, K.; Okuda, M.; Inokuma, H. Detection of Babesia canis rossi, B. canis vogeli, and Hepatozoon canis in dogs in a village of eastern Sudan by using a screening PCR and sequencing methodologies. Clin. Diagn. Lab. Immunol. 2005, 12, 1343–1346. [Google Scholar]
- Wahlang, L.; Lakshmanan, B.; Thomas, N.; Bosewell, A.; Jose, J.K.; Sunanda, C.K.; Aravindakshan, T.V. Comparative analysis of conventional and real time PCR for detection of haemoparasites in dogs. Indian J. Biotechnol. 2019, 18, 9–15. [Google Scholar]
- Alabí, A.; Monti, G.; Otth, C.; Sepúlveda-García, P.; Sánchez-Hidalgo, M.; Mello, V.V.C.; Müller, A. Molecular survey and genetic diversity of hemoplasmas in rodents from Chile. Microorganisms 2020, 8, 1493. [Google Scholar] [CrossRef]
- Hornok, S.; Boldogh, S.A.; Takács, N.; Kontschán, J.; Szekeres, S.; Sós, E.; Sándor, A.D.; Wang, Y.; Tuska-Szalay, B. Molecular epidemiological study on ticks and tick-borne protozoan parasites (Apicomplexa: Cytauxzoon and Hepatozoon spp.) from wild cats (Felis silvestris), Mustelidae and red squirrels (Sciurus vulgaris) in central Europe, Hungary. Parasit. Vectors 2022, 21, 174. [Google Scholar] [CrossRef]
- Tila, H.; Khan, M.; Almutairi, M.M.; Alouffi, A.; Ahmed, H.; Tanaka, T.; Tsai, K.H.; Ali, A. First report on detection of Hepatozoon ayorgbor in Rhipicephalus haemaphysaloides and Hepatozoon colubri in Haemaphysalis sulcata and Hyalomma anatolicum: Risks of spillover of Hepatozoon spp. from wildlife to domestic animals. Front. Vet. Sci. 2023, 10, 1255482. [Google Scholar] [CrossRef] [PubMed]
- Chisu, V.; Giua, L.; Bianco, P.; Masala, G.; Sechi, S.; Cocco, R.; Piredda, I. Molecular Survey of Hepatozoon canis Infection in Domestic Dogs from Sardinia, Italy. Vet. Sci. 2023, 31, 640. [Google Scholar] [CrossRef] [PubMed]
- Dos Anjos Pacheco, T.; Lee, D.A.B.; Maia, M.O. Molecular survey of Hepatozoon spp. and piroplasmids in rodents and marsupials from midwestern Brazil, with evidence of a novel Piroplasmida clade (“South American Rodentia”) in the echimyid rodent Thrichomys pachyurus. Parasitol. Res. 2025, 124, 19. [Google Scholar] [CrossRef]
- Solano-Gallego, L.; Baneth, G. Babesiosis in dog and cats—Expanding parasitological and clinical spectra. Vet. Parasitol. 2011, 181, 48–60. [Google Scholar] [CrossRef]
- Sorenson, M.D.; Ast, J.C.; Dimcheff, D.E.; Yuri, T.; Mindell, D.P. Primers for a PCR based approach to mitochondrial genome sequencing in birds and other vertebrates. Mol. Phyl. Evol. 1999, 12, 105–114. [Google Scholar] [CrossRef]
- Avise, J. Phylogeography: The History and Formation of Species; Harvard University Press: Cambridge, MA, USA, 2000. [Google Scholar]
- Weck, B.C.; Serpa, M.C.A.; Ramos, V.N.; Luz, H.R.; Costa, F.B.; Ramirez, D.G.; Benatti, H.R.; Piovezan, U.; Szabó, M.P.J.; Marcili, A.; et al. Novel genotypes of Hepatozoon spp. in small mammals, Brazil. Parasit. Vectors 2022, 15, 87. [Google Scholar] [CrossRef]
- Uiterwijk, M.; Vojta, L.; Šprem, N.; Beck, A.; Jurković, D.; Kik, M.; Duscher, G.G.; Hodžić, A.; Reljić, S.; Sprong, H.; et al. Diversity of Hepatozoon species in wild mammals and ticks in Europe. Parasit. Vectors 2023, 16, 27. [Google Scholar] [CrossRef]
- Harrison, D.L.; Bates, P.J.J. The Mammals of Arabia, 2nd ed.; Harrison Zoological Museum Publication: Kent, UK, 1991. [Google Scholar]
- Herbreteau, V.; Jittapalapong, S.; Rerkamnuaychoke, W.; Chaval, Y.; Cosson, J.F.; Morand, S. Protocols for Field and Laboratory Rodent Studies; Kasetsart University: Bankok, Thailand, 2011. [Google Scholar]
- Alotaibi, B.H.; Amor, N.; Merella, P.; Mohammed, O.B.; Alagaili, A.N. Genetic diversity of wild rodents and detection of Coxiella burnetii, the causative agent of Q fever, in Saudi Arabia. Vet. Res. Commun. 2022, 46, 769–780. [Google Scholar] [CrossRef]
- Qamar, W.; Khan, M.R.; Arafah, A. Optimization of conditions to extract high quality DNA for PCR analysis from whole blood using SDS-proteinase K method. Saudi J. Biol. Sci. 2017, 24, 1465–1469. [Google Scholar] [CrossRef]
- Palumbi, S.; Martin, A.; Romano, S.; McMillan, W.O.; Stice, L.; Grabowski, G. The Simple Fool’s Guide to PCR, 2nd ed.; University of Hawaii: Honolulu, HI, USA, 1991. [Google Scholar]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; UGENE Team. Uni-pro UGENE: A unifed bioinformatics toolkit. Bioinformatics 2012, 15, 1166–1167. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Gubbels, J.M.; de Vos, A.P.; Van der Weide, M.; Viseras, J.; Schouls, L.M.; de Vries, E. Simultaneous detection of bovine Theileria and Babesia species by reverse line blot hybridization. J. Clin. Microbiol. 1999, 37, 1782–1789. [Google Scholar] [CrossRef]
- Matjila, P.T.; Leisewitz, A.L.; Oosthuizen, M.C.; Jongejan, F.; Penzhorn, B.L. Detection of a Theileria species in dogs in South Africa. Vet. Parasitol. 2008, 157, 34–40. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Demoner, L.D.C.; Magro, N.M.; da Silva, M.R.L.; de Paula Antunes, J.M.A.; Calabuig, C.I.P.; O’Dwyer, L.H. Hepatozoon spp. infections in wild rodents in an area of endemic canine hepatozoonosis in southeastern Brazil. Ticks Tick-Borne Dis. 2016, 7, 859–864. [Google Scholar] [CrossRef]
- Han, H.; Wu, Y.; Dong, H. First report of Hepatozoon (Apicomplexa: Adeleorina) from king ratsnakes (Elaphe carinata) in Shanghai, with description of a new species. Acta Parasit. 2015, 60, 266–274. [Google Scholar] [CrossRef]
- Tomé, B.; Maia, J.P.M.C.; Harris, D.J. Molecular assessment of apicomplexan parasites in the snake Psammophis from North Africa: Do multiple parasite lineages reflect the final vertebrate host diet? J. Parasitol. 2013, 99, 883–887. [Google Scholar] [CrossRef]
- Jameie, F.; Nasiri, V.; Paykari, H. Morphological detection and molecular characterization of Hepatozoon spp. from venomous terrestrial snakes in Iran. Exp. Parasitol. 2022, 239, 108309. [Google Scholar] [CrossRef]
- Parodi, P.; Bazzano, V.; Armúa-Fernández, M.T.; Félix, M.L.; Carvalho, L.A.; Freire, J.; Venzal, J.M. Molecular survey of Piroplasmida, Hepatozoon spp. and Anaplasmataceae in anemic and thrombocytopenic dogs from Uruguay. Vet. Parasitol. Reg. Stud. Rep. 2024, 51, 101027. [Google Scholar] [CrossRef]
- Baker, E.; Dennis, M.; Jensen, A.; Garrett, K.B.; Cleveland, C.A.; Yabsley, M.J.; Brown, J.D.; Why, K.V.; Gerhold, R. Survey for Babesia spp. in wildlife in the eastern United States. Int. J. Parasitol. Parasites Wildl. 2024, 25, 101015. [Google Scholar] [CrossRef]
- Panda, R.; Nehra, A.K.; Ram, H.; Karikalan, M.; Garg, R.; Nala, R.R.; Pawde, A.M. Phylogenetic analysis and haplotype networking of Hepatozoon felis infecting wild animals in Gir National Park, Gujarat, India. Parasitol. Res. 2024, 123, 92. [Google Scholar] [CrossRef]
- Bazzano, V.; Félix, M.L.; Parodi, P. Phylogenetic analysis of Hepatozoon spp. (Apicomplexa: Hepatozoidae) infecting Philodryas patagoniensis (Serpentes: Dipsadidae) in Uruguay. Parasitol. Res. 2020, 119, 1093–1100. [Google Scholar] [CrossRef]
- Hodžić, A.; Alić, A.; Beck, R.; Beck, A.; Huber, D.; Otranto, D.; Baneth, G.; Duscher, G.G. Hepatozoon martis n. sp. (Adeleorina: Hepatozoidae): Morphological and pathological features of a Hepatozoon species infecting martens (family Mustelidae). Ticks Tick-Borne Dis. 2018, 9, 912–920. [Google Scholar] [CrossRef]
- Shalaby, I.; Gherbawy, Y.; Jamjoom, M.; Banaja, A. Prevalence and genotyping of Cryptosporidium in stool samples collected from children in Taif City (Saudi Arabia). Trop. Biomed. 2014, 31, 215–224. [Google Scholar]
- Hawash, Y.; Ismail, K.H.; Abdel-Wahab, M. Shift in parasitic infections during the Corona pandemic: A hospital-based retrospective study. Trop. Biomed. 2021, 38, 94–101. [Google Scholar]
- Abdel-Baki, A.S.; Al-Quraishy, S.; Zhang, J.Y. Redescription of Haemogregarina garnhami (Apicomplexa: Adeleorina) from the blood of Psammophis schokari (Serpentes: Colubridae) as Hepatozoon garnhami n. comb. based on molecular, morphometric and morphologic characters. Acta Parasitol. 2014, 59, 294–300. [Google Scholar] [CrossRef]
- Mohammed, O.B.; Amor, N.; Omer, S.A.; Alagaili, A.N. Seroprevalence of Toxoplasma gondii and Neospora caninum in Dromedary camels (Camelus dromedarius) from Saudi Arabia. Rev. Bras. Parasitol. Vet. 2020, 30, e019119. [Google Scholar] [CrossRef]
- Metwally, D.M.; Al-Damigh, M.A.; Al-Turaiki, I.M.; El-Khadragy, M.F. Molecular Characterization of Sarcocystis Species Isolated from Sheep and Goats in Riyadh, Saudi Arabia. Animals 2019, 9, 256. [Google Scholar] [CrossRef]
- Metwally, D.M.; Al-Otaibi, T.T.; Al-Turaiki, I.M.; El-Khadragy, M.F.; Alajmi, R.A. Identification of Sarcocystis spp. in One-humped Camels (Camelus dromedarius) from Riyadh and Dammam, Saudi Arabia, via Histological and Phylogenetic Approaches. Animals 2020, 10, 1108. [Google Scholar] [CrossRef]
- Moafa, H.N.; Altemani, A.H.; Alaklabi, A. The Prevalence of Toxoplasma gondii in Saudi Arabia (1994–2023): A Systematic Review and Meta-Analysis. J. Epidemiol. Glob. Health 2024, 14, 1413–1452. [Google Scholar] [CrossRef]
- Amr, Z.S. Mammals of Jordan: Jordan Country Study on Biological Diversity; United Nations Environment Programme: Amman, Jordan, 2000; pp. 1–100. [Google Scholar]
- Alanazi, A.D.; Al-Mohammed, H.I.; Alyousif, M.S.; Said, A.E.; Salim, B.; Abdel-Shafy, S. Species diversity and seasonal distribution of hard ticks (Acari: Ixodidae) infesting mammalian hosts in various districts of Riyadh Province, Saudi Arabia. J. Med. Entomol. 2019, 56, 1027–1032. [Google Scholar] [CrossRef]
- Bajer, A.; Pawelczyk, A.; Behnke, J.M.; Gilbert, F.S.; Sinski, E. Factors affecting the component community structure of haemoparasites in bank voles (Clethrionomys glareolus) from the Mazury Lake District region of Poland. Parasitology 2001, 122, 43–54. [Google Scholar] [CrossRef]
- Pawelczyk, A.; Bajer, A.; Behnke, J.M.; Gilbert, F.S.; Sinski, E. Factors affecting the component community structure of haemoparasites in common voles (Microtus arvalis) from the Mazury Lake District region of Poland. Parasitol. Res. 2004, 92, 270–284. [Google Scholar] [CrossRef]
- Aktas, M.; Özübek, S.; Altay, K.; Balkaya, I.; Utuk, A.E.; Kirbas, A. A molecular and parasitological survey of Hepatozoon canis in domestic dogs in Turkey. Vet. Parasitol. 2015, 209, 264–267. [Google Scholar] [CrossRef]
- Díaz-Sáncheza, A.A.; Hofmann-Lehmannb, R.; Melib, M.L.; Roblejo-Ariasa, L.; Fonseca-Rodríguezd, O.; Pérez Castilloa, A.; Vega Cañizaresa, E.; Riveroa, E.L.; Chiltonc, N.B.; Corona-Gonzáleza, B. Molecular detection and characterization of Hepatozoon canis in stray dogs from Cuba. Parasitol. Int. 2021, 80, 102200. [Google Scholar] [CrossRef]
- Santos, E.C.F.; Moura–Martiniano, N.O.; Vilela, R.V.; Lucio, C.S.; Silva, A.F.; Oliveira, S.V.; Gazeta, G.S. Hepatozoon Infecting Bats in the Southeastern Brazilian Rainforest. J. Wildl. Dis. 2020, 56, 693–697. [Google Scholar] [CrossRef]
- Karadjian, G.; Chavatte, J.M.; Landau, I. Systematic revision of the adeleid haemogregarines, with creation of Bartazoon ng, reassignment of Hepatozoon argantis Garnham, 1954 to Hemolivia, and molecular data on Hemolivia stellata. Parasite 2015, 22, 2015031. [Google Scholar] [CrossRef][Green Version]
- Maia, J.P.; Harris, D.J.; Carranza, S.; Goméz-Díaz, E. Assessing the diversity, host-specificity and infection patterns of apicomplexan parasites in reptiles from Oman, Arabia. Parasitology 2016, 143, 1730–1747. [Google Scholar] [CrossRef]
- Hamšíková, Z.; Silaghi, C.; Rudolf, I.; Venclíková, K.; Mahríková, L.; Slovák, M. Molecular detection and phylogenetic analysis of Hepatozoon spp. in questing Ixodes ricinus ticks and rodents from Slovakia and Czech Republic. Parasitol. Res. 2016, 115, 3897–3904. [Google Scholar] [CrossRef]
- Sousa, K.C.; Fernandes, M.P.; Herrera, H.M.; Benevenute, J.L.; Santos, F.M.; Rocha, F.L. Molecular detection of Hepatozoon spp. in domestic dogs and wild mammals in southern Pantanal, Brazil with implications in the transmission route. Vet. Parasitol. 2017, 237, 37–46. [Google Scholar] [CrossRef]
- Perles, L.; Roque, A.L.R.; D’Andrea, P.S.; Lemos, E.R.S.; Santos, A.F.; Morales, A.C. Genetic diversity of Hepatozoon spp. in rodents from Brazil. Sci. Rep. 2019, 9, 10122. [Google Scholar] [CrossRef]
- Modrý, D.; Beck, R.; Hrazdilová, K.; Baneth, G. A review of methods for detection of Hepatozoon infection in carnivores and arthropod vectors. Vector Borne Zoonotic Dis. 2017, 17, 66–72. [Google Scholar] [CrossRef]
- Alabí, A.; Monti, G.; Otth, C.; Sepúlveda-García, P.; Perles, L.; Machado, R.Z.; André, M.R.; Bittencourt, P.; Müller, A. Genetic diversity of Hepatozoon spp. in rodents from Chile. Rev. Bras. Parasitol. Vet. 2021, 30, e012721. [Google Scholar] [CrossRef]
- Stumpf, M.P. Haplotype diversity and SNP frequency dependence in the description of genetic variation. Eur. J. Hum. Genet. 2004, 12, 469–477. [Google Scholar] [CrossRef]
- Harris, D.J.; Maia, J.P.; Perera, A. Molecular characterization of Hepatozoon species in reptiles from the Seychelles. J. Parasitol. 2011, 97, 106–110. [Google Scholar] [CrossRef]
- Laakkonen, J.; Sukura, A.; Oksanen, A.; Henttonen, H.; Soveri, T. Haemogregarines of the genus Hepatozoon (Apicomplexa: Adeleina) in rodents from northern Europe. Folia Parasitol. 2001, 48, 263–267. [Google Scholar] [CrossRef]
- Karbowiak, G.; Rychlik, L.; Nowakowski, W.; Wita, I. Natural infections of small mammals with blood parasites on the borderland of boreal and temperate forest zones. Acta Theriol. 2005, 50, 31–42. [Google Scholar] [CrossRef]
- Kamani, J.; Harrus, S.; Nachum-Biala, Y.; Gutiérrez, R.; Mumcuoglu, K.Y.; Baneth, G. Prevalence of Hepatozoon and Sarcocystis spp. in rodents and their ectoparasites in Nigeria. Acta Trop. 2018, 187, 124–128. [Google Scholar] [CrossRef]
- Aziz, K.J.; Hamadamin, B.Q. Epidemiological and molecular study of Theileria spp. in sheep and goats in Erbil, Iraq. Trop. Anim. Health Prod. 2025, 57, 80. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, Q.; Jiao, F.C. Detection of piroplasms infection in sheep, dogs and hedgehogs in Central China. Infect. Dis. Poverty 2014, 3, 18. [Google Scholar] [CrossRef]
- Mangombi, J.B.; N’dilimabaka, N.; Lekana-Douki, J.B.; Banga, O.; Maghendji-Nzondo, S.; Bourgarel, M. First investigation of pathogenic bacteria, protozoa and viruses in rodents and shrews in context of forest-savannah-urban areas interface in the city of Franceville (Gabon). PLoS ONE 2021, 16, e0248244. [Google Scholar] [CrossRef]
- Sivakumar, T.; Hayashida, K.; Sugimoto, C.; Yokoyama, N. Evolution and genetic diversity of Theileria. Infect. Genet. Evol. 2014, 27, 250–263. [Google Scholar] [CrossRef]
| Rodent Species | Localities | Geographical Coordinates | Hepatozoon spp. PCR Positive % | Theileria spp. PCR Positive % |
|---|---|---|---|---|
| A. niloticus (N = 32) | Baljurashi (n = 32) | 19°51′34″ N 41°33′26″ E | 31.25% | - |
| G. cheesmani (N = 34) | Baljurashi (n = 34) | 19°51′34″ N 41°33′26″ E | 26.47% | - |
| G. dasyurus (N = 6) | Baljurashi (n = 6) | 19°51′34″ N 41°33′26″ E | - | - |
| G. nanus (N = 14) | 28.57% | 21.43% | ||
| Al Darb (n = 4) | 17°44′00.2″ N 42°15′23.3″ E | - | 75.00% | |
| Bish (n = 2) | 17°34′41.4″ N 42°31′03.2″ E | - | - | |
| Ahad Al Msarha (n = 3) | 16°42′34.7″ N 42°56′12.7″ E | 100.00% | - | |
| Wadi Sabya (n = 2) | 17°09′30.5″ N 42°54′45.1″ E | - | - | |
| Wadi Reem (n = 1) | 23°57′52.2″ N 39°27′27.2″ E | - | - | |
| Al Radha (n = 2) | 17°31′05.3″ N 42°24′40.2″ E | 50.00% | - | |
| R. rattus (N = 25) | 32.00% | 24.00% | ||
| Samtah (n = 2) | 16°35′29″ N 42°56′22″ E | 100.00% | - | |
| Sabya (n = 3) | 17°8′56.3″ N 42°37′33.3″ E | - | - | |
| Al Aridhah (n = 4) | 17°01′36.8″ N 43°02′42.1″ E | 25.00% | - | |
| Fifa (n = 2) | 17°15′0″ N 43°06′0″ E | - | - | |
| Al Dair (n = 4) | 17°20′43.4″ N 43°07′47.0″ E | 25.00% | - | |
| Baljurashi (n = 10) | 19°51′34″ N 41°33′26″ E | 40.00% | 60.00% |
| Species | Host/Geographic Origin | Geographic Origin | Accession Numbers | References | |
|---|---|---|---|---|---|
| Hepatozoon sp. | Akodon sp. | Brazil | KU667308 | [44] | |
| Oligoryzomys flavescens | Brazil | KU667309 | |||
| Hepatozoon bashtari | Echis coloratus | Saudi Arabia | MN497412 | [13] | |
| Hepatozoon sp. | Elaphe carinata | Saudi Arabia | KF939625 | [45] | |
| Psammophis schokari | North Africa | KC696569 | [46] | ||
| Pseudocerastes fieldi | Iran | MZ412878 | [47] | ||
| Hepatozoon americanum | Canis lupus familiaris | Uruguay | OR814214 | [48] | |
| Canis lupus familiaris | Uruguay | OR814215 | |||
| Hepatozoon ophisauri | Ondatra zibethicus | USA | PP234622 | [49] | |
| Hepatozoon canis | Fox | Spain | AY150067 | [1] | |
| Dusicyon thous | Brazil | AY461375 | |||
| Hepatozoon felis | Asiatic lion | India | ON075470 | [50] | |
| Domestic cat | Uruguay | MT210597 | [51] | ||
| Domestic cat | Uruguay | MT210598 | |||
| Hepatozoon martis | Martes martes | Bosnia and Herzegovina | MG136687 | [52] | |
| Martes foina | Croatia | MG136688 | |||
| Hepatozoon pyramidumi | Echis pyramidum | Saudi Arabia | MT025290 | [12] | |
| Hepatozoon sp. | Hap_1 | G. cheesmani | Saudi Arabia | PV342392 | Present study |
| Hap_2 | G. nanus | PV342388 | Present study | ||
| Hap_3 | PV342393 | ||||
| Hap_4 | R. rattus | PV342391 | Present study | ||
| Hap_5 | PV342394 | ||||
| Hap_6 | A. niloticus | PV342389 | Present study | ||
| Hap_7 | PV342390 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farjallah, S.; Alagaili, A.N.; AlOsaimi, B.H.; Merella, P.; Mohammed, O.B.; Amor, N. First Molecular Evidence and Phylogeny of Hepatozoon sp. and Theileria sp. in Saudi Rodents. Vet. Sci. 2025, 12, 608. https://doi.org/10.3390/vetsci12070608
Farjallah S, Alagaili AN, AlOsaimi BH, Merella P, Mohammed OB, Amor N. First Molecular Evidence and Phylogeny of Hepatozoon sp. and Theileria sp. in Saudi Rodents. Veterinary Sciences. 2025; 12(7):608. https://doi.org/10.3390/vetsci12070608
Chicago/Turabian StyleFarjallah, Sarra, Abdulaziz Nasser Alagaili, Bandar H. AlOsaimi, Paolo Merella, Osama B. Mohammed, and Nabil Amor. 2025. "First Molecular Evidence and Phylogeny of Hepatozoon sp. and Theileria sp. in Saudi Rodents" Veterinary Sciences 12, no. 7: 608. https://doi.org/10.3390/vetsci12070608
APA StyleFarjallah, S., Alagaili, A. N., AlOsaimi, B. H., Merella, P., Mohammed, O. B., & Amor, N. (2025). First Molecular Evidence and Phylogeny of Hepatozoon sp. and Theileria sp. in Saudi Rodents. Veterinary Sciences, 12(7), 608. https://doi.org/10.3390/vetsci12070608







