Influence of Sub-Inhibitory Concentrations of Sanitizers and Oxacillin on the Resistance of Methicillin-Resistant Staphylococcus spp.
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganisms and Antimicrobial Agents
2.2. Determination of Minimum Inhibitory Concentration (MIC)
2.3. Effect of Sub-MIC Antimicrobials on Growth Characteristics, Susceptibility to Sanitizers and OXA
2.4. Disk Diffusion Assay
2.5. Biofilm Quantification Assay
2.6. Whole Genome Sequencing (WGS)
2.7. Statistical Analysis
3. Results
3.1. Determination of the MIC of Antimicrobial Agents Against Staphylococcus spp. Before and After Application of Sub-MIC Doses
3.2. Agar Diffusion Disk Analysis
3.3. Quantification of Biofilm Formation
3.4. WGS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Silva Cândido, T.J.; da Silva, A.C.; de Matos, L.G.; da Silva do Nascimento, M.; Camargo, C.H.; Cobo Zanella, R.; Mores Rall, V.L.; Cirone Silva, N.C. Enterotoxigenic Potential and Molecular Typing of Staphylococcus sp. Isolated from Organic and Conventional Fresh Minas Cheese in the State of São Paulo, Brazil. Int. Dairy J. 2020, 102, 104605. [Google Scholar] [CrossRef]
- Dantas, S.T.A.; Silva, L.B.B.; Takume, L.T.S.; Rossi, B.F.; Bonsaglia, E.C.R.; Fernandes Júnior, A.; Pantoja, J.C.F.; dos Santos, M.V.; Gonçalves, J.L.; Ribon, A.O.B.; et al. Diversity of Staphylococcus aureus Enterotoxin Genes and Its Potential Impact on Severity of Mastitis in Dairy Cows. Microb. Pathog. 2025, 198, 107119. [Google Scholar] [CrossRef]
- Fontana, C.; Favaro, M. Coagulase-positive and coagulase-negative staphylococci in human disease. In Pet-to-Man Travelling Staphylococci: A World in Progress; Elsevier: Amsterdam, The Netherlands, 2018; pp. 25–42. [Google Scholar] [CrossRef]
- Abreu, A.C.d.S.; Crippa, B.L.; de Souza, V.V.M.A.; Nuñez, K.V.M.; de Almeida, J.M.; Rodrigues, M.X.; Silva, N.C.C. Assessment of Sanitiser Efficacy against Staphylococcus spp. Isolated from Minas Frescal Cheese Producers in São Paulo, Brazil. Int. Dairy J. 2021, 123, 105171. [Google Scholar] [CrossRef]
- Rainard, P.; Foucras, G.; Fitzgerald, J.R.; Watts, J.L.; Koop, G.; Middleton, J.R. Knowledge Gaps and Research Priorities in Staphylococcus aureus Mastitis Control. Transbound. Emerg. Dis. 2018, 65, 149–165. [Google Scholar] [CrossRef]
- Pires, S.M.; Redondo, H.G.; Pessoa, J.; Jakobsen, L.S.; Thomsen, S.T. Risk Ranking of Foodborne Diseases in Denmark: Reflections on a National Burden of Disease Study. Food Control 2024, 158, 110199. [Google Scholar] [CrossRef]
- Crippa, B.L.; De Matos, L.G.; Souza, F.N.; Silva, N.C.C. Non-Aureus Staphylococci and Mammaliicocci (NASM): Their Role in Bovine Mastitis and One Health. J. Dairy Res. 2024, 91, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Kerro Dego, O.; Vidlund, J. Staphylococcal Mastitis in Dairy Cows. Front. Vet. Sci. 2024, 11, 1356259. [Google Scholar] [CrossRef]
- Persson Waller, K.; Myrenås, M.; Börjesson, S.; Kim, H.; Widerström, M.; Monsen, T.; Sigurðarson Sandholt, A.K.; Östlund, E.; Cha, W. Genotypic Characterization of Staphylococcus chromogenes and Staphylococcus simulans from Swedish Cases of Bovine Subclinical Mastitis. J. Dairy. Sci. 2023, 106, 7991–8004. [Google Scholar] [CrossRef]
- Souza, F.N.; Santos, K.R.; Ferronatto, J.A.; Ramos Sanchez, E.M.; Toledo-Silva, B.; Heinemann, M.B.; De Vliegher, S.; Della Libera, A.M.M.P. Bovine-Associated Staphylococci and Mammaliicocci Trigger T-Lymphocyte Proliferative Response and Cytokine Production Differently. J. Dairy. Sci. 2023, 106, 2772–2783. [Google Scholar] [CrossRef]
- Crippa, B.L.; Rodrigues, R.d.S.; de Melo Tavares, R.; Morasi, R.M.; de Almeida, J.M.; Yamatogi, R.S.; Silva, N.C.C. Why Non-aureus Staphylococcus (NAS) Isolated from Bovine Milk Should Be a Concern for the Rise of Superbugs. Microbe 2025, 7, 100376. [Google Scholar] [CrossRef]
- Ariza-Miguel, J.; Oniciuc, E.A.; Sanz, I.; Fernández-Natal, I.; Hernández, M.; Rodríguez-Lázaro, D. Evaluation of Two Commercially Available Chromogenic Media for Confirmation of Methicillin-Resistant Staphylococcus aureus from Human, Animal, and Food Samples. Int. J. Food Microbiol. 2015, 209, 26–28. [Google Scholar] [CrossRef]
- Gutiérrez, D.; Delgado, S.; Vázquez-Sánchez, D.; Martínez, B.; Cabo, M.L.; Rodríguez, A.; Herrera, J.J.; García, P. Incidence of Staphylococcus aureus and Analysis of Associated Bacterial Communities on Food Industry Surfaces. Appl. Environ. Microbiol. 2012, 78, 8547–8554. [Google Scholar] [CrossRef]
- Mc Carlie, S.; Boucher, C.E.; Bragg, R.R. Molecular Basis of Bacterial Disinfectant Resistance. Drug Resist. Updates 2020, 48, 100672. [Google Scholar] [CrossRef]
- Oniciuc, E.A.; Likotrafiti, E.; Alvarez-Molina, A.; Prieto, M.; López, M.; Alvarez-Ordóñez, A. Food Processing as a Risk Factor for Antimicrobial Resistance Spread along the Food Chain. Curr. Opin. Food Sci. 2019, 30, 21–26. [Google Scholar] [CrossRef]
- Barcelos, M.M.; Martins, L.; Grenfell, R.C.; Juliano, L.; Anderson, K.L.; dos Santos, M.V.; Gonçalves, J.L. Comparison of Standard and On-Plate Extraction Protocols for Identification of Mastitis-Causing Bacteria by MALDI-TOF MS. Braz. J. Microbiol. 2019, 50, 849–857. [Google Scholar] [CrossRef]
- de Almeida, J.M.; Maffei, J.T.; Gebara, C.; Minafra, C.; Toledo-Silva, B.; Gonçalves, M.C.; Langoni, H.; Neto, A.T.; Souza, F.N.; Silva, N.C.C. Exploring Probiotic Potential and Antimicrobial Properties of Lactic Acid Bacteria from Cow’s Milk. Appl. Food Res. 2024, 4, 100461. [Google Scholar] [CrossRef]
- Clinical & Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Clinical & Laboratory Standards Institute: Wayne, PA, USA, 2024; Available online: https://clsi.org/shop/standards/m07/ (accessed on 10 September 2025).
- dos Santos, D.L.S.; Almeida, N.A.; de Almeida, J.M.; Oliveira, M.E.A.S.; Rocha, L.d.O.; Cirone Silva, N.C. Antimicrobial Activity Evaluation of Combinations of Essential Oils, Thymol and R-Limonene against Food-Borne Pathogens and Spoilage Agents. Food Biosci. 2024, 62, 105035. [Google Scholar] [CrossRef]
- Karatzas, K.A.G.; Webber, M.A.; Jorgensen, F.; Woodward, M.J.; Piddock, L.J.V.; Humphrey, T.J. Prolonged Treatment of Salmonella Enterica serovar Typhimurium with Commercial Disinfectants Selects for Multiple Antibiotic Resistance, Increased Efflux and Reduced Invasiveness. J. Antimicrob. Chemother. 2007, 60, 947–955. [Google Scholar] [CrossRef] [PubMed]
- Clinical & Laboratory Standards Institute. M100-Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; Clinical & Laboratory Standards Institute: Wayne, PA, USA, 2018; Available online: https://fda.report/Standards/37761 (accessed on 10 September 2025).
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 15.0; The European Committee on Antimicrobial Susceptibility Testing: Växjö, Sweden, 2022; Available online: https://www.eucast.org (accessed on 10 September 2025).
- Stepanović, S.; Vuković, D.; Dakić, I.; Savić, B.; Švabić-Vlahović, M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 2000, 40, 175–179. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Lu, J.; Langmead, B. Improved Metagenomic Analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Buzón-Durán, L.; Alonso-Calleja, C.; Riesco-Peláez, F.; Capita, R. Effect of Sub-Inhibitory Concentrations of Biocides on the Architecture and Viability of MRSA Biofilms. Food Microbiol. 2017, 65, 294–301. [Google Scholar] [CrossRef] [PubMed]
- Bland, R.; Brown, S.R.B.; Waite-Cusic, J.; Kovacevic, J. Probing Antimicrobial Resistance and Sanitizer Tolerance Themes and Their Implications for the Food Industry through the Listeria monocytogenes Lens. Compr. Rev. Food Sci. Food Saf. 2022, 21, 1777–1802. [Google Scholar] [CrossRef]
- dos Santos, D.L.S.; de Miranda, J.F.; Rodrigues, R.A.F.; Prata, A.S.; Silva, N.C.C. Evaluation of the Stability and Antimicrobial Activity of Emulsions Loaded with Thymol for the Post-Harvest Sanitization of Lettuce (Lactuca Sativa L). Food Biosci. 2025, 66, 106176. [Google Scholar] [CrossRef]
- Aodah, A.H.; Bakr, A.A.; Booq, R.Y.; Rahman, M.J.; Alzahrani, D.A.; Alsulami, K.A.; Alshaya, H.A.; Alsuabeyl, M.S.; Alyamani, E.J.; Tawfik, E.A. Preparation and Evaluation of Benzalkonium Chloride Hand Sanitizer as a Potential Alternative for Alcohol-Based Hand Gels. Saudi Pharm. J. 2021, 29, 807–814. [Google Scholar] [CrossRef]
- Bonsaglia, E.C.R.; Calvo, G.H.; Sordelli, D.O.; Silva, N.C.C.; Rall, V.L.M.; Casas, A.; Buzzola, F. The Impact of Low-Level Benzalkonium Chloride Exposure on Staphylococcus spp. Strains and Control by Photoinactivation. Antibiotics 2023, 12, 1244. [Google Scholar] [CrossRef]
- Lee, J.; Iwasaki, T.; Ohtani, S.; Matsui, H.; Nejima, R.; Mori, Y.; Kagaya, F.; Yagi, A.; Yoshimura, A.; Hanaki, H.; et al. Benzalkonium Chloride Resistance in Staphylococcus epidermidis on the Ocular Surface of Glaucoma Patients under Long-Term Administration of Eye Drops. Transl. Vis. Sci. Technol. 2020, 9, 9. [Google Scholar] [CrossRef]
- Ríos-Castillo, A.G.; Umaña, F.F.; Rodríguez-Jerez, J.J. Long-Term Antibacterial Efficacy of Disinfectants Based on Benzalkonium Chloride and Sodium Hypochlorite Tested on Surfaces against Resistant Gram-Positive Bacteria. Food Control 2018, 93, 219–225. [Google Scholar] [CrossRef]
- Speck, S.; Wenke, C.; Feßler, A.T.; Kacza, J.; Geber, F.; Scholtzek, A.D.; Hanke, D.; Eichhorn, I.; Schwarz, S.; Rosolowski, M.; et al. Borderline Resistance to Oxacillin in Staphylococcus aureus after Treatment with Sub-Lethal Sodium Hypochlorite Concentrations. Heliyon 2020, 6, e04070. [Google Scholar] [CrossRef]
- Huet, A.A.; Raygada, J.L.; Mendiratta, K.; Seo, S.M.; Kaatz, G.W. Multidrug Efflux Pump Overexpression in Staphylococcus aureus after Single and Multiple in Vitro Exposures to Biocides and Dyes. Microbiology 2008, 154, 3144–3153. [Google Scholar] [CrossRef] [PubMed]
- Shang, W.; Rao, Y.; Zheng, Y.; Yang, Y.; Hu, Q.; Hu, Z.; Yuan, J.; Peng, H.; Xiong, K.; Tan, L.; et al. β-Lactam Antibiotics Enhance the Pathogenicity of Methicillin-Resistant Staphylococcus aureus via SarA-Controlled Lipoprotein-like Cluster Expression. mBio 2019, 10, e00880-19. [Google Scholar] [CrossRef] [PubMed]
- Scholtzek, A.D.; Hanke, D.; Walther, B.; Eichhorn, I.; Stöckle, S.D.; Klein, K.S.; Gehlen, H.; Lübke-Becker, A.; Schwarz, S.; Feßler, A.T. Molecular Characterization of Equine Staphylococcus aureus Isolates Exhibiting Reduced Oxacillin Susceptibility. Toxins 2019, 11, 535. [Google Scholar] [CrossRef]
- Mcdougal, L.K.; Thornsberry, C. The role of beta-lactamase in staphylococcal resistance to penicillinase-resistant penicillins and cephalosporins. J. Clin. Microbiol. 1986, 23, 832–839. [Google Scholar] [CrossRef]
- Zuniga, E.; Melville, P.A.; Saidenberg, A.B.S.; Laes, M.A.; Gonsales, F.F.; Salaberry, S.R.S.; Gregori, F.; Brandão, P.E.; dos Santos, F.G.B.; Lincopan, N.E.; et al. Occurrence of Genes Coding for MSCRAMM and Biofilm-Associated Protein Bap in Staphylococcus spp. Isolated from Bovine Subclinical Mastitis and Relationship with Somatic Cell Counts. Microb. Pathog. 2015, 89, 1–6. [Google Scholar] [CrossRef]
- Jian, Y.; Li, T.; Zhao, L.; Zhao, N.; Liu, Y.; Lv, H.; Wang, Y.; Liu, Q.; Li, M. Regulation of Bla System in ST59-Related Oxacillin-Susceptible MecA-Positive Staphylococcus aureus. J. Antimicrob. Chemother. 2022, 77, 604–614. [Google Scholar] [CrossRef]
- Perault, A.; Turlan, C.; Eynard, N.; Vallé, Q.; Bousquet-Mélou, A.; Giraud, E. Repeated Exposure of Escherichia coli to High Ciprofloxacin Concentrations Selects GyrB Mutants That Show Fluoroquinolone-Specific Hyperpersistence. Front. Microbiol. 2022, 13, 908296. [Google Scholar] [CrossRef]
- Chatterjee, A.; Poon, R.; Chatterjee, S.S. Stp1 Loss of Function Promotes β-Lactam Resistance in Staphylococcus aureus That Is Independent of Classical Genes. Antimicrob. Agents Chemother. 2020, 64, e02222-19. [Google Scholar] [CrossRef]
- Lade, H.; Kim, J.S. Molecular Determinants of β-Lactam Resistance in Methicillin-Resistant Staphylococcus aureus (MRSA): An Updated Review. Antibiotics 2023, 12, 1362. [Google Scholar] [CrossRef]
- Juma, M.A.; Kumburu, H.; Wadugu, B.; Kuchaka, D.; Shayo, M.; Kimu, P.; Kanje, L.E.; Beti, M.; van Zwetselaar, M.; Mmbaga, B.; et al. Whole Genome Sequencing-Based Characterization and Determination of Quinolone Resistance among Methicillin-Resistant and Methicillin-Susceptible S. aureus Isolates from Patients Attending Regional Referral Hospitals in Tanzania. BMC Genom. 2024, 25, 1130. [Google Scholar] [CrossRef] [PubMed]
- Huynh, T.Q.; Tran, V.N.; Thai, V.C.; Nguyen, H.A.; Giang Nguyen, N.T.; Tran, M.K.; Truc Nguyen, T.P.; Le, C.A.; Ngan Ho, L.T.; Surian, N.U.; et al. Genomic Alterations Involved in Fluoroquinolone Resistance Development in Staphylococcus aureus. PLoS ONE 2023, 18, e0287973. [Google Scholar] [CrossRef] [PubMed]
- Sultan, A.M.; Ahmed, M.A. Distribution of chlorhexidine resistance genes among Staphylococcus aureus clinical isolates: The challenge of antiseptic resistance. Germs 2022, 12, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Archer, G.L. Roles of CcrA and CcrB in Excision and Integration of Staphylococcal Cassette Chromosome Mec, a Staphylococcus aureus Genomic Island. J. Bacteriol. 2010, 192, 3204–3212. [Google Scholar] [CrossRef]
- Hassanzadeh, S.; Ganjloo, S.; Pourmand, M.R.; Mashhadi, R.; Ghazvini, K. Epidemiology of efflux pumps genes mediating resistance among Staphylococcus aureus; a systematic review. Microb. Pathog. 2020, 139, 103850. [Google Scholar] [CrossRef]
- Hicks, N.D.; Giffen, S.R.; Culviner, P.H.; Chao, M.C.; Dulberger, C.L.; Liu, Q.; Stanley, S.; Brown, J.; Sixsmith, J.; Wolf, I.D.; et al. Mutations in DnaA and a Cryptic Interaction Site Increase Drug Resistance in Mycobacterium Tuberculosis. PLoS Pathog. 2020, 16, e1009063. [Google Scholar] [CrossRef]
- Kerek, Á.; Szabó, Á.; Jerzsele, Á. Antimicrobial Susceptibility Profiles of Staphylococcus aureus Isolates from Domestic Pigeons in Hungary in 2022. Antibiotics 2025, 14, 525. [Google Scholar] [CrossRef]
- Fowoyo, P.T.; Ogunbanwo, S.T. Antimicrobial Resistance in Coagulase-Negative Staphylococci from Nigerian Traditional Fermented Foods. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 4. [Google Scholar] [CrossRef]
- Jin, X.; Liu, S.; Zhang, Z.; Liu, T.; Li, N.; Liang, Y.; Zheng, J.; Peng, N. Enrofloxacin-Induced Transfer of Multiple-Antibiotic Resistance Genes and Emergence of Novel Resistant Bacteria in Red Swamp Crayfish Guts and Pond Sediments. J. Hazard. Mater. 2023, 443, 130261. [Google Scholar] [CrossRef] [PubMed]
- Silva Júnior, A.E.; Vasconcelos, P.C.; Saraiva, M.M.S.; Filho, L.S.; Silva, N.M.V.; Givisiez, P.E.N.; Oliveira, C.J.B. Antimicrobial Susceptibility Profiles of Staphylococcus spp. Contaminating Raw Goat Milk. Vet. World 2021, 14, 1074–1079. [Google Scholar] [CrossRef]
- Scott, N.; Seeraj, C.; Satram, B.; Sandy, N.M.; Seuradge, K.; Seerattan, B.; Seeram, I.; Stewart-Johnson, A.; Adesiyun, A. Occurrence of Methicillin-Resistant Staphylococcus aureus in Pets and Their Owners in Rural and Urban Communities in Trinidad. J. Infect. Dev. Ctries. 2022, 16, 1458–1465. [Google Scholar] [CrossRef]
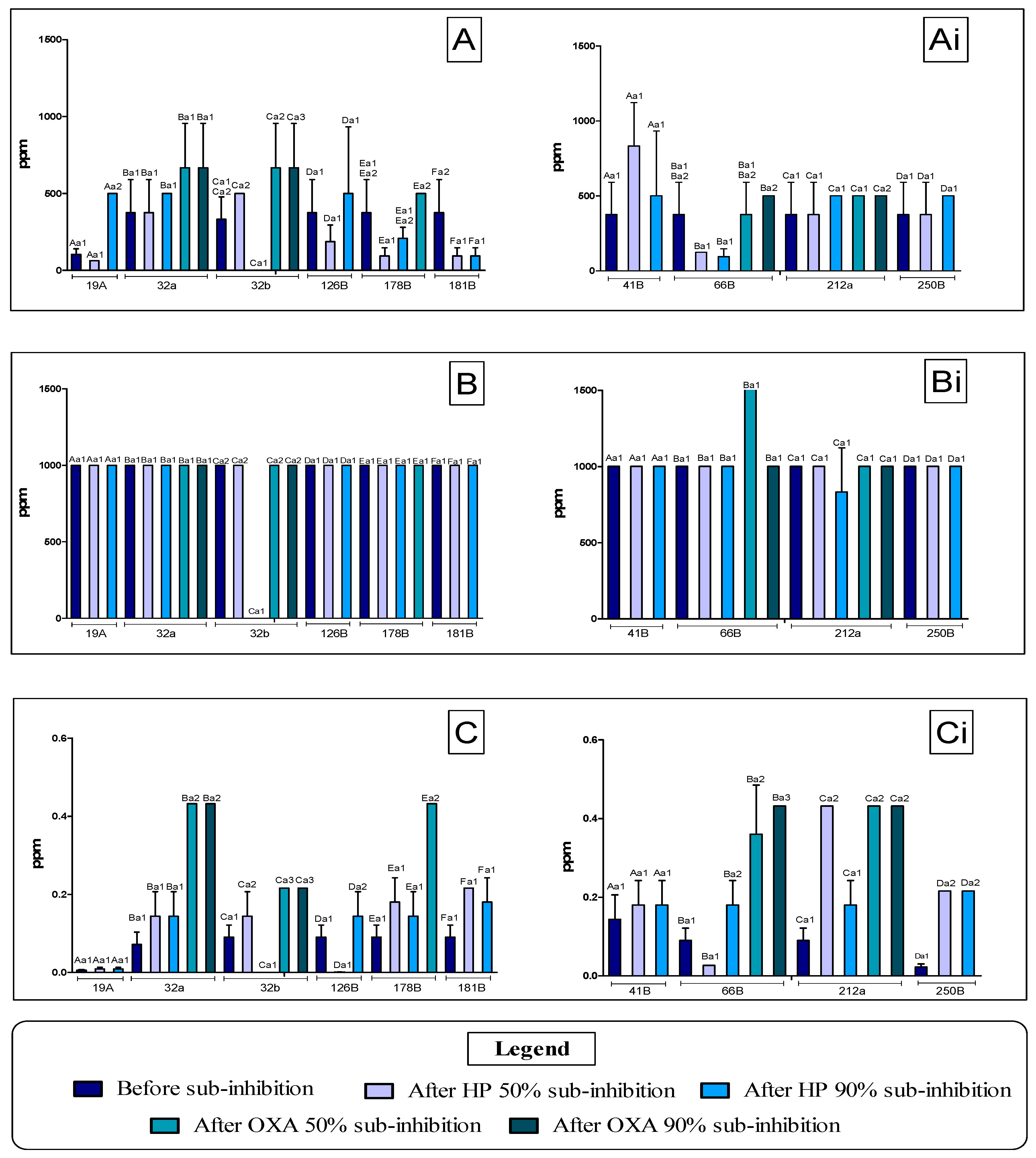
| Antimicrobial Agent | MIC 50 (ppm) | MIC 90 (ppm) | ¼ MIC 50 (ppm) | ¼ MIC 90 (ppm) |
|---|---|---|---|---|
| BAC | 125 | 250 | 31.25 (Treatment 1) | 62.5 (Treatment 2) |
| HP | 1000 | 1000 | 250 (Treatment 3) | 250 (Treatment 4) |
| OXA | 0.027 | 0.108 | 0.00675 (Treatment 5) | 0.027 (Treatment 6) |
| Resistance Profile of Strains Before Sub-Inhibition | Sub-Inhibitory Treatment with HP | Sub-Inhibitory Treatment with OXA | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Isolated | Classification | ¼ MIC50 | ¼ MIC90 | ¼ MIC50 | ¼ MIC90 | ||||
| Isolated | Classification | Isolated | Classification | Isolated | Classification | Isolated | Classification | ||
| 19A | WA d | 19A1 | NA a | 19A2 | NA a | - | - | - | - |
| 32A | NA d | 32A1 | NA a | 32A2 | WA b | 32A3 | WA d | 32A4 b | NA |
| 32B | WA e | 32B1 | NA b | 32B2 | NA a | 32B3 | NA a | 32B4 b | NA |
| 41B | WA f | 41B1 | NA c | 41B2 | WA c | - | - | - | - |
| 66B | WA a | 66B1 | NA c | 66B2 | NA a | 66B3 | NA b | 66B4 a | NA |
| 126B | NA f | 126B1 | WA f | 126B2 | NA a | - | - | - | - |
| 178B | NA f | 178B1 | WA e | 178B2 | WA d | 178B3 | NA b | - | - |
| 181B | WA g | 181B1 | WA d | 181B2 | WA b | - | - | - | - |
| 212A | WA b | 212A1 | WA d | 212A2 | NA b | 212A3 | NA c | 212A4 c | NA |
| 250B | WA c | 250B1 | WA d | 250B2 | NA b | - | - | - | - |
| Isolates | Mutation Position | Gene | Protein Name or Function | NCBI Library |
|---|---|---|---|---|
| 32a–32a4 | 4276 | recF | DNA replication/repair protein | ASM609437v1 ASM4594097v1 ASM1932966v1 |
| gyrB | DNA topoisomerase (ATP-hydrolyzing) subunit B | |||
| 12,099 | serS | Serine--tRNA ligase | ASM609437v1 ASM4594097v1 ASM1932966v1 | |
| sph | Sphingomyelin phosphodiesterase | |||
| 13,655 | sph | Sphingomyelin phosphodiesterase | ASM609437v1 ASM4594241v1 ASM4594097v1 | |
| 20,005 | rplI | 50S ribosomal protein L9 | ASM609437v1 ASM4594097v1 ASM1932966v1 | |
| 49,380 | blaR1 | Beta-lactam sensor/signal transducer BlaR1 | ASM4594097v1 ASM4594241v1 ASM4594006v1 | |
| 58,217 | galU | uncharacterized gene | ASM1932966v1 | |
| 93,331; 93,351 | mecR1 | Beta-lactam sensor/signal transducer MecR1 | ASM1932966v1 | |
| 96,456 | cstB | Uncharacterized gene | ASM1932966v1 | |
| 32,667 | - | IS3 family transposase | ASM1932942v1 | |
| 42,855 | - | DUF6038 family protein | ASM609437v1 | |
| 60,993 | - | SdrD B-like domain-containing protein | ASM609437v1 | |
| 32,673; 32,823; 33,258; 33,273; 33,303 | - | IS3 family transposase | ASM1932966v1 | |
| 42,855 | - | DUF6038 family protein | ASM609437v1 | |
| 197,627 | - | Metal ABC transporter solute-binding protein, Zn/Mn family | ASM609437v2 | |
| 181B–181B2 | 33,653 | hsdR, hsdM, and hsdS | Type I restriction endonuclease subunit R | ASM4594097v1 |
| 76,949; 76,966; 77,007 | - | DUF5906 domain-containing protein | ASM1932966v1 | |
| 77,402 | ccrB | Cassette chromosome recombinase CcrB | ASM4594097v1 ASM4594006v1 ASM1932942v0 | |
| 77,648 | ccrB | Cassette chromosome recombinase CcrB | ASM1932942v1 ASM1932966v ASM4594097v1 | |
| 212a–212A4 | 86; 136; 138; 152; 159; 166; 186; 217; 229; 250; 256; 268; 276; 292; 343; 479; 694; 725; 734; 739; 776; 912 | dnaA | Chromosomal replication initiator protein DnaA | ASM4857110v1 ASM1146687v1 ASM4857109v1 |
| 1531; 1537 2072 | dnaN | DNA polymerase III subunit beta | ASM1146687v1 ASM4857110v1 ASM4857108v1 | |
| 3372; 3378; 3387; 3438 | - | DsbA family protein | ASM1146687v1 | |
| 4000; 4035; 4235; 4351 | recF | DNA replication/repair protein RecF | ASM1146687v1 ASM4857110v1 ASM4857108v1 | |
| 5545 | gyrB | DNA topoisomerase (ATP-hydrolyzing) subunit B | ASM1146687v1 ASM4857110v1 ASM4857108v1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Betim, M.E.; Santos, D.L.S.d.; Lopes, T.d.S.; Crippa, B.L.; Bonsaglia, É.R.; Dantas, S.T.A.; Rall, V.L.M.; Buzzola, F.; Galvão, J.A.; Gebara, C.; et al. Influence of Sub-Inhibitory Concentrations of Sanitizers and Oxacillin on the Resistance of Methicillin-Resistant Staphylococcus spp. Vet. Sci. 2025, 12, 979. https://doi.org/10.3390/vetsci12100979
Betim ME, Santos DLSd, Lopes TdS, Crippa BL, Bonsaglia ÉR, Dantas STA, Rall VLM, Buzzola F, Galvão JA, Gebara C, et al. Influence of Sub-Inhibitory Concentrations of Sanitizers and Oxacillin on the Resistance of Methicillin-Resistant Staphylococcus spp. Veterinary Sciences. 2025; 12(10):979. https://doi.org/10.3390/vetsci12100979
Chicago/Turabian StyleBetim, Maria Eugênia, Daniel Lucino Silva dos Santos, Thiago dos Santos Lopes, Bruna Lourenço Crippa, Érika Romão Bonsaglia, Stéfani Thais Alves Dantas, Vera Lúcia Mores Rall, Fernanda Buzzola, Julia Arantes Galvão, Clarice Gebara, and et al. 2025. "Influence of Sub-Inhibitory Concentrations of Sanitizers and Oxacillin on the Resistance of Methicillin-Resistant Staphylococcus spp." Veterinary Sciences 12, no. 10: 979. https://doi.org/10.3390/vetsci12100979
APA StyleBetim, M. E., Santos, D. L. S. d., Lopes, T. d. S., Crippa, B. L., Bonsaglia, É. R., Dantas, S. T. A., Rall, V. L. M., Buzzola, F., Galvão, J. A., Gebara, C., Thaler, A., Neto, & Silva, N. C. C. (2025). Influence of Sub-Inhibitory Concentrations of Sanitizers and Oxacillin on the Resistance of Methicillin-Resistant Staphylococcus spp. Veterinary Sciences, 12(10), 979. https://doi.org/10.3390/vetsci12100979








