Multivariate Single-Step GWAS Reveals Pleiotropic Genomic Regions and Candidate Genes Associated with Male Scrotal Circumference and Female Fertility Traits in Retinta Beef Cattle
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Phenotypic Data and Pedigree
2.3. Genotyping and Quality Control
2.4. Single-Step GBLUP Analysis
2.5. Genome-Wide Association Study Analysis
2.6. Functional Analysis
3. Results and Discussion
3.1. Phenotypic Values
3.2. Heritability Estimates
3.3. Genetic Correlations Between SC and Female Fertility Traits
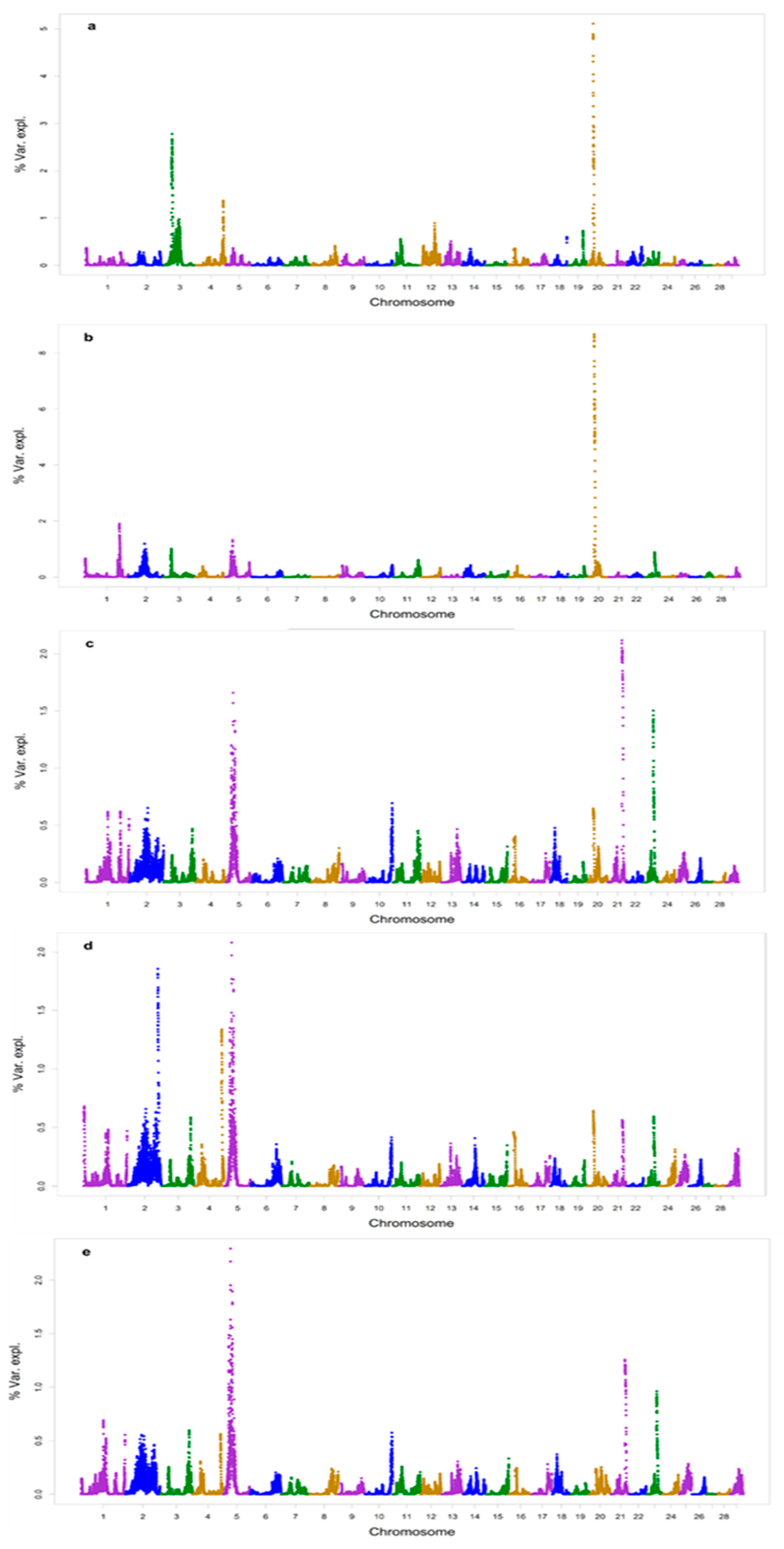
3.4. Genome-Wide Association Study
3.5. Common Genes Between SC and Female Fertility Traits
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eenennaam, A.L.V. Considerations Related to Breed or Biological Type. Vet. Clin. Food Anim. Pract. 2013, 29, 493–516. [Google Scholar] [CrossRef] [PubMed]
- Krupová, Z.; Krupa, E.; Wolfová, M. Economic Weights of Current and New Breeding Objective Traits in Aberdeen Angus. Czech J. Anim. Sci. 2020, 65, 77–85. [Google Scholar] [CrossRef]
- Burns, B.M.; Fordyce, G.; Holroyd, R.G. A Review of Factors That Impact on the Capacity of Beef Cattle Females to Conceive, Maintain a Pregnancy and Wean a Calf—Implications for Reproductive Efficiency in Northern Australia. Anim. Reprod. Sci. 2010, 122, 1–22. [Google Scholar] [CrossRef]
- Summers, A.F.; Rosasco, S.L.; Scholljegerdes, E.J. BEEF SPECIES-RUMINANT NUTRITION CACTUS BEEF SYMPOSIUM: Influence of Management Decisions during Heifer Development on Enhancing Reproductive Success and Cow Longevity1. J. Anim. Sci. 2019, 97, 1407–1414. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.; Hammond, K.; Parnell, P.F.; MacKinnon, M.J.; Sivarajasingam, S. Estimates of Heritability and Repeatability for Reproductive Traits in Australian Beef Cattle. Livest. Prod. Sci. 1990, 25, 15–30. [Google Scholar] [CrossRef]
- Cammack, K.M.; Thomas, M.G.; Enns, R.M. Reproductive Traits and Their Heritabilities in Beef Cattle. Prof. Anim. Sci. 2009, 25, 517–528. [Google Scholar] [CrossRef]
- Johnston, D.J. Genetic Improvement of Reproduction in Beef Cattle. In Proceedings of the 10th World Congress on Genetics Applied to Livestock Production (WCGALP), Vancouver, BC, Canada, 17–22 August 2014; American Society of Animal Science: Champaign, IL, USA, 2014. [Google Scholar]
- Martínez-Velázquez, G.; Gregory, K.E.; Bennett, G.L.; Van Vleck, L.D. Genetic Relationships between Scrotal Circumference and Female Reproductive Traits1. J. Anim. Sci. 2003, 81, 395–401. [Google Scholar] [CrossRef]
- Reding, J.J.; van der Westhuizen, R.R.; Berry, D.P.; van Marle-Köster, E. Understanding the Underlying Genetic Mechanisms for Age at First Calving, Inter-Calving Period and Scrotal Circumference in Bonsmara Cattle. BMC Genom. 2023, 24, 480. [Google Scholar] [CrossRef]
- Jiménez, J.M.; Morales, R.M.; Demyda-Peyrás, S.; Molina, A. Genetic Relationships between Male and Female Reproductive Traits in Retinta Beef Cattle. Livest. Sci. 2025, 291, 105610. [Google Scholar] [CrossRef]
- Vargas, C.A.; Elzo, M.A.; Chase, C.C.; Chenoweth, P.J.; Olson, T.A. Estimation of Genetic Parameters for Scrotal Circumference, Age at Puberty in Heifers, and Hip Height in Brahman Cattle. J. Anim. Sci. 1998, 76, 2536–2541. [Google Scholar] [CrossRef]
- Morris, C.A.; Wilson, J.A.; Bennett, G.L.; Cullen, N.G.; Hickey, S.M.; Hunter, J.C. Genetic Parameters for Growth, Puberty, and Beef Cow Reproductive Traits in a Puberty Selection Experiment. N. Z. J. Agric. Res. 2000, 43, 83–91. [Google Scholar] [CrossRef]
- Terakado, A.P.N.; Boligon, A.A.; Baldi, F.; Silva, J.A.I.V.; Albuquerque, L.G. Genetic Associations between Scrotal Circumference and Female Reproductive Traits in Nelore Cattle1. J. Anim. Sci. 2015, 93, 2706–2713. [Google Scholar] [CrossRef]
- Siddiqui, M.A.R.; Bhattacharjee, J.; Das, Z.C.; Islam, M.M.; Islam, M.A.; Haque, M.A.; Parrish, J.J.; Shamsuddin, M. Crossbred Bull Selection for Bigger Scrotum and Shorter Age at Puberty with Potentials for Better Quality Semen. Reprod. Domest. Anim. Zuchthyg. 2008, 43, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Regatieri, I.C.; Boligon, A.A.; Costa, R.B.; de Souza, F.R.P.; Baldi, F.; Takada, L.; Venturini, G.C.; de Camargo, G.M.F.; Fernandes, G.A.; Tonhati, H.; et al. Association between Single Nucleotide Polymorphisms and Sexual Precocity in Nellore Heifers. Anim. Reprod. Sci. 2017, 177, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Morales, R.; Menéndez-Buxadera, A.; Avilés, C.; Molina, A. Direct and Maternal Genetic Effects for Preweaning Growth in Retinta Cattle Estimated by a Longitudinal Approach throughout the Calving Trajectory of the Cow. J. Anim. Breed. Genet. Z. Tierz. Zucht. 2013, 130, 425–434. [Google Scholar] [CrossRef]
- ARCA. Datos Censales Raza Retinta. Available online: https://servicio.mapa.gob.es/arca/flujos.html?_flowId=datosCensalesRaza-flow&tipoOperacion=CONSULTA&formatoPagina=0&id=50218 (accessed on 1 September 2025).
- Pérez-González, J.; Frantz, A.C.; Torres-Porras, J.; Castillo, L.; Carranza, J. Population Structure, Habitat Features and Genetic Structure of Managed Red Deer Populations. Eur. J. Wildl. Res. 2012, 58, 933–943. [Google Scholar] [CrossRef]
- Rodero-Serrano, E.; Demyda-Peyrás, S.; González-Martinez, A.; Rodero-Franganillo, A.; Moreno-Millán, M. The Rob(1;29) Chromosome Translocation in Endangered Andalusian Cattle Breeds. Livest. Sci. 2013, 158, 32–39. [Google Scholar] [CrossRef]
- Morales, R.; Phocas, F.; Solé, M.; Demyda-Peyrás, S.; Menéndez-Buxadera, A.; Molina, A. Breeding Beef Cattle for an Extended Productive Life: Evaluation of Selection Criteria in the Retinta Breed. Livest. Sci. 2017, 204, 115–121. [Google Scholar] [CrossRef]
- Jiménez, J.M.; Morales, R.M.; Menéndez-Buxadera, A.; Demyda-Peyrás, S.; Laseca, N.; Molina, A. Estimation of the Genetic Components of (Co)Variance and Preliminary Genome-Wide Association Study for Reproductive Efficiency in Retinta Beef Cattle. Animals 2023, 13, 501. [Google Scholar] [CrossRef]
- Valera Córdoba, M.; Jiménez, J.M.; Molina Alcalá, A.; Delgado, C.; Rodero Franganillo, A. Circunferencia escrotal como predictor de la capacidad reproductiva en razas de vacuno de carne autóctono: Curvas de crecimiento en el vacuno retinto. Arch. Zootec. 2000, 49, 229–240. [Google Scholar]
- Legarra, A.; Aguilar, I.; Misztal, I. A Relationship Matrix Including Full Pedigree and Genomic Information. J. Dairy Sci. 2009, 92, 4656–4663. [Google Scholar] [CrossRef]
- Christensen, O.F.; Lund, M.S. Genomic Prediction When Some Animals Are Not Genotyped. Genet. Sel. Evol. 2010, 42, 2. [Google Scholar] [CrossRef]
- Legarra, A.; Christensen, O.F.; Aguilar, I.; Misztal, I. Single Step, a General Approach for Genomic Selection. Livest. Sci. 2014, 166, 54–65. [Google Scholar] [CrossRef]
- Wang, W.; Lu, N.; Xia, Y.; Gu, A.; Wu, B.; Liang, J.; Zhang, W.; Wang, Z.; Su, J.; Wang, X. FAS and FASLG Polymorphisms and Susceptibility to Idiopathic Azoospermia or Severe Oligozoospermia. Reprod. Biomed. Online 2009, 18, 141–147. [Google Scholar] [CrossRef]
- Georges, M.; Charlier, C.; Hayes, B. Harnessing Genomic Information for Livestock Improvement. Nat. Rev. Genet. 2019, 20, 135–156. [Google Scholar] [CrossRef]
- Costa, R.B.; Camargo, G.M.F.; Diaz, I.D.P.S.; Irano, N.; Dias, M.M.; Carvalheiro, R.; Boligon, A.A.; Baldi, F.; Oliveira, H.N.; Tonhati, H.; et al. Genome-Wide Association Study of Reproductive Traits in Nellore Heifers Using Bayesian Inference. Genet. Sel. Evol. GSE 2015, 47, 67. [Google Scholar] [CrossRef] [PubMed]
- Stegemiller, M.R.; Murdoch, G.K.; Rowan, T.N.; Davenport, K.M.; Becker, G.M.; Hall, J.B.; Murdoch, B.M. Genome-Wide Association Analyses of Fertility Traits in Beef Heifers. Genes 2021, 12, 217. [Google Scholar] [CrossRef]
- Dias, M.S.; Pedrosa, V.B.; Rocha da Cruz, V.A.; Silva, M.R.; Batista Pinto, L.F. Genome-Wide Association and Functional Annotation Analysis for the Calving Interval in Nellore Cattle. Theriogenology 2024, 218, 214–222. [Google Scholar] [CrossRef] [PubMed]
- McDaneld, T.G.; Kuehn, L.A.; Thomas, M.G.; Snelling, W.M.; Smith, T.P.L.; Pollak, E.J.; Cole, J.B.; Keele, J.W. Genomewide Association Study of Reproductive Efficiency in Female Cattle. J. Anim. Sci. 2014, 92, 1945–1957. [Google Scholar] [CrossRef] [PubMed]
- Purfield, D.C.; Evans, R.D.; Carthy, T.R.; Berry, D.P. Genomic Regions Associated With Gestation Length Detected Using Whole-Genome Sequence Data Differ Between Dairy and Beef Cattle. Front. Genet. 2019, 10, 1068. [Google Scholar] [CrossRef]
- Irano, N.; de Camargo, G.M.F.; Costa, R.B.; Terakado, A.P.N.; Magalhães, A.F.B.; Silva, R.M.d.O.; Dias, M.M.; Bignardi, A.B.; Baldi, F.; Carvalheiro, R.; et al. Genome-Wide Association Study for Indicator Traits of Sexual Precocity in Nellore Cattle. PLoS ONE 2016, 11, e0159502. [Google Scholar] [CrossRef]
- Casas, E.; Lunstra, D.D.; Stone, R.T. Quantitative Trait Loci for Male Reproductive Traits in Beef Cattle. Anim. Genet. 2004, 35, 451–453. [Google Scholar] [CrossRef]
- Fortes, M.R.S.; Lehnert, S.A.; Bolormaa, S.; Reich, C.; Fordyce, G.; Corbet, N.J.; Whan, V.; Hawken, R.J.; Reverter, A. Finding Genes for Economically Important Traits: Brahman Cattle Puberty. Anim. Prod. Sci. 2012, 52, 143–150. [Google Scholar] [CrossRef]
- Melo, T.P.; Fortes, M.R.S.; Bresolin, T.; Mota, L.F.M.; Albuquerque, L.G.; Carvalheiro, R. Multitrait Meta-Analysis Identified Genomic Regions Associated with Sexual Precocity in Tropical Beef Cattle. J. Anim. Sci. 2018, 96, 4087–4099. [Google Scholar] [CrossRef]
- Morales, R.M.; Calvo-Rubio, G.A.; Ziadi, C.; Vargas-Pérez, M.Á.; Demyda-Peyrás, S.; Molina, A. Weighted Single-Step GWAS Reveals Genomic Regions Associated with Female Fertility in the Spanish Retinta Beef Cattle. Animals 2025, 15, 2665. [Google Scholar] [CrossRef] [PubMed]
- Thermo Fisher Scientific. Axiom CNV Summary Tool User Manual; Thermo Fisher Scientific: Waltham, MA, USA, 2015. [Google Scholar]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Marschner, I.C. Glm2: Fitting Generalized Linear Models with Convergence Problems. R J. 2011, 3, 12. [Google Scholar] [CrossRef]
- R-Core-Team. R: A Language and Environment for Statistical Computing V4.4.2 “Pile of Leaves”. 2024. R Foundation for Statistical Computing: Vienna, Austria, 2024. Available online: https://www.r-project.org/ (accessed on 10 July 2025).
- Aguilar, I.; Misztal, I.; Johnson, D.L.; Legarra, A.; Tsuruta, S.; Lawlor, T.J. Hot Topic: A Unified Approach to Utilize Phenotypic, Full Pedigree, and Genomic Information for Genetic Evaluation of Holstein Final Score1. J. Dairy Sci. 2010, 93, 743–752. [Google Scholar] [CrossRef]
- VanRaden, P.M. Efficient Methods to Compute Genomic Predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef]
- Lourenco, D.; Tsuruta, S.; Aguilar, I.; Masuda, Y.; Bermann, M.; Legarra, A.; Misztal, I. Recent Updates in the BLUPF90 Software Suite. In Proceedings of the 12th World Congress of Genetics Applied to Livestock Production, Rotterdam, The Netherlands, 3–8 July 2022; Wageningen Academic Publishers: Wageningen, The Netherlands, 2022; pp. 1530–1533. [Google Scholar]
- Wang, H.; Misztal, I.; Aguilar, I.; Legarra, A.; Muir, W.M. Genome-Wide Association Mapping Including Phenotypes from Relatives without Genotypes. Genet. Res. 2012, 94, 73–83. [Google Scholar] [CrossRef]
- Aguilar, I.; Misztal, I.; Tsuruta, S.; Legarra, A.; Wang, H. PREGSF90–POSTGSF90: Computational Tools for the Implementation of Single-Step Genomic Selection and Genome-Wide Association with Ungenotyped Individuals in BLUPF90 Programs. In Proceedings of the 10th World Congress of Genetics Applied to Livestock Production (WCGALP), Vancouver, BC, Canada, 17–22 August 2014. [Google Scholar]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- UniProt Consortium. THSD7B—Thrombospondin Type-1 Domain-Containing Protein 7B—Bos Taurus (Bovine). Available online: https://www.uniprot.org/uniprotkb/F1MF39/entry (accessed on 24 July 2025).
- Bonamy, M.; de Iraola, J.J.; Baldo, A.; Prando, A.; Giovambattista, G.; Munilla, S. Early Rather than Late Scrotal Circumference Measurements Better Reflect Female Precocity in Beef Cattle. Livest. Sci. 2018, 218, 79–84. [Google Scholar] [CrossRef]
- Palomares, R.A.; Wolfe, D.F. Factors That Affect The Scrotal Circumference Of The Bull And Its Impact On Herd Reproductive Performance. A Review. Clin. Theriogenology 2011, 3, 115–127. [Google Scholar]
- Chenoweth, P.J.; Hopkins, F.M.; Spitzer, J.C.; Larsen, R.E. Guidelines for Using the Bull Breeding Soundness Evaluation Form. Clin. Theriogenology 2010, 2, 43–50. Available online: https://clinicaltheriogenology.net/index.php/CT/article/view/11373/17883 (accessed on 20 July 2025).
- Federation BIF Guidelines Wiki. Available online: https://guidelines.beefimprovement.org/index.php/Guidelines_for_Uniform_Beef_Improvement_Programs (accessed on 3 September 2025).
- ICAR Section 3; ICAR Guidelines for Beef Cattle Production Recording. The International Committee for Animal Recording (ICAR): Utrecht, The Netherlands, 2018.
- Van Melis, M.H.; Eler, J.P.; Rosa, G.J.M.; Ferraz, J.B.S.; Figueiredo, L.G.G.; Mattos, E.C.; Oliveira, H.N. Additive Genetic Relationships between Scrotal Circumference, Heifer Pregnancy, and Stayability in Nellore Cattle. J. Anim. Sci. 2010, 88, 3809–3813. [Google Scholar] [CrossRef]
- Schmidt, P.; Ferreira, I.A.; Silveira, D.D.; Campos, G.S.; Souza, F.R.P.; Carvalheiro, R.; Boligon, A.A. Reproductive Performance of Cows and Genetic Correlation with Weight Gains and Principal Components of Traits Used in Selection of Nelore Cattle. Livest. Sci. 2019, 229, 77–84. [Google Scholar] [CrossRef]
- Novotná, A.; Brzáková, M.; Birovaš, A.; Veselá, Z. Genetic Evaluation of the Scrotal Circumference of Beef Bulls in the Czech Republic. Czech J. Anim. Sci. 2022, 67, 349–355. [Google Scholar] [CrossRef]
- Gutiérrez, J.P.; Goyache, F. Estimation of Genetic Parameters of Type Traits in Asturiana de Los Valles Beef Cattle Breed. J. Anim. Breed. Genet. 2002, 119, 93–100. [Google Scholar] [CrossRef]
- Giess, L.K.; Boldt, R.J.; Culbertson, M.; Thomas, M.G.; Speidel, S.E.; Enns, R.M. 16 Genetic Parameter Estimates for Age at First Calving in Simmental and Red Angus Heifers. J. Anim. Sci. 2022, 100, 9–10. [Google Scholar] [CrossRef]
- González-Murray, R.A.; Martínez, P.G.; Vigíl, V.; Yazar- Gunes, H.; Sánchez-Castro, M.A.; Enns, R.M.; Speidel, S.E.; Thomas, M.G. Heterosis Effects on Age at First Calving in a Multibreed Beef Cattle Herd in Panama. Transl. Anim. Sci. 2021, 5, S185–S188. [Google Scholar] [CrossRef]
- López-Paredes, J.; Pérez-Cabal, M.A.; Jiménez-Montero, J.A.; Alenda, R. Influence of Age at First Calving in a Continuous Calving Season on Productive, Functional, and Economic Performance in a Blonde d’Aquitaine Beef Population1. J. Anim. Sci. 2018, 96, 4015–4027. [Google Scholar] [CrossRef] [PubMed]
- Brzáková, M.; Čítek, J.; Svitáková, A.; Veselá, Z.; Vostrý, L. Genetic Parameters for Age at First Calving and First Calving Interval of Beef Cattle. Animals 2020, 10, 2122. [Google Scholar] [CrossRef] [PubMed]
- Buzanskas, M.E.; Pires, P.S.; Chud, T.C.S.; Bernardes, P.A.; Rola, L.D.; Savegnago, R.P.; Lôbo, R.B.; Munari, D.P. Parameter Estimates for Reproductive and Carcass Traits in Nelore Beef Cattle. Theriogenology 2017, 92, 204–209. [Google Scholar] [CrossRef]
- Molina Alcalá, A.; Valera Córdoba, M.; Pérez, J.A.; Álvarez, F.; Jiménez, J.M.; Muñoz Vilches, P.; Tapia Román, N.; Cámara, M.C. Parámetros reproductivos en el ganado vacuno de raza retinta explotado en dehesa: Edad al primer parto e intervalo entre partos. Feagas 2002, 22, 63–70. [Google Scholar]
- Smith, B.A.; Brinks, J.S.; Richardson, G.V. Estimation of Genetic Parameters among Reproductive and Growth Traits in Yearling Heifers. J. Anim. Sci. 1989, 67, 2881–2885. [Google Scholar] [CrossRef]
- Berry, D.P.; Evans, R.D. Genetics of Reproductive Performance in Seasonal Calving Beef Cows and Its Association with Performance Traits. J. Anim. Sci. 2014, 92, 1412–1422. [Google Scholar] [CrossRef]
- Lopez, B.I.; Son, J.-H.; Seo, K.; Lim, D. Estimation of Genetic Parameters for Reproductive Traits in Hanwoo (Korean Cattle). Animals 2019, 9, 715. [Google Scholar] [CrossRef]
- Veselá, Z.; Vostrý, L.; Svitáková, A. Genetic Analysis of Female Fertility Traits in Beef Cattle in the Czech Republic. Interbull Bull. 2013. Available online: https://journal.interbull.org/index.php/ib/article/view/1785 (accessed on 1 September 2025).
- Cortes, O.; Carleos, C.; Baro, J.a.; Fernández, M.a.; Villa, J.; Menéndez-Buxadera, A.; Cañon, J. Realized Genetic Parameters of Growth and Reproductive Traits After 25 Years of Selection in the Asturiana de los Valles Beef Cattle Breed. Actas Iberoam. Conserv. Anim. 2015, 5, 78–86. [Google Scholar]
- Koots, K.R.; Gibson, J.P.; Wilton, J.W. Analyses of Published Genetic Parameter Estimates for Beef Production Traits. 2. Phenotypic and Genetic Correlations. Anim. Breed. Abstr. 1994, 62, 825–853. [Google Scholar]
- Ziadi, C.; Muñoz-Mejías, E.; Sánchez, M.; López, M.D.; González-Casquet, O.; Molina, A. Genetic Analysis of Reproductive Efficiency in Spanish Goat Breeds Using a Random Regression Model as a Strategy for Improving Female Fertility. Ital. J. Anim. Sci. 2021, 20, 1681–1688. [Google Scholar] [CrossRef]
- Laseca, N.; Demyda-Peyrás, S.; Valera, M.; Ramón, M.; Escribano, B.; Perdomo-González, D.I.; Molina, A. A Genome-Wide Association Study of Mare Fertility in the Pura Raza Español Horse. Anim. Int. J. Anim. Biosci. 2022, 16, 100476. [Google Scholar] [CrossRef]
- Falconer, D.S. Introduction to Quantitative Genetics, 4th ed.; Pearson Education Limited: London, UK, 1996. [Google Scholar]
- Toelle, V.D.; Robison, O.W. Estimates of Genetic Correlations between Testicular Measurements and Female Reproductive Traits in Cattle. J. Anim. Sci. 1985, 60, 89–100. [Google Scholar] [CrossRef]
- Gama, M.P.M.D.; Paz, C.C.P.D.; Junior, M.L.S.; Ventura, H.T.; Bernardes, P.A.; Faro, L.E. Genetic Associations between Growth Traits and Age at First Calving in Guzera Cattle Using Random Regression Models. Livest. Sci. 2021, 250, 104585. [Google Scholar] [CrossRef]
- Santana, M.L.; Eler, J.P.; Bignardi, A.B.; Ferraz, J.B.S. Two-Trait Random Regression Model to Estimate the Genetic Association of Scrotal Circumference with Female Reproductive Performance in Nelore Cattle. Theriogenology 2015, 83, 1534–1540. [Google Scholar] [CrossRef]
- Johnston, D.J.; Corbet, N.J.; Barwick, S.A.; Wolcott, M.L.; Holroyd, R.G. Genetic Correlations of Young Bull Reproductive Traits and Heifer Puberty Traits with Female Reproductive Performance in Two Tropical Beef Genotypes in Northern Australia. Anim. Prod. Sci. 2013, 54, 74–84. [Google Scholar] [CrossRef]
- Santoso, S.; Herdis, H.; Arifiantini, R.I.; Gunawan, A.; Sumantri, C. Correlation among testosterone concentrations, scrotal circumference, libido, and sperm quantity in pasundan bulls. Jurnal Veteriner 2021, 22, 389–397. [Google Scholar] [CrossRef]
- Siqueira, J.; Oba, E.; Pinho, R.; Guimarães, S.; Neto, T.; Guimarães, J. Sexual Precocity of Nellore Bulls That Are Offspring of Super Precocious, Precocious and Normal Cows in Extensive Farming Conditions. Reprod. Domest. Anim. 2014, 49, 851–857. [Google Scholar] [CrossRef]
- Pérez Osorio, J.; Cardona Álvarez, J.A.; Gómez León, V.E.; Otero Arroyo, R.J. Relação entre o perímetro escrotal e parâmetros da qualidade do sêmen em machos da raça Guzerá, da puberdade até os 36 meses de idade. Rev. Cienc. Agric. 2016, 13, 29–38. [Google Scholar] [CrossRef]
- Wahyudi, I.; Qalfin, M.; Susanti, R.; Widiatningrum, T. Relationship between Scrotal Circumference and Quality of Semen Production in Bulls: A Meta-Analysis Review. J. Sain Peternak. Indones. 2022, 17, 159–169. [Google Scholar] [CrossRef]
- Corbet, N.J.; Burns, B.M.; Johnston, D.J.; Wolcott, M.L.; Corbet, D.H.; Venus, B.K.; Li, Y.; McGowan, M.R.; Holroyd, R.G. Male Traits and Herd Reproductive Capability in Tropical Beef Cattle. 2. Genetic Parameters of Bull Traits. Anim. Prod. Sci. 2012, 53, 101–113. [Google Scholar] [CrossRef]
- Islam, M.R.; Husain, S.S.; Hoque, M.A.; Talukder, M.K.; Rahman, M.S.; Ali, M.Y. Computer Assisted Sperm Analysis of Brahman Crossbred Breeding Bull Semen. Bangladesh J. Anim. Sci. 2017, 46, 1–9. [Google Scholar] [CrossRef]
- Tiezzi, F.; Maltecca, C. Accounting for Trait Architecture in Genomic Predictions of US Holstein Cattle Using a Weighted Realized Relationship Matrix. Genet. Sel. Evol. 2015, 47, 24. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Lourenco, D.; Aguilar, I.; Legarra, A.; Misztal, I. Weighting Strategies for Single-Step Genomic BLUP: An Iterative Approach for Accurate Calculation of GEBV and GWAS. Front. Genet. 2016, 7, 151. [Google Scholar] [CrossRef]
- Martinez-Castillero, M.; Then, C.; Altarriba, J.; Srihi, H.; López-Carbonell, D.; Díaz, C.; Martinez, P.; Hermida, M.; Varona, L. Detection of Genomic Regions with Pleiotropic Effects for Growth and Carcass Quality Traits in the Rubia Gallega Cattle Breed. Animals 2021, 11, 1682. [Google Scholar] [CrossRef]
- Laseca, N.; Molina, A.; Perdomo-González, D.; Ziadi, C.; Azor, P.J.; Valera, M. Exploring the Genetic Landscape of Vitiligo in the Pura Raza Español Horse: A Genomic Perspective. Animals 2024, 14, 2420. [Google Scholar] [CrossRef] [PubMed]
- Guiraldelli, M.F.; Eyster, C.; Wilkerson, J.L.; Dresser, M.E.; Pezza, R.J. Mouse HFM1/Mer3 Is Required for Crossover Formation and Complete Synapsis of Homologous Chromosomes during Meiosis. PLoS Genet. 2013, 9, e1003383. [Google Scholar] [CrossRef]
- Tang, D.; Xu, C.; Geng, H.; Gao, Y.; Cheng, H.; Ni, X.; He, X.; Cao, Y. A Novel Homozygous Mutation in the Meiotic Gene MSH4 Leading to Male Infertility Due to Non-Obstructive Azoospermia. Am. J. Transl. Res. 2020, 12, 8185–8191. [Google Scholar] [PubMed]
- Tang, D.; Lv, M.; Gao, Y.; Cheng, H.; Li, K.; Xu, C.; Geng, H.; Li, G.; Shen, Q.; Wang, C.; et al. Novel Variants in Helicase for Meiosis 1 Lead to Male Infertility Due to Non-Obstructive Azoospermia. Reprod. Biol. Endocrinol. 2021, 19, 129. [Google Scholar] [CrossRef]
- Kadri, N.K.; Harland, C.; Faux, P.; Cambisano, N.; Karim, L.; Coppieters, W.; Fritz, S.; Mullaart, E.; Baurain, D.; Boichard, D.; et al. Coding and Noncoding Variants in HFM1, MLH3, MSH4, MSH5, RNF212, and RNF212B Affect Recombination Rate in Cattle. Genome Res. 2016, 26, 1323–1332. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, W.; Jiang, H.; Wu, B.-L. Primary Ovarian Insufficiency Collaboration Mutations in HFM1 in Recessive Primary Ovarian Insufficiency. N. Engl. J. Med. 2014, 370, 972–974. [Google Scholar] [CrossRef]
- Tang, F.; Gao, Y.; Li, K.; Tang, D.; Hao, Y.; Lv, M.; Wu, H.; Cheng, H.; Fei, J.; Jin, Z.; et al. Novel Deleterious Splicing Variant in HFM1 Causes Gametogenesis Defect and Recurrent Implantation Failure: Concerning the Risk of Chromosomal Abnormalities in Embryos. J. Assist. Reprod. Genet. 2023, 40, 1689–1702. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.; Hong, Z.; Ma, B.; He, X.; Ma, L.; Wang, M.; Zhang, Y. Identification of Compound Heterozygous Variants in MSH4 as a Novel Genetic Cause of Diminished Ovarian Reserve. Reprod. Biol. Endocrinol. RBE 2023, 21, 76. [Google Scholar] [CrossRef]
- Ma, L.; O’Connell, J.R.; VanRaden, P.M.; Shen, B.; Padhi, A.; Sun, C.; Bickhart, D.M.; Cole, J.B.; Null, D.J.; Liu, G.E.; et al. Cattle Sex-Specific Recombination and Genetic Control from a Large Pedigree Analysis. PLoS Genet. 2015, 11, e1005387. [Google Scholar] [CrossRef]
- Oliveira Junior, G.A.; Pinheiro, V.G.; Fonseca, P.A.S.; Costa, C.B.; Pioltine, E.M.; Botigelli, R.C.; Razza, E.M.; Ereno, R.L.; Ferraz, J.B.S.; Seneda, M.M.; et al. Genomic and phenotypic analyses of antral follicle count in Aberdeen Angus cows. Livest. Sci. 2021, 249, 7. [Google Scholar] [CrossRef]
- Llavanera, M.; Delgado-Bermúdez, A.; Fernandez-Fuertes, B.; Recuero, S.; Mateo, Y.; Bonet, S.; Barranco, I.; Yeste, M. GSTM3, but Not IZUMO1, Is a Cryotolerance Marker of Boar Sperm. J. Anim. Sci. Biotechnol. 2019, 10, 61. [Google Scholar] [CrossRef]
- Llavanera, M.; Delgado-Bermúdez, A.; Mateo-Otero, Y.; Padilla, L.; Romeu, X.; Roca, J.; Barranco, I.; Yeste, M. Exploring Seminal Plasma GSTM3 as a Quality and In Vivo Fertility Biomarker in Pigs-Relationship with Sperm Morphology. Antioxidants 2020, 9, 741. [Google Scholar] [CrossRef]
- Dickinson, S.E.; Griffin, B.A.; Elmore, M.F.; Kriese-Anderson, L.; Elmore, J.B.; Dyce, P.W.; Rodning, S.P.; Biase, F.H. Transcriptome Profiles in Peripheral White Blood Cells at the Time of Artificial Insemination Discriminate Beef Heifers with Different Fertility Potential. BMC Genom. 2018, 19, 129. [Google Scholar] [CrossRef]
- Bartocci, A.; Pollard, J.W.; Stanley, E.R. Regulation of Colony-Stimulating Factor 1 during Pregnancy. J. Exp. Med. 1986, 164, 956–961. [Google Scholar] [CrossRef]
- Cohen, P.E.; Chisholm, O.; Arceci, R.J.; Stanley, E.R.; Pollard, J.W. Absence of Colony-Stimulating Factor-1 in Osteopetrotic (Csfmop/Csfmop) Mice Results in Male Fertility Defects. Biol. Reprod. 1996, 55, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Pollard, J.W. Role of Colony-Stimulating Factor-1 in Reproduction and Development. Mol. Reprod. Dev. 1997, 46, 54–60; discussion 60–61. [Google Scholar] [CrossRef]
- Cohen, P.E.; Pollard, J.W. Use of the Osteopetrotic Mouse for Studying Macrophages in the Reproductive Tract. In Proceedings of the Immunobiology of Reproduction; Hunt, J.S., Ed.; Springer: New York, NY, USA, 1994; pp. 104–122. [Google Scholar]
- Shafiei, S.; Tajik, P.; Ghasemzadeh-nava, H.; Movahedin, M.; Talebkhan Garoussi, M.; Qasemi-Panahi, B.; Rahimi Feyli, P. Isolation of Bovine Spermatogonial Cells and Co-Culture with Prepubertal Sertoli Cells in the Presence of Colony Stimulating Factor-1. Iran. J. Vet. Med. 2013, 7, 83–90. [Google Scholar] [CrossRef]
- Lee, R.S.F.; Li, N.; Ledgard, A.M.; Pollard, J.W. Dynamic Regulation of Expression of Colony-Stimulating Factor 1 in the Reproductive Tract of Cattle During the Estrous Cycle and in Pregnancy1. Biol. Reprod. 2003, 69, 518–528. [Google Scholar] [CrossRef][Green Version]
- Zhang, L.; Zhen, J.; Huang, Q.; Liu, H.; Li, W.; Zhang, S.; Min, J.; Li, Y.; Shi, L.; Woods, J.; et al. Mouse Spermatogenesis-Associated Protein 1 (SPATA1), an IFT20 Binding Partner, Is an Acrosomal Protein. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2020, 249, 543–555. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Plessis, C.; Prunier, J.; Martin, H.; Labrecque, R.; Sirard, M.A. DNA Methylation Profiles in Bovine Sperm Are Associated with Daughter Fertility. Epigenetics 2023, 18, 2280889. [Google Scholar] [CrossRef]
- Luo, Z.; Zhu, J.; Fang, Z.; Xu, R.; Wan, R.; He, Y.; Chen, Y.; Chen, S.; Wang, Q.; Liu, Q.; et al. Exercise-Augmented THSD7B Exhibited a Positive Prognostic Implication and Tumor-Suppressed Functionality in Pan-Cancer. Front. Immunol. 2024, 15, 1440226. [Google Scholar] [CrossRef]
- Forutan, M.; Engle, B.N.; Chamberlain, A.J.; Ross, E.M.; Nguyen, L.T.; D’Occhio, M.J.; Snr, A.C.; Kho, E.A.; Fordyce, G.; Speight, S.; et al. Genome-Wide Association and Expression Quantitative Trait Loci in Cattle Reveals Common Genes Regulating Mammalian Fertility. Commun. Biol. 2024, 7, 724. [Google Scholar] [CrossRef]
- Madureira, G.; Mion, B.; Van Winters, B.; Peñagaricano, F.; Li, J.; Ribeiro, E.S. Endometrial Responsiveness to Interferon-Tau and Its Association with Subsequent Reproductive Performance in Dairy Heifers. J. Dairy Sci. 2024, 107, 7371–7391. [Google Scholar] [CrossRef]
- Zhu, Q.; Sun, J.; An, C.; Li, X.; Xu, S.; He, Y.; Zhang, X.; Liu, L.; Hu, K.; Liang, M. Mechanism of LncRNA Gm2044 in Germ Cell Development. Front. Cell Dev. Biol. 2024, 12, 1410914. [Google Scholar] [CrossRef]
- Ma, D.-D.; Wang, D.-H.; Yang, W.-X. Kinesins in Spermatogenesis. Biol. Reprod. 2017, 96, 267–276. [Google Scholar] [CrossRef]
- Fortes, M.R.S.; Porto-Neto, L.R.; Satake, N.; Nguyen, L.T.; Freitas, A.C.; Melo, T.P.; Scalez, D.C.B.; Hayes, B.; Raidan, F.S.S.; Reverter, A.; et al. X Chromosome Variants Are Associated with Male Fertility Traits in Two Bovine Populations. Genet. Sel. Evol. GSE 2020, 52, 46. [Google Scholar] [CrossRef] [PubMed]
| Trait | Mean ± SD | Min | Max | CV (%) |
|---|---|---|---|---|
| SC (cm) | 33.98 ± 2.27 | 25.76 | 42.85 | 6.67 |
| AFC (months) | 35.07 ± 5.18 | 20.15 | 48.99 | 14.77 |
| CI12 (months) | 15.06 ± 4.01 | 9.0 | 24.93 | 26.63 |
| ACI (months) | 14.59 ± 2.50 | 10.99 | 24.93 | 17.16 |
| RE (%) | 75.70 ± 13.55 | 20 | 100 | 17.90 |
| Trait | (SE) | (SE) | (SE) | (SE) |
|---|---|---|---|---|
| SC | 1.82 (0.35) | 1.24 (0.24) | 1.98 (0.26) | 0.36 (0.07) |
| AFC | 4.80 (0.31) | 15.69 (0.36) | 22.31 (0.28) | 0.11 (0.01) |
| CI12 | 2.54 (0.20) | 3.07 (0.14) | 11.15 (0.19) | 0.15 (0.01) |
| ACI | 2.63 (0.15) | 2.70 (0.10) | 8.38 (0.13) | 0.19 (0.01) |
| RE | 50.64 (2.13) | 73.17 (1.81) | 138.26 (1.74) | 0.19 (0.01) |
| Trait | SC | AFC | CI12 | ACI | RE |
|---|---|---|---|---|---|
| SC | - | −0.229 * (0.114) | −0.049 (0.121) | −0.149 (0.110) | 0.031 (0.104) |
| AFC | 0.000 (1.001) | - | 0.099 (0.052) | 0.063 (0.044) | −0.195 * (0.038) |
| CI12 | 0.014 (0.979) | −0.005 (1.008) | - | 0.806 * (0.024) | −0.410 * (0.036) |
| ACI | −0.010 (1.014) | 0.044 (0.936) | 0.582 (0.332) | - | −0.698 * (0.020) |
| RE | 0.009 (0.986) | −0.470 (2.020) | −0.381 (1.755) | −0.685 (3.002) | - |
| ID | Size | Chr | Start | End | Var | Genes |
|---|---|---|---|---|---|---|
| 2_60 | 23 | 2 | 60,087,290 | 61,080,508 | 1.118 | THSD7B |
| 3_33 | 73 | 3 | 33,000,242 | 33,997,602 | 1.818 | LAMTOR5, SLC16A4, RBM15, KCNC4, SLC6A17, UBL4B, ALX3, STRIP1, AHCYL1, CSF1, EPS8L3, GSTM3, GSTM1, GSTM5, AMPD2, GNAT2, GPR61, AMIGO1, CYB561D1, ATXN7L2, SYPL2 |
| 3_40 | 20 | 3 | 40,208,518 | 41,195,205 | 1.114 | COL11A1 |
| 3_43 | 18 | 3 | 43,619,460 | 44,610,516 | 1.594 | PLPPR4, PLPPR5, U6, SNX7 |
| 3_46 | 16 | 3 | 46,086,314 | 46,922,624 | 1.515 | DPYD, PTBP2, U3, U6 |
| 3_47 | 28 | 3 | 47,820,211 | 48,808,060 | 2.512 | RWDD3, TLCD4, ALG14, CNN3, SLC44A3 |
| 3_50 | 11 | 3 | 50,309,627 | 51,172,850 | 2.712 | TMED5, MTF2, DIPK1A, SNORA66, SNORD21, EVI5, 5S_rRNA, GFI1, RPAP2, GLMN, C3H1orf146 |
| 3_51 | 14 | 3 | 51,867,500 | 52,816,409 | 2.564 | CDC7, HFM1, ZNF644, BARHL2 |
| 3_53 | 20 | 3 | 53,000,242 | 53,955,669 | 2.473 | ZNF326, LRRC8D, LRRC8C, LRRC8B, GBP6 |
| 3_54 | 9 | 3 | 54,676,435 | 55,537,894 | 1.112 | KYAT3, GTF2B, PKN2 |
| 3_56 | 18 | 3 | 56,292,899 | 57,283,356 | 2.561 | LMO4, SNORA62, HS2ST1, SELENOF, SH3GLB1 |
| 3_57 | 14 | 3 | 57,811,390 | 58,733,254 | 2.156 | COL24A1, ZNHIT6, CCN1 |
| 3_58 | 23 | 3 | 58,933,414 | 59,926,660 | 4.492 | DDAH1, DNAI3, MCOLN3, MCOLN2, LPAR3, SSX2IP, CTBS, SPATA1, GNG5, RPF1, UOX, DNASE2B, SAMD13, PRKACB |
| 3_60 | 24 | 3 | 60,147,732 | 61,129,959 | 6.332 | TTLL7 |
| 3_61 | 28 | 3 | 61,758,600 | 62,738,616 | 7.530 | ADGRL2 |
| 3_63 | 18 | 3 | 63,429,032 | 64,428,546 | 3.140 | U6 |
| 3_65 | 29 | 3 | 65,163,724 | 66,162,056 | 6.773 | ADGRL4, IFI44, IFI44L, bta-mir-10184 |
| 3_66 | 22 | 3 | 66,198,115 | 67,124,807 | 1.582 | PTGFR, GIPC2, U6, DNAJB4, U6, FUBP1, NEXN, MIGA1, U6, U6, USP33, ZZZ3, AK5 |
| 3_67 | 29 | 3 | 67,421,386 | 68,402,961 | 3.846 | PIGK, ST6GALNAC5, ST6GALNAC3 |
| 3_68 | 24 | 3 | 68,432,145 | 69,421,920 | 1.812 | U1, U6, ASB17, MSH4, RABGGTB, ACADM, SLC44A5 |
| 3_70 | 20 | 3 | 70,167,658 | 71,141,852 | 1.612 | ERICH3, TNNI3K, FPGT, LRRIQ3 |
| 3_74 | 13 | 3 | 74,365,922 | 75,196,565 | 1.238 | CTH, ANKRD13C, SRSF11, LRRC40, LRRC7 |
| 3_76 | 16 | 3 | 76,035,716 | 76,958,680 | 1.050 | DEPDC1, RPE65 |
| Traits | Type | Gene Stable ID | |||
|---|---|---|---|---|---|
| SC, AFC, CI12 | 1.118 | 5.478 | 2.9 | protein_coding | ENSBTAG00000021755 |
| protein_coding | ENSBTAG00000044055 (THSD7B) | ||||
| lncRNA | ENSBTAG00000066098 | ||||
| lncRNA | ENSBTAG00000074523 | ||||
| lncRNA | ENSBTAG00000067793 | ||||
| lncRNA | ENSBTAG00000070030 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ziadi, C.; Morales, R.M.; Vargas-Pérez, M.Á.; Calvo-Rubio, G.A.; Demyda-Peyrás, S.; Molina, A. Multivariate Single-Step GWAS Reveals Pleiotropic Genomic Regions and Candidate Genes Associated with Male Scrotal Circumference and Female Fertility Traits in Retinta Beef Cattle. Vet. Sci. 2025, 12, 977. https://doi.org/10.3390/vetsci12100977
Ziadi C, Morales RM, Vargas-Pérez MÁ, Calvo-Rubio GA, Demyda-Peyrás S, Molina A. Multivariate Single-Step GWAS Reveals Pleiotropic Genomic Regions and Candidate Genes Associated with Male Scrotal Circumference and Female Fertility Traits in Retinta Beef Cattle. Veterinary Sciences. 2025; 12(10):977. https://doi.org/10.3390/vetsci12100977
Chicago/Turabian StyleZiadi, Chiraz, Rosa María Morales, María Ángeles Vargas-Pérez, Gabriel Anaya Calvo-Rubio, Sebastián Demyda-Peyrás, and Antonio Molina. 2025. "Multivariate Single-Step GWAS Reveals Pleiotropic Genomic Regions and Candidate Genes Associated with Male Scrotal Circumference and Female Fertility Traits in Retinta Beef Cattle" Veterinary Sciences 12, no. 10: 977. https://doi.org/10.3390/vetsci12100977
APA StyleZiadi, C., Morales, R. M., Vargas-Pérez, M. Á., Calvo-Rubio, G. A., Demyda-Peyrás, S., & Molina, A. (2025). Multivariate Single-Step GWAS Reveals Pleiotropic Genomic Regions and Candidate Genes Associated with Male Scrotal Circumference and Female Fertility Traits in Retinta Beef Cattle. Veterinary Sciences, 12(10), 977. https://doi.org/10.3390/vetsci12100977






