Combined Genome-Wide Association Study and Haplotype Analysis Identifies Candidate Genes Affecting Growth Traits of Inner Mongolian Cashmere Goats
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
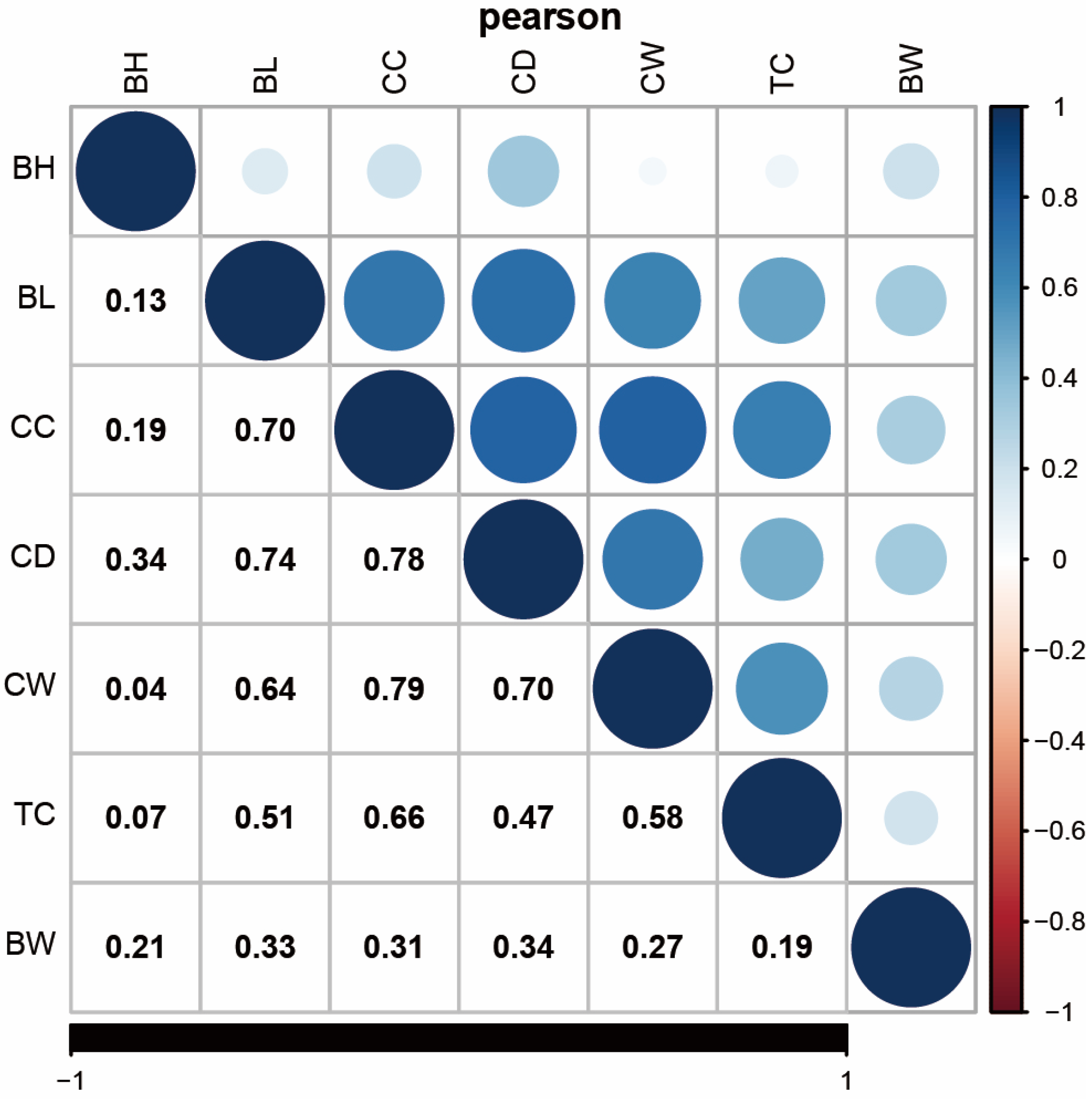
2.1. Phenotypic Statistics and Correlation Analysis
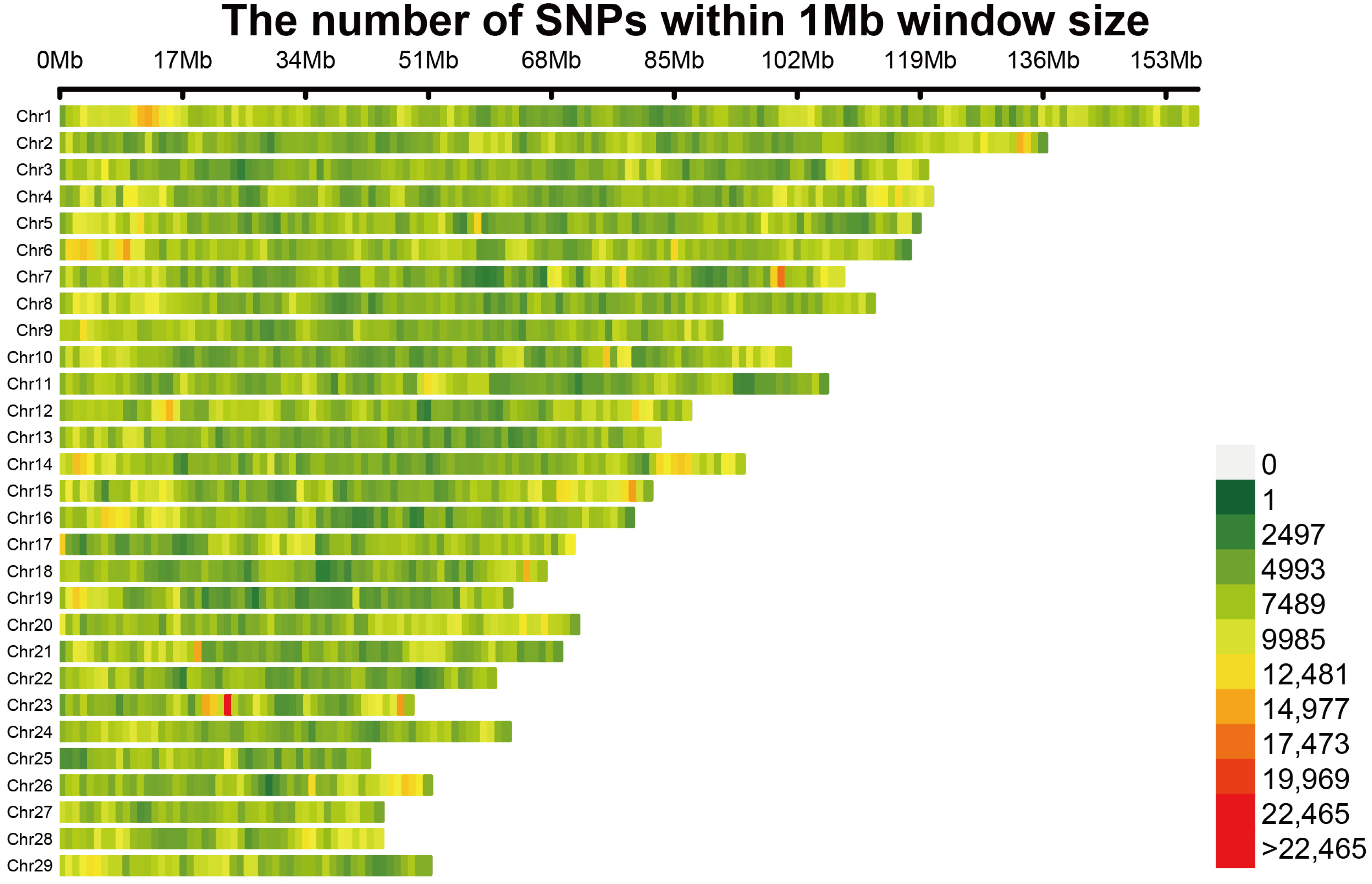
2.2. Genotyping
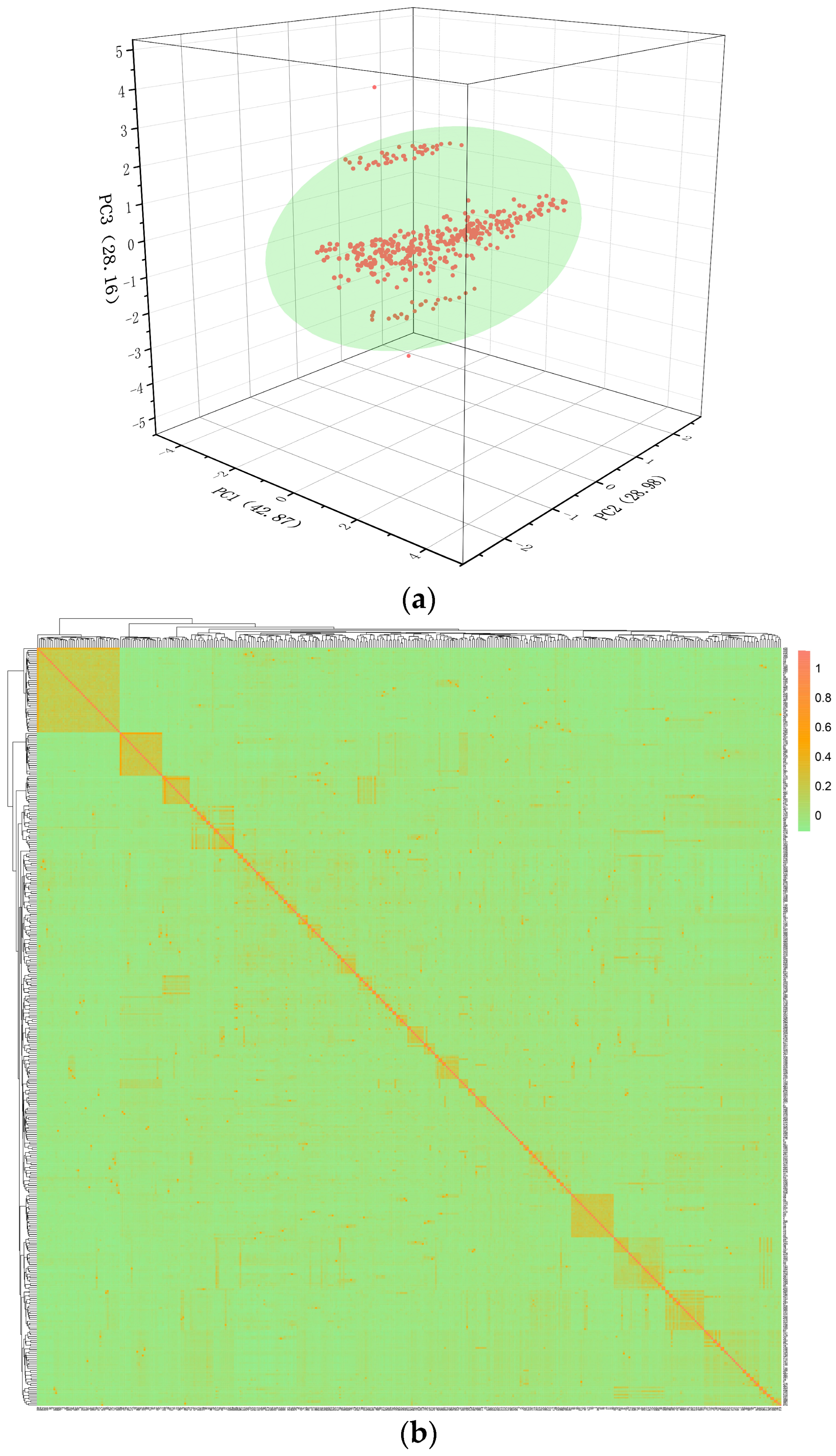
2.3. Quality Control and Population Stratification Assessment
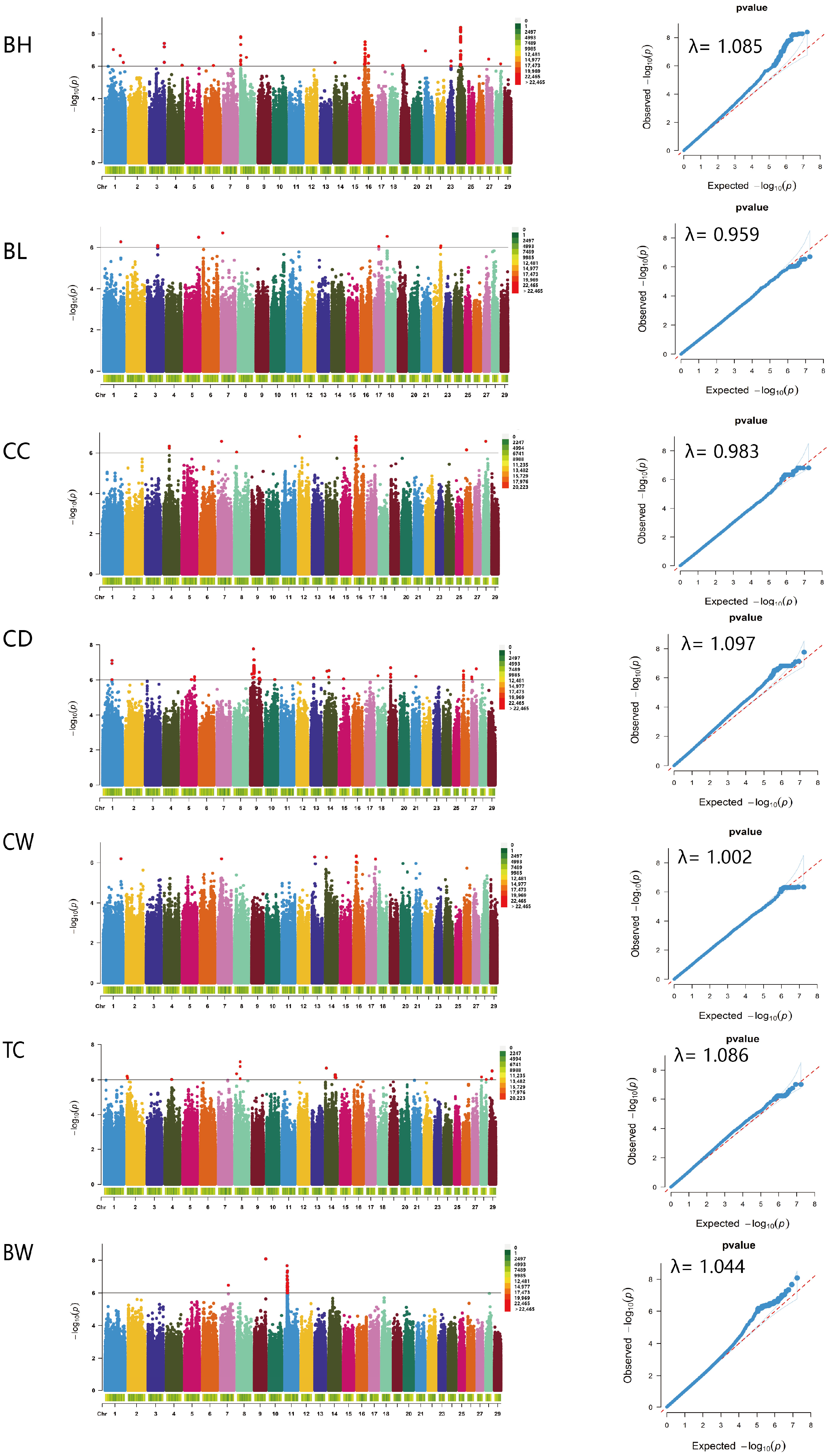
2.4. Genome-Wide Association Analysis
2.5. Gene Annotation and Enrichment Analysis
2.6. Population Genetic Parameter Estimation
2.7. Association Analysis of Haplotype Combination and Growth Traits
3. Results
3.1. Phenotypic Statistics and Correlation Analysis
3.2. Genotyping
3.3. Genetic Relationship Analysis and PCA Analysis
3.4. Genome-Wide Association Analysis
3.5. Population Genetic Parameter Estimation
3.6. Association Analysis of Haplotype Combination and Growth Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BH | Body height |
| BL | Body length |
| CC | Chest circumference |
| CD | Chest depth |
| CW | Chest width |
| TC | Tube circumference |
| BW | Body weight |
| GWAS | Genome-wide associated studies |
| SNPs | Single nucleotide polymorphisms |
| LD | Linkage disequilibrium |
| IMCGs | Inner Mongolian cashmere goats |
| HWE | Hardy–Weinberg equilibrium test |
| Ho | Homozygosity |
| He | Heterozygosity |
| PIC | Polymorphism information content |
| Ne | Effective allele numbers |
| MAF | Minor allele frequency |
| IBS | Identical by state |
References
- Yang, R.; Zhou, D.; Tan, X.; Zhao, Z.; Lv, Y.; Tian, X.; Ren, L.; Wang, Y.; Li, J.; Zhao, Y.; et al. Genome-Wide Association Study of Body Conformation Traits in Tashi Goats (Capra hircus). Animals 2024, 14, 1145. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Cao, X.; Hanif, Q.; Pi, L.; Hu, L.; Huang, Y.; Lan, X.; Lei, C.; Chen, H. Integrating Genome-Wide CNVs Into QTLs and High Confidence GWAScore Regions Identified Positional Candidates for Sheep Economic Traits. Front. Genet. 2020, 11, 569. [Google Scholar] [CrossRef] [PubMed]
- Ncube, K.T.; Dzomba, E.F.; Hadebe, K.; Soma, P.; Frylinck, L.; Muchadeyi, F.C. Carcass Quality Profiles and Associated Genomic Regions of South African Goat Populations Investigated Using Goat SNP50K Genotypes. Animals 2022, 12, 364. [Google Scholar] [CrossRef] [PubMed]
- Al-Mamun, H.A.; Kwan, P.; Clark, S.A.; Ferdosi, M.H.; Tellam, R.; Gondro, C. Genome-Wide Association Study of Body Weight in Australian Merino Sheep Reveals an Orthologous Region on OAR6 to Human and Bovine Genomic Regions Affecting Height and Weight. Genet. Sel. Evol. 2015, 47, 66. [Google Scholar] [CrossRef]
- Zhuang, Z.; Xu, L.; Yang, J.; Gao, H.; Zhang, L.; Gao, X.; Li, J.; Zhu, B. Weighted Single-Step Genome-Wide Association Study for Growth Traits in Chinese Simmental Beef Cattle. Genes 2020, 11, 189. [Google Scholar] [CrossRef]
- Tam, V.; Patel, N.; Turcotte, M.; Bossé, Y.; Paré, G.; Meyre, D. Benefits and Limitations of Genome-Wide Association Studies. Nat. Rev. Genet. 2019, 20, 467–484. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, J.; Zhao, F.; Ren, H.; Xu, L.; Lu, J.; Zhang, S.; Zhang, X.; Wei, C.; Lu, G.; et al. Genome-Wide Association Studies for Growth and Meat Production Traits in Sheep. PLoS ONE 2013, 8, e66569. [Google Scholar] [CrossRef]
- Islam, R.; Liu, X.; Gebreselassie, G.; Abied, A.; Ma, Q.; Ma, Y. Genome-Wide Association Analysis Reveals the Genetic Locus for High Reproduction Trait in Chinese Arbas Cashmere Goat. Genes Genom. 2020, 42, 893–899. [Google Scholar] [CrossRef]
- Abdoli, R.; Mirhoseini, S.Z.; Ghavi Hossein-Zadeh, N.; Zamani, P.; Moradi, M.H.; Ferdosi, M.H.; Gondro, C. Genome-Wide Association Study of First Lambing Age and Lambing Interval in Sheep. Small Rumin. Res. 2019, 178, 43–45. [Google Scholar] [CrossRef]
- Peng, W.-F.; Xu, S.-S.; Ren, X.; Lv, F.-H.; Xie, X.-L.; Zhao, Y.-X.; Zhang, M.; Shen, Z.-Q.; Ren, Y.-L.; Gao, L.; et al. A Genome-Wide Association Study Reveals Candidate Genes for the Supernumerary Nipple Phenotype in Sheep (Ovis aries). Anim. Genet. 2017, 48, 570–579. [Google Scholar] [CrossRef]
- Ghaffarilaleh, V.; Javanmard, A.; Saberivand, A.; Asadzadeh, N.; Masoudi, R.; Barfourooshi, H.J.; Rashidi, A.; Eghbalsaied, S. Variation and Frequency of Supernumerary Teats, Litter Size, Histological Features and the Fibroblast Growth Factor 2 (FGF-2) Gene Expression Pattern in Goats. Theriogenology 2022, 179, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Martin, P.; Palhière, I.; Tosser-Klopp, G.; Rupp, R. Heritability and Genome-Wide Association Mapping for Supernumerary Teats in French Alpine and Saanen Dairy Goats. J. Dairy Sci. 2016, 99, 8891–8900. [Google Scholar] [CrossRef]
- Nazari-Ghadikolaei, A.; Mehrabani-Yeganeh, H.; Miarei-Aashtiani, S.R.; Staiger, E.A.; Rashidi, A.; Huson, H.J. Genome-Wide Association Studies Identify Candidate Genes for Coat Color and Mohair Traits in the Iranian Markhoz Goat. Front. Genet. 2018, 9, 105. [Google Scholar] [CrossRef]
- Wang, F.H.; Zhang, L.; Gong, G.; Yan, X.C.; Zhang, L.T.; Zhang, F.T.; Liu, H.F.; Lv, Q.; Wang, Z.Y.; Wang, R.J.; et al. Genome-Wide Association Study of Fleece Traits in Inner Mongolia Cashmere Goats. Anim. Genet. 2021, 52, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Luo, H.; Huang, X.; Wei, C.; Di, J.; Tian, Y.; Fu, X.; Li, B.; Liu, G.E.; Fang, L.; et al. Integration of a Single-Step Genome-Wide Association Study with a Multi-Tissue Transcriptome Analysis Provides Novel Insights into the Genetic Basis of Wool and Weight Traits in Sheep. Genet. Sel. Evol. 2021, 53, 56. [Google Scholar] [CrossRef]
- Zhu, C.; Li, N.; Cheng, H.; Ma, Y. Genome Wide Association Study for the Identification of Genes Associated with Tail Fat Deposition in Chinese Sheep Breeds. Biol. Open 2021, 10, bio054932. [Google Scholar] [CrossRef]
- Niciura, S.C.M.; Benavides, M.V.; Okino, C.H.; Ibelli, A.M.G.; Minho, A.P.; Esteves, S.N.; Chagas, A.C.d.S. Genome-Wide Association Study for Haemonchus Contortus Resistance in Morada Nova Sheep. Pathogens 2022, 11, 939. [Google Scholar] [CrossRef]
- Schafer, A.J.; Hawkins, J.R. DNA Variation and the Future of Human Genetics. Nat. Biotechnol. 1998, 16, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Weiss, K.M.; Terwilliger, J.D. How Many Diseases Does It Take to Map a Gene with SNPs? Nat. Genet. 2000, 26, 151–157. [Google Scholar] [CrossRef]
- Liu, N.; Zhang, K.; Zhao, H. Haplotype-Association Analysis. Adv. Genet. 2008, 60, 335–405. [Google Scholar]
- Chalbi, S.; Dettori, M.L.; Djemali, M.; Vacca, G.M.; Petretto, E.; Pazzola, M.; Bedhiaf-Romdhani, S. Haplotype Structure of MSTN, IGF1, and BMP2 Genes in Tunisian Goats (Capra hircus) and Their Association with Morphometric Traits. Trop. Anim. Health Prod. 2022, 55, 2. [Google Scholar] [CrossRef] [PubMed]
- Na, R.; Ni, W.; E, G.; Zeng, Y.; Han, Y.; Huang, Y. SNP Screening of the MSTN Gene and Correlation Analysis between Genetic Polymorphisms and Growth Traits in Dazu Black Goat. Anim. Biotechnol. 2021, 32, 558–565. [Google Scholar] [CrossRef]
- Huang, Y.-Z.; Zhan, Z.-Y.; Li, X.-Y.; Wu, S.-R.; Sun, Y.-J.; Xue, J.; Lan, X.-Y.; Lei, C.-Z.; Zhang, C.-L.; Jia, Y.-T.; et al. SNP and Haplotype Analysis Reveal IGF2 Variants Associated with Growth Traits in Chinese Qinchuan Cattle. Mol. Biol. Rep. 2014, 41, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Uemoto, Y.; Kikuchi, T.; Egawa, S.; Kohira, K.; Saito, T.; Sakuma, H.; Miyashita, S.; Arata, S.; Kojima, T.; et al. SNP- and Haplotype-Based Genome-Wide Association Studies for Growth, Carcass, and Meat Quality Traits in a Duroc Multigenerational Population. BMC Genet. 2016, 17, 60. [Google Scholar] [CrossRef]
- Luo, Y.; Xu, Q.; Xue, M.; Wang, Y.; Yang, X.; Chan, S.; Tang, Q.; Wang, F.; Sun, R.; Chao, Z.; et al. Novel Haplotype in the HHEX Gene Promoter Associated with Body Length in Pigs. Genes 2023, 14, 511. [Google Scholar] [CrossRef] [PubMed]
- Onzima, R.B.; Upadhyay, M.R.; Doekes, H.P.; Brito, L.F.; Bosse, M.; Kanis, E.; Groenen, M.A.M.; Crooijmans, R.P.M.A. Genome-Wide Characterization of Selection Signatures and Runs of Homozygosity in Ugandan Goat Breeds. Front. Genet. 2018, 9, 318. [Google Scholar] [CrossRef] [PubMed]
- Gáspárdy, A.; Holly, V.; Zenke, P.; Maróti-Agóts, Á.; Sáfár, L.; Bali Papp, Á.; Kovács, E. The Response of Prion Genic Variation to Selection for Scrapie Resistance in Hungarian Indigenous Sheep Breeds. Acta Vet. Hung. 2018, 66, 562–572. [Google Scholar] [CrossRef]
- Djaout, A.; Chiappini, B.; Gaouar, S.B.S.; Afri-Bouzebda, F.; Conte, M.; Chekkal, F.; El-Bouyahiaoui, R.; Boukhari, R.; Agrimi, U.; Vaccari, G. Biodiversity and Selection for Scrapie Resistance in Sheep: Genetic Polymorphism in Eight Breeds of Algeria. J. Genet. 2018, 97, 453–461. [Google Scholar] [CrossRef]
- Calderón-Chagoya, R.; Vega-Murillo, V.E.; García-Ruiz, A.; Ríos-Utrera, Á.; Martínez-Velázquez, G.; Montaño-Bermúdez, M. Discovering Genomic Regions Associated with Reproductive Traits and Frame Score in Mexican Simmental and Simbrah Cattle Using Individual SNP and Haplotype Markers. Genes 2023, 14, 2004. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce Framework for Analyzing next-Generation DNA Sequencing Data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Tao, L.; He, X.Y.; Pan, L.X.; Wang, J.W.; Gan, S.Q.; Chu, M.X. Genome-Wide Association Study of Body Weight and Conformation Traits in Neonatal Sheep. Anim. Genet. 2020, 51, 336–340. [Google Scholar] [CrossRef]
- Jiang, J.; Cao, Y.; Shan, H.; Wu, J.; Song, X.; Jiang, Y. The GWAS Analysis of Body Size and Population Verification of Related SNPs in Hu Sheep. Front. Genet. 2021, 12, 642552. [Google Scholar] [CrossRef]
- Kijas, J.W.; Hadfield, T.; Naval Sanchez, M.; Cockett, N. Genome-Wide Association Reveals the Locus Responsible for Four-Horned Ruminant. Anim. Genet. 2016, 47, 258–262. [Google Scholar] [CrossRef] [PubMed]
- Greyvenstein, O.F.C.; Reich, C.M.; van Marle-Koster, E.; Riley, D.G.; Hayes, B.J. Polyceraty (Multi-Horns) in Damara Sheep Maps to Ovine Chromosome 2. Anim. Genet. 2016, 47, 263–266. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Zhou, Z.; Pu, Y.; Chen, X.; Ma, Y.; Jiang, L. Mapping the Four-Horned Locus and Testing the Polled Locus in Three Chinese Sheep Breeds. Anim. Genet. 2016, 47, 623–627. [Google Scholar] [CrossRef]
- Ma, X.; Guan, L.; Xuan, J.; Wang, H.; Yuan, Z.; Wu, M.; Liu, R.; Zhu, C.; Wei, C.; Zhao, F.; et al. Effect of Polymorphisms in the CAMKMT Gene on Growth Traits in Ujumqin Sheep. Anim. Genet. 2016, 47, 618–622. [Google Scholar] [CrossRef]
- Valencia, C.P.L.; Franco, L.Á.Á.; Herrera, D.H. Association of Single Nucleotide Polymorphisms in the CAPN, CAST, LEP, GH, and IGF-1 Genes with Growth Parameters and Ultrasound Characteristics of the Longissimus Dorsi Muscle in Colombian Hair Sheep. Trop. Anim. Health Prod. 2022, 54, 82. [Google Scholar] [CrossRef]
- Cheng, J.; Wang, W.; Zhang, D.; Zhang, Y.; Li, X.; Zhao, Y.; Xu, D.; Zhao, L.; Li, W.; Wang, J.; et al. Identification of Polymorphic Loci in OSMR and GHR Genes and Analysis of Their Association with Growth Traits in Sheep. Anim. Biotechnol. 2023, 34, 2546–2553. [Google Scholar] [CrossRef]
- Bayraktar, M.; Shoshin, O. Estimation of the Associations between GH and DGAT1 Genes and Growth Traits by Using Decision Tree in Awassi Sheep. Anim. Biotechnol. 2022, 33, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; He, S.-G.; Li, W.-R.; Luo, L.-Y.; Yan, Z.; Mo, D.-X.; Wan, X.; Lv, F.-H.; Yang, J.; Xu, Y.-X.; et al. Genomic Analyses of Wild Argali, Domestic Sheep, and Their Hybrids Provide Insights into Chromosome Evolution, Phenotypic Variation, and Germplasm Innovation. Genome Res. 2022, 32, 1669–1684. [Google Scholar] [CrossRef]
- NCI-NHGRI Working Group on Replication in Association Studies; Chanock, S.J.; Manolio, T.; Boehnke, M.; Boerwinkle, E.; Hunter, D.J.; Thomas, G.; Hirschhorn, J.N.; Abecasis, G.; Altshuler, D.; et al. Replicating Genotype-Phenotype Associations. Nature 2007, 447, 655–660. [Google Scholar] [PubMed]
- Wu, X.; Fang, M.; Liu, L.; Wang, S.; Liu, J.; Ding, X.; Zhang, S.; Zhang, Q.; Zhang, Y.; Qiao, L.; et al. Genome Wide Association Studies for Body Conformation Traits in the Chinese Holstein Cattle Population. BMC Genom. 2013, 14, 897. [Google Scholar] [CrossRef] [PubMed]
- Easa, A.A.; Selionova, M.; Aibazov, M.; Mamontova, T.; Sermyagin, A.; Belous, A.; Abdelmanova, A.; Deniskova, T.; Zinovieva, N. Identification of Genomic Regions and Candidate Genes Associated with Body Weight and Body Conformation Traits in Karachai Goats. Genes 2022, 13, 1773. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Guo, T.; Zhao, H.; Qiao, G.; Han, M.; Liu, J.; Yuan, C.; Wang, T.; Li, F.; Yue, Y.; et al. Genome-Wide Association Study Using Individual Single-Nucleotide Polymorphisms and Haplotypes for Erythrocyte Traits in Alpine Merino Sheep. Front. Genet. 2020, 11, 848. [Google Scholar] [CrossRef]
- Abdel-Latif, A.; Zuba-Surma, E.K.; Case, J.; Tiwari, S.; Hunt, G.; Ranjan, S.; Vincent, R.J.; Srour, E.F.; Bolli, R.; Dawn, B. TGF-Beta1 Enhances Cardiomyogenic Differentiation of Skeletal Muscle-Derived Adult Primitive Cells. Basic Res. Cardiol. 2008, 103, 514–524. [Google Scholar] [CrossRef]
- Baselga, J.; Rothenberg, M.L.; Tabernero, J.; Seoane, J.; Daly, T.; Cleverly, A.; Berry, B.; Rhoades, S.K.; Ray, C.A.; Fill, J.; et al. TGF-Beta Signalling-Related Markers in Cancer Patients with Bone Metastasis. Biomarkers 2008, 13, 217–236. [Google Scholar] [CrossRef]
- Liu, I.M.; Schilling, S.H.; Knouse, K.A.; Choy, L.; Derynck, R.; Wang, X.-F. TGFbeta-Stimulated Smad1/5 Phosphorylation Requires the ALK5 L45 Loop and Mediates the pro-Migratory TGFbeta Switch. EMBO J. 2009, 28, 88–98. [Google Scholar] [CrossRef]
- Stepień-Wyrobiec, O.; Hrycek, A.; Wyrobiec, G. Transforming Growth Factor Beta (TGF-Beta): Its Structure, Function, and Role in the Pathogenesis of Systemic Lupus Erythematosus. Postep. Hig. I Med. Dosw. (Online) 2008, 62, 688–693. [Google Scholar]
- Warzecha, Z.; Dembiński, A.; Ceranowicz, P.; Dembiński, M.; Kownacki, P.; Konturek, S.J.; Tomaszewska, R.; Stachura, J.; Hładki, W.; Pawlik, W.W. Immunohistochemical Expression of FGF-2, PDGF-A, VEGF and TGF Beta RII in the Pancreas in the Course of Ischemia/Reperfusion-Induced Acute Pancreatitis. J. Physiol. Pharmacol. 2004, 55, 791–810. [Google Scholar]
- Song, K.; Cornelius, S.C.; Danielpour, D. Development and Characterization of DP-153, a Nontumorigenic Prostatic Cell Line That Undergoes Malignant Transformation by Expression of Dominant-Negative Transforming Growth Factor Beta Receptor Type II. Cancer Res. 2003, 63, 4358–4367. [Google Scholar] [PubMed]
- Li, H.; Deeb, N.; Zhou, H.; Mitchell, A.D.; Ashwell, C.M.; Lamont, S.J. Chicken Quantitative Trait Loci for Growth and Body Composition Associated with Transforming Growth Factor-Beta Genes. Poult. Sci. 2003, 82, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Chen, S.; Yang, N. Expression and Methylation of FGF2, TGF-β and Their Downstream Mediators during Different Developmental Stages of Leg Muscles in Chicken. PLoS ONE 2013, 8, e79495. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tang, S.; Ou, J.; Sun, D.; Zhang, Y.; Xu, G.; Zhang, Y. A Novel 62-Bp Indel Mutation in the Promoter Region of Transforming Growth Factor-Beta 2 (TGFB2) Gene Is Associated with Body Weight in Chickens. Anim. Genet. 2011, 42, 108–112. [Google Scholar] [CrossRef]
- de Mello, F.; Streit, D.P.; Sabin, N.; Gabillard, J.-C. Dynamic Expression of Tgf-Β2, Tgf-Β3 and Inhibin βA during Muscle Growth Resumption and Satellite Cell Differentiation in Rainbow Trout (Oncorhynchus mykiss). Gen. Comp. Endocrinol. 2015, 210, 23–29. [Google Scholar] [CrossRef]
- Yin, Y.; Zhang, Y.; Hua, Z.; Wu, A.; Pan, X.; Yang, J.; Wang, X. Muscle Transcriptome Analysis Provides New Insights into the Growth Gap between Fast- and Slow-Growing Sinocyclocheilus Grahami. Front. Genet. 2023, 14, 1217952. [Google Scholar] [CrossRef]
- Darzi Niarami, M.; Masoudi, A.A.; Vaez Torshizi, R. Association of Single Nucleotide Polymorphism of GHSR and TGFB2 Genes with Growth and Body Composition Traits in Sire and Dam Lines of a Broiler Chicken. Anim. Biotechnol. 2014, 25, 13–22. [Google Scholar] [CrossRef]
- Zhu, J.; Chen, C.; Yang, B.; Guo, Y.; Ai, H.; Ren, J.; Peng, Z.; Tu, Z.; Yang, X.; Meng, Q.; et al. A Systems Genetics Study of Swine Illustrates Mechanisms Underlying Human Phenotypic Traits. BMC Genom. 2015, 16, 88. [Google Scholar] [CrossRef]
- Seo, H.-S.; Serra, R. Tgfbr2 Is Required for Development of the Skull Vault. Dev. Biol. 2009, 334, 481–490. [Google Scholar] [CrossRef]
- Wu, Y.; Dey, R.; Han, A.; Jayathilaka, N.; Philips, M.; Ye, J.; Chen, L. Structure of the MADS-Box/MEF2 Domain of MEF2A Bound to DNA and Its Implication for Myocardin Recruitment. J. Mol. Biol. 2010, 397, 520–533. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Zhang, S.; Wu, W. Analysis of MEF2A Mutations in a Chinese Population with Premature Coronary Artery Disease. Genet. Test. Mol. Biomark. 2013, 17, 352–355. [Google Scholar] [CrossRef] [PubMed]
- Juszczuk-Kubiak, E.; Flisikowski, K.; Wicińska, K. Nucleotide Sequence and Variations of the Bovine Myocyte Enhancer Factor 2C (MEF2C) Gene Promoter in Bos Taurus Cattle. Mol. Biol. Rep. 2011, 38, 1269–1276. [Google Scholar] [CrossRef][Green Version]
- Wang, L.; Fan, C.; Topol, S.E.; Topol, E.J.; Wang, Q. Mutation of MEF2A in an Inherited Disorder with Features of Coronary Artery Disease. Science 2003, 302, 1578–1581. [Google Scholar] [CrossRef] [PubMed]
- Postigo, A.A.; Dean, D.C. Differential Expression and Function of Members of the Zfh-1 Family of Zinc Finger/Homeodomain Repressors. Proc. Natl. Acad. Sci. USA 2000, 97, 6391–6396. [Google Scholar] [CrossRef]
- Ahn, H.-J.; Cha, Y.; Moon, S.-H.; Jung, J.-E.; Park, K.-S. Ell3 Enhances Differentiation of Mouse Embryonic Stem Cells by Regulating Epithelial-Mesenchymal Transition and Apoptosis. PLoS ONE 2012, 7, e40293. [Google Scholar] [CrossRef][Green Version]
- Gehrke, L.J.; Upadhyay, M.; Heidrich, K.; Kunz, E.; Klaus-Halla, D.; Weber, F.; Zerbe, H.; Seichter, D.; Graf, A.; Krebs, S.; et al. A de Novo Frameshift Mutation in ZEB2 Causes Polledness, Abnormal Skull Shape, Small Body Stature and Subfertility in Fleckvieh Cattle. Sci. Rep. 2020, 10, 17032. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, X.; Li, F.; La, Y.; Li, G.; Zhang, Y.; Li, X.; Zhao, Y.; Song, Q.; Wang, W. The Association of Polymorphisms in the Ovine PPARGC1B and ZEB2 Genes with Body Weight in Hu Sheep. Anim. Biotechnol. 2022, 33, 90–97. [Google Scholar] [CrossRef]
- Di Filippo, E.S.; Costamagna, D.; Giacomazzi, G.; Cortés-Calabuig, Á.; Stryjewska, A.; Huylebroeck, D.; Fulle, S.; Sampaolesi, M. Zeb2 Regulates Myogenic Differentiation in Pluripotent Stem Cells. Int. J. Mol. Sci. 2020, 21, 2525. [Google Scholar] [CrossRef]
- Tani, S.; Okada, H.; Onodera, S.; Chijimatsu, R.; Seki, M.; Suzuki, Y.; Xin, X.; Rowe, D.W.; Saito, T.; Tanaka, S.; et al. Stem Cell-Based Modeling and Single-Cell Multiomics Reveal Gene-Regulatory Mechanisms Underlying Human Skeletal Development. Cell Rep. 2023, 42, 112276. [Google Scholar] [CrossRef]
- D’Avanzo, N.; Cheng, W.W.L.; Xia, X.; Dong, L.; Savitsky, P.; Nichols, C.G.; Doyle, D.A. Expression and Purification of Recombinant Human Inward Rectifier K+ (KCNJ) Channels in Saccharomyces Cerevisiae. Protein Expr. Purif. 2010, 71, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Utsunomiya, Y.T.; Xu, L.; Hay, E.H.A.; Bickhart, D.M.; Alexandre, P.A.; Rosen, B.D.; Schroeder, S.G.; Carvalheiro, R.; de Rezende Neves, H.H.; et al. Genome-Wide CNV Analysis Reveals Variants Associated with Growth Traits in Bos Indicus. BMC Genom. 2016, 17, 419. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Peng, W.; Cao, X.; Huang, Y.; Lan, X.; Lei, C.; Chen, H. Differential Expression of KCNJ12 Gene and Association Analysis of Its Missense Mutation with Growth Traits in Chinese Cattle. Animals 2019, 9, 273. [Google Scholar] [CrossRef]
- Koska, J.; Stefan, N.; Dubois, S.; Trinidad, C.; Considine, R.V.; Funahashi, T.; Bunt, J.C.; Ravussin, E.; Permana, P.A. mRNA Concentrations of MIF in Subcutaneous Abdominal Adipose Cells Are Associated with Adipocyte Size and Insulin Action. Int. J. Obes. 2009, 33, 842–850. [Google Scholar] [CrossRef]
- Engelman, J.A.; Lisanti, M.P.; Scherer, P.E. Specific Inhibitors of P38 Mitogen-Activated Protein Kinase Block 3T3-L1 Adipogenesis. J. Biol. Chem. 1998, 273, 32111–32120. [Google Scholar] [CrossRef] [PubMed]
- Kyosseva, S.V. Mitogen-Activated Protein Kinase Signaling. Int. Rev. Neurobiol. 2004, 59, 201–220. [Google Scholar]
- Wang, K.; Wu, P.; Chen, D.; Zhou, J.; Yang, X.; Jiang, A.; Xiao, W.; Qiu, X.; Zeng, Y.; Xu, X.; et al. Detecting the Selection Signatures in Chinese Duroc, Landrace, Yorkshire, Liangshan, and Qingyu Pigs. Funct. Integr. Genom. 2021, 21, 655–664. [Google Scholar] [CrossRef]
- Vanvanhossou, S.F.U.; Scheper, C.; Dossa, L.H.; Yin, T.; Brügemann, K.; König, S. A Multi-Breed GWAS for Morphometric Traits in Four Beninese Indigenous Cattle Breeds Reveals Loci Associated with Conformation, Carcass and Adaptive Traits. BMC Genom. 2020, 21, 783. [Google Scholar] [CrossRef]



| Traits | SNP | Chr | Position | Base Mutation | BETA | p Value | r2 (%) | Distance (bp) | Gene |
|---|---|---|---|---|---|---|---|---|---|
| BH | chr14_5226699 | 14 | 5,226,699 | A > G | −1.831 | 5.98 × 10−7 | 2.526 | −56,540 | E2F5 |
| chr21_6110120 | 21 | 6,110,120 | C > A | 1.754 | 1.13 × 10−7 | 2.769 | within | MEF2A | |
| CC | chr4_44148386 | 4 | 44,148,386 | G > A | 2.274 | 4.65 × 10−7 | 2.513 | −98,274 | IGFBP3 |
| chr4_44149271 | 4 | 44,149,271 | A > C | 2.274 | 4.65 × 10−7 | 2.513 | −84,250 | IGFBP3 | |
| chr4_44149291 | 4 | 44,149,291 | G > A | 2.274 | 4.65 × 10−7 | 2.513 | −84,270 | IGFBP3 | |
| chr4_44149517 | 4 | 44,149,517 | G > A | 2.274 | 4.65 × 10−7 | 2.513 | −84,496 | IGFBP3 | |
| chr4_44150099 | 4 | 44,150,099 | C > T | 2.274 | 4.65 × 10−7 | 2.513 | −85,078 | IGFBP3 | |
| chr4_44151090 | 4 | 44,151,090 | G > A | 2.264 | 5.76 × 10−7 | 2.472 | −86,069 | IGFBP3 | |
| chr7_29685358 | 7 | 29,685,358 | A→C | −1.907 | 2.66 × 10−7 | 2.686 | within | MSH3 | |
| chr26_12222407 | 26 | 12,222,407 | G→A | 3.430 | 7.11 × 10−7 | 2.441 | within | BAG3 | |
| chr28_20591571 | 28 | 20,591,571 | A→G | 3.119 | 2.66 × 10−7 | 2.626 | within | CCAR1 | |
| CD | chr26_12637966 | 26 | 12,637,966 | G→T | −1.100 | 3.10 × 10−7 | 2.683 | −7966 | BAG3 |
| chr26_12637982 | 26 | 12,637,982 | G→A | −1.100 | 3.10 × 10−7 | 2.683 | 408,077 | BAG3 | |
| chr26_12640600 | 26 | 12,640,600 | C→T | −0.664 | 8.41 × 10−7 | 2.518 | 410,695 | BAG3 | |
| CW | chr7_29685358A | 7 | 29,685,358 | A > C | −0.897 | 6.46 × 10−7 | 2.547 | within | MSH3 |
| chr16_20378402 | 16 | 20,378,402 | C > T | 1.996 | 5.03 × 10−7 | 2.494 | within | TGFB2 | |
| chr16_20379075 | 16 | 20,379,075 | A > G | 1.996 | 5.03 × 10−7 | 2.494 | within | TGFB2 | |
| chr16_20384805 | 16 | 20,384,805 | C > G | 1.996 | 5.03 × 10−7 | 2.494 | within | TGFB2 | |
| chr16_20385646 | 16 | 20,385,646 | C > T | 1.996 | 5.03 × 10−7 | 2.494 | within | TGFB2 | |
| chr16_20388796 | 16 | 20,388,796 | C > T | 1.996 | 5.03 × 10−7 | 2.494 | within | TGFB2 | |
| chr16_20391015 | 16 | 20,391,015 | C > T | 1.996 | 5.03 × 10−7 | 2.494 | within | TGFB2 | |
| chr16_20392873 | 16 | 20,392,873 | A > G | 1.996 | 5.03 × 10−7 | 2.494 | within | TGFB2 | |
| chr16_20392878 | 16 | 20,392,878 | C > T | 1.996 | 5.03 × 10−7 | 2.494 | within | TGFB2 | |
| chr16_20395972 | 16 | 20,395,972 | A > G | 1.974 | 4.60 × 10−7 | 2.510 | within | TGFB2 | |
| chr16_20397072T | 16 | 20,397,072 | T > C | 1.974 | 4.60 × 10−7 | 2.510 | within | TGFB2 | |
| chr16_20399832 | 16 | 20,399,832 | G > A | 1.996 | 5.03 × 10−7 | 2.494 | within | TGFB2 | |
| chr16_20400420 | 16 | 20,400,420 | G > A | 1.996 | 5.03 × 10−7 | 2.494 | within | TGFB2 | |
| chr16_20533282 | 16 | 20,533,282 | G > A | 2.059 | 9.36 × 10−7 | 2.378 | −93,711 | TGFB2 | |
| BW | chr2_84197799 | 2 | 84,197,799 | G→A | 1.689 | 9.68 × 10−7 | 2.366 | 21,935 | ZEB2 |
| chr2_84199224 | 2 | 84,199,224 | A→C | 1.689 | 9.68 × 10−7 | 2.366 | −23,360 | ZEB2 | |
| chr2_84205973 | 2 | 84,205,973 | G→A | 1.686 | 6.73 × 10−7 | 2.433 | −30,109 | ZEB2 | |
| chr2_84220465 | 2 | 84,220,465 | G→A | 1.686 | 6.73 × 10−7 | 2.433 | −44,601 | ZEB2 | |
| chr2_84226486 | 2 | 84,226,486 | G→A | 1.686 | 6.73 × 10−7 | 2.433 | −50,622 | ZEB2 | |
| chr2_84230899 | 2 | 84,230,899 | C→T | 1.686 | 6.73 × 10−7 | 2.433 | −55,035 | ZEB2 | |
| chr10_88442808 | 10 | 88,442,808 | G→A | −0.999 | 6.22 × 10−7 | 2.547 | 355002, within | HACD3, MEGF11 | |
| chr10_88442873 | 10 | 88,442,873 | A→C | −0.984 | 9.09 × 10−7 | 2.472 | 354937, within | HACD3, MEGF11 | |
| chr17_355945 | 17 | 355,945 | G→A | 2.036 | 1.82 × 10−7 | 2.686 | within | MIF | |
| chr19_34727083 | 19 | 34,727,083 | A→C | 1.690 | 3.379 × 10−8 | 2.994 | 178913, 248058 | MAP2K3, KCNJ12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ao, X.; Rong, Y.; Han, M.; Wang, X.; Xia, Q.; Shang, F.; Liu, Y.; Lv, Q.; Wang, Z.; Su, R.; et al. Combined Genome-Wide Association Study and Haplotype Analysis Identifies Candidate Genes Affecting Growth Traits of Inner Mongolian Cashmere Goats. Vet. Sci. 2024, 11, 428. https://doi.org/10.3390/vetsci11090428
Ao X, Rong Y, Han M, Wang X, Xia Q, Shang F, Liu Y, Lv Q, Wang Z, Su R, et al. Combined Genome-Wide Association Study and Haplotype Analysis Identifies Candidate Genes Affecting Growth Traits of Inner Mongolian Cashmere Goats. Veterinary Sciences. 2024; 11(9):428. https://doi.org/10.3390/vetsci11090428
Chicago/Turabian StyleAo, Xiaofang, Youjun Rong, Mingxuan Han, Xinle Wang, Qincheng Xia, Fangzheng Shang, Yan Liu, Qi Lv, Zhiying Wang, Rui Su, and et al. 2024. "Combined Genome-Wide Association Study and Haplotype Analysis Identifies Candidate Genes Affecting Growth Traits of Inner Mongolian Cashmere Goats" Veterinary Sciences 11, no. 9: 428. https://doi.org/10.3390/vetsci11090428
APA StyleAo, X., Rong, Y., Han, M., Wang, X., Xia, Q., Shang, F., Liu, Y., Lv, Q., Wang, Z., Su, R., Zhang, Y., & Wang, R. (2024). Combined Genome-Wide Association Study and Haplotype Analysis Identifies Candidate Genes Affecting Growth Traits of Inner Mongolian Cashmere Goats. Veterinary Sciences, 11(9), 428. https://doi.org/10.3390/vetsci11090428





