Different Immune Control of Gram-Positive and Gram-Negative Mammary Infections in Dairy Cows
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Samples
2.3. Bacteriological Analysis and Somatic Cell Count
2.4. Bacterial Killing Test
- -
- Live cell control: Bacteria were reacted with 50% BHI in saline for 2 h at 37 °C, centrifuged at 10,400× g for 3 min at 4 °C, resuspended in the same volume of saline, and kept at room temperature for 60 min. Cells were again centrifuged, washed with sterile saline, and resuspended in 1 mL of saline.
- -
- Dead cell control: Bacteria were reacted with BHI/saline for 2 h at 37 °C, centrifuged at 10,400× g for 3 min at 4 °C, resuspended in 70% isopropyl alcohol, and kept at room temperature for 60 min. Cells were again centrifuged, washed with sterile saline, and resuspended in 1 mL of saline.
2.5. NAGase Test
2.6. Statistical Analysis
- Negative: quarters with negative bacteriological analysis and SCC < 100,000;
- Inflamed: quarters with negative bacteriological analysis and SCC between 100,000 and 200,000;
- Latent: quarters with positive bacteriological analysis and SCC < 200,000;
- Subclinical: quarters with SCC > 200,000.
3. Results
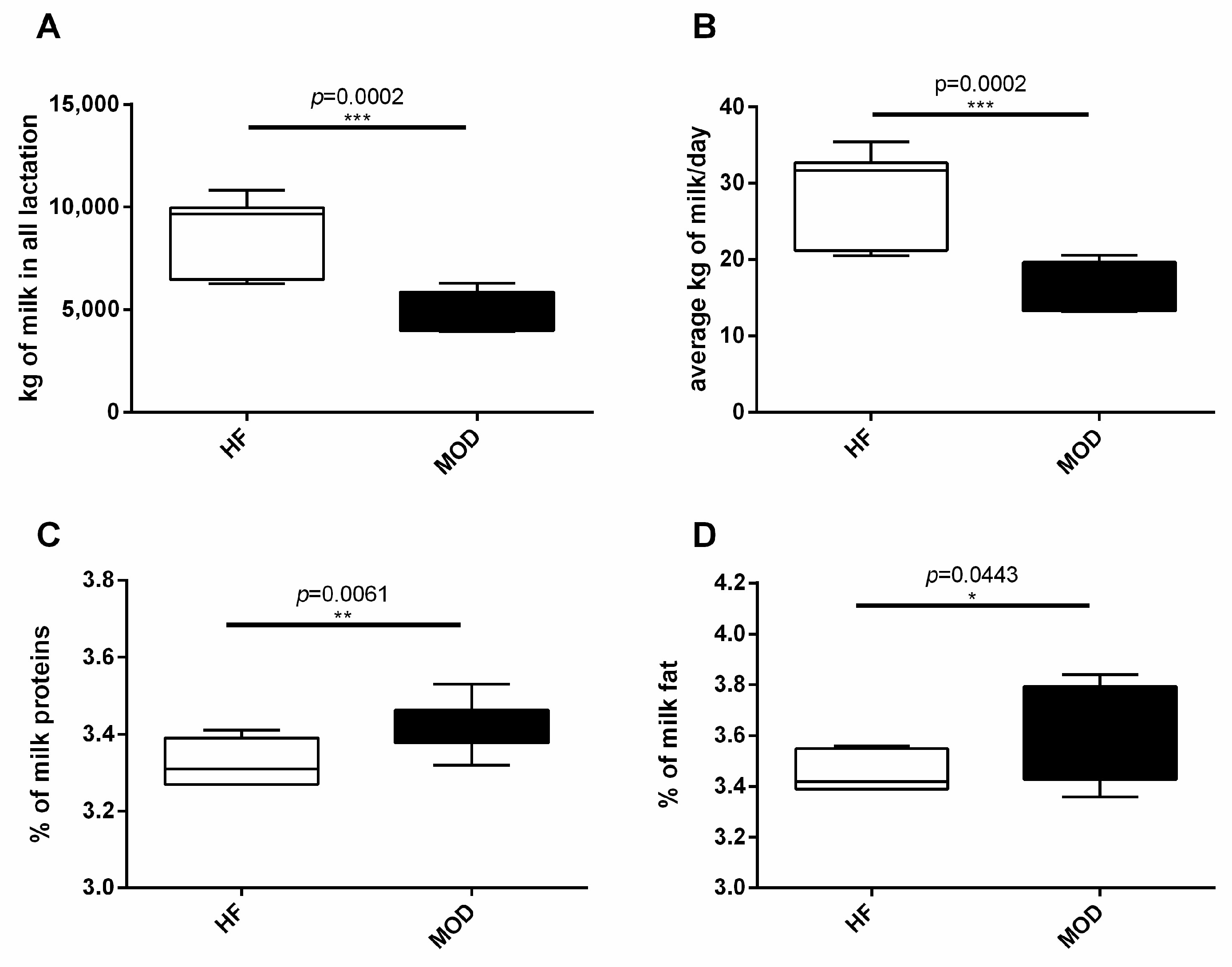
3.1. MOD Group Produced Less but Higher Quality Milk Compared to HF
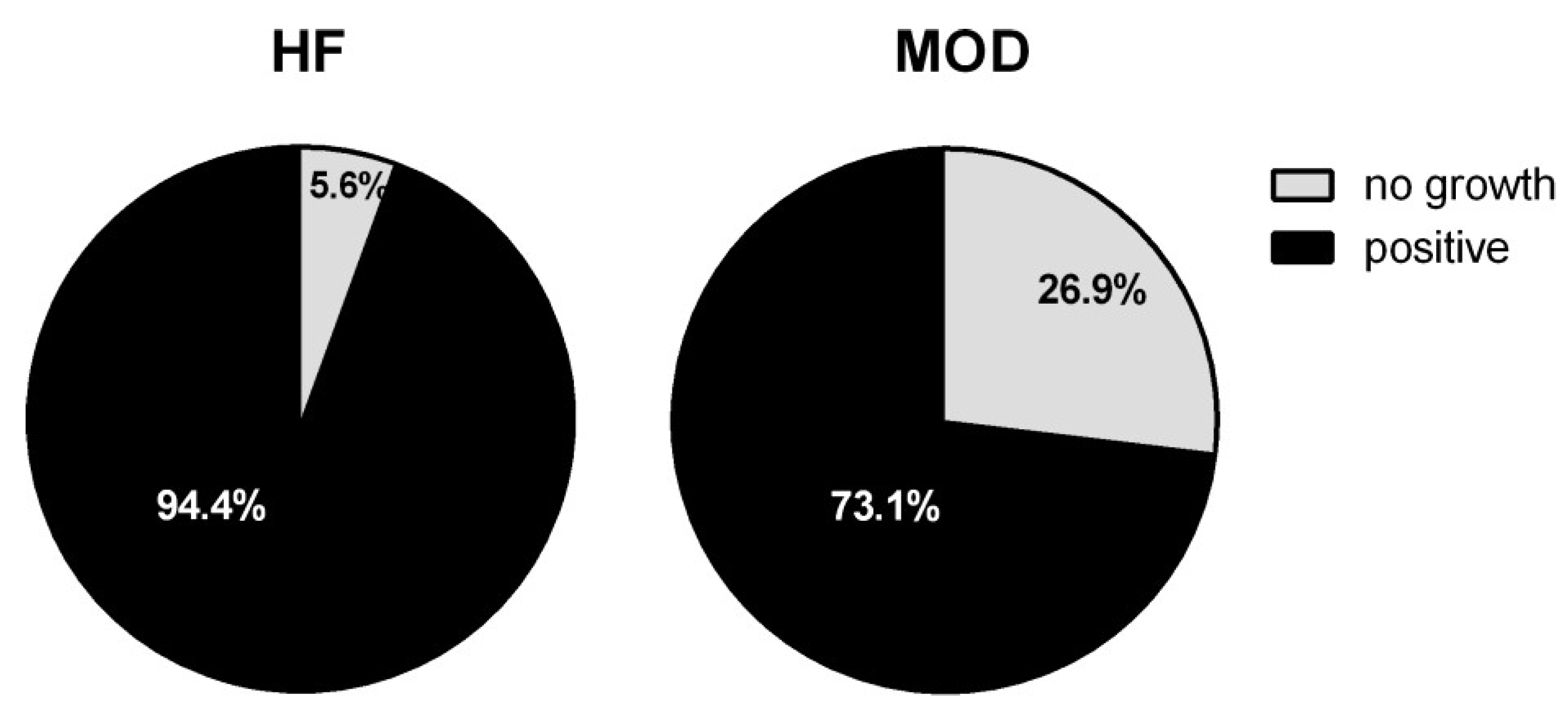
3.2. Bacteriological Analyses
3.3. Similar E. coli Bacterial Killing Activity of the Milk Was Shown by Cows with Allegedly Low and High Susceptibility to Mastitis
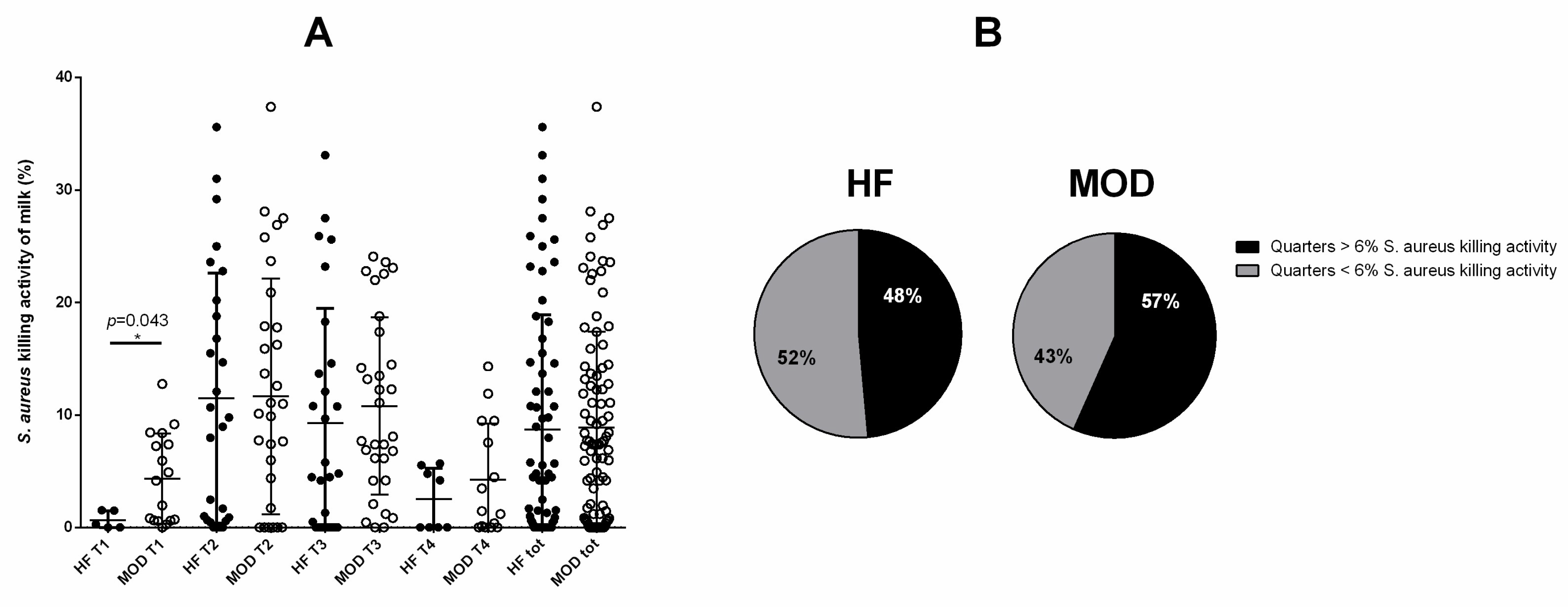
3.4. A Tendency to Greater S. aureus Killing Activity of the Milk Was Shown by Cows with Allegedly Low Susceptibility to Mastitis
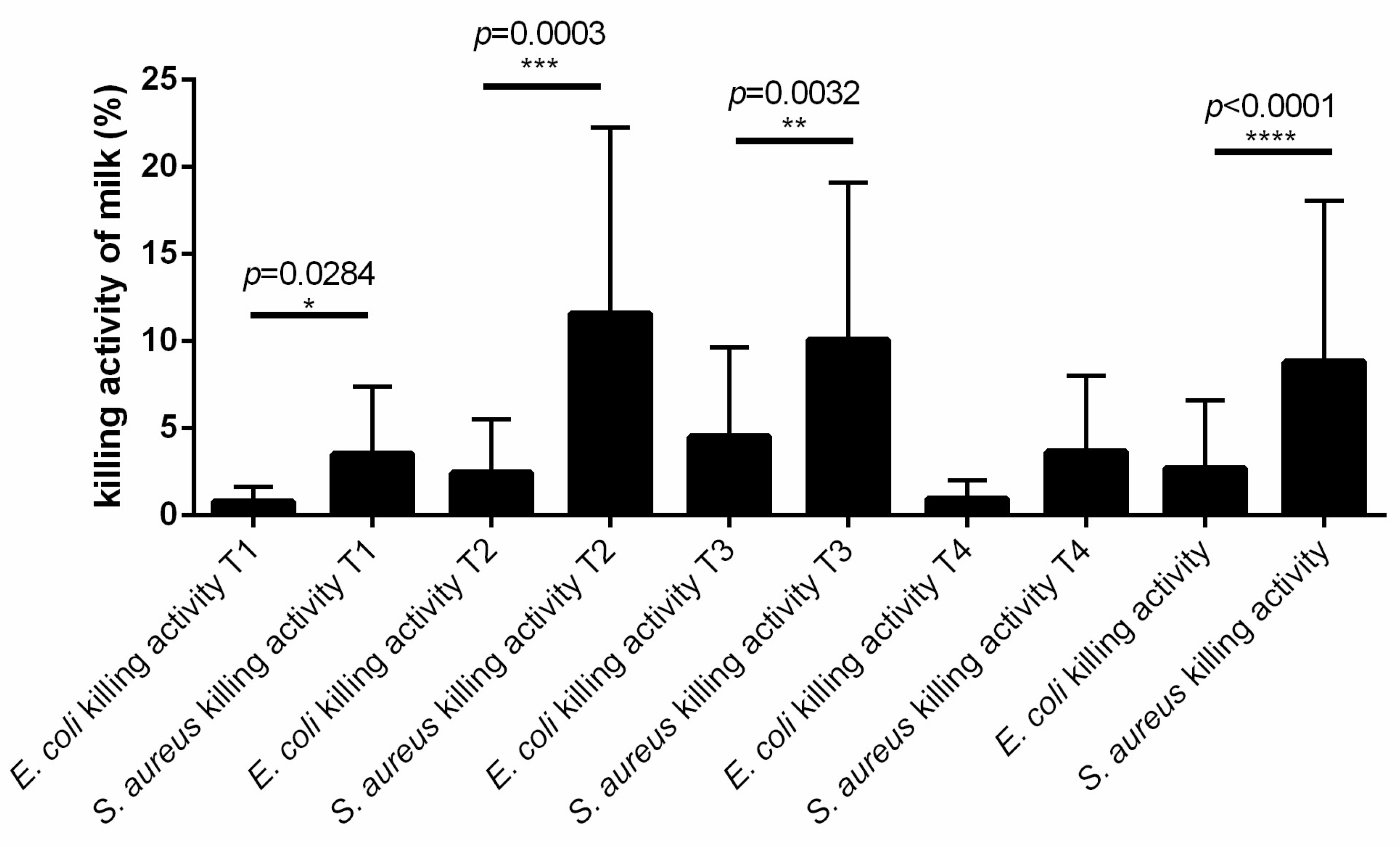
3.5. The Killing Activity of Cow Milk Is More Effective against S. aureus Than E. coli
3.6. NAGase Activity Is Involved in the Control of S. aureus and It Is Stimulated by Milk Microbiota or IMI
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ruegg, P.L. A 100-Year Review: Mastitis Detection, Management, and Prevention. J. Dairy Sci. 2017, 100, 10381–10397. [Google Scholar] [CrossRef] [PubMed]
- Rainard, P.; Riollet, C. Innate Immunity of the Bovine Mammary Gland. Vet. Res. 2006, 37, 369–400. [Google Scholar] [CrossRef] [PubMed]
- Schukken, Y.H.; Günther, J.; Fitzpatrick, J.; Fontaine, M.C.; Goetze, L.; Holst, O.; Leigh, J.; Petzl, W.; Schuberth, H.J.; Sipka, A.; et al. Host-Response Patterns of Intramammary Infections in Dairy Cows. Veter-Immunol. Immunopathol. 2011, 144, 270–289. [Google Scholar] [CrossRef] [PubMed]
- De Vliegher, S.; Fox, L.K.; Piepers, S.; McDougall, S.; Barkema, H.W. Invited Review: Mastitis in Dairy Heifers: Nature of the Disease, Potential Impact, Prevention, and Control. J. Dairy Sci. 2012, 95, 1025–1040. [Google Scholar] [CrossRef] [PubMed]
- Jamali, H.; Barkema, H.W.; Jacques, M.; Lavallée-Bourget, E.M.; Malouin, F.; Saini, V.; Stryhn, H.; Dufour, S. Invited Review: Incidence, Risk Factors, and Effects of Clinical Mastitis Recurrence in Dairy Cows. J. Dairy Sci. 2018, 101, 4729–4746. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen Recognition and Innate Immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [PubMed]
- Turin, L.; Riva, F. Toll-Like Receptor Family in Domestic Animal Species. Crit. Rev. Immunol. 2008, 28, 513–538. [Google Scholar] [CrossRef] [PubMed]
- Shuster, D.E.; Lee, E.K.; Kehrli, M.E. Bacterial Growth, Inflammatory Cytokine Production, and Neutrophil Re-cruitment during Coliform Mastitis in Cows within Ten Days after Calving, Compared with Cows at Midlacta-tion. Am. J. Vet. Res. 1996, 57, 1569–1575. [Google Scholar] [CrossRef]
- Brightbill, H.D.; Modlin, R.L. Toll-like Receptors: Molecular Mechanisms of the Mammalian Immune Response. Immunology 2000, 101, 1–10. [Google Scholar] [CrossRef]
- Paape, M.J.; Bannerman, D.D.; Zhao, X.; Lee, J.W. The Bovine Neutrophil: Structure and Function in Blood and Milk. Veter-Res. 2003, 34, 597–627. [Google Scholar]
- Sordillo, L.M.; Streicher, K.L. Mammary Gland Immunity and Mastitis Susceptibility. J. Mammary Gland. Biol. Neoplasia 2002, 7, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Wellnitz, O.; Bruckmaier, R.M. The Innate Immune Response of the Bovine Mammary Gland to Bacterial Infec-tion. Vet. J. 2012, 192, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Rainard, P.; Cunha, P.; Martins, R.P.; Gilbert, F.B.; Germon, P.; Foucras, G. Type 3 Immunity: A Perspective for the Defense of the Mammary Gland against Infections. Veter-Res. 2020, 51, 129. [Google Scholar] [CrossRef] [PubMed]
- Leemans, J.C.; Heikens, M.; Van Kessel, K.P.M.; Florquin, S.; Van der Poll, T. Lipoteichoic Acid and Peptidoglycan from Staphylococcus aureus Synergistically Induce Neutrophil Influx into the Lungs of Mice. Clin. Diagn. Lab. Immunol. 2003, 10, 950. [Google Scholar] [CrossRef]
- Rainard, P.; Gilbert, F.B.; Germon, P. Immune Defenses of the Mammary Gland Epithelium of Dairy Ruminants. Front. Immunol. 2022, 13, 1031785. [Google Scholar] [CrossRef]
- Giagu, A.; Penati, M.; Traini, S.; Dore, S.; Addis, M.F. Milk Proteins as Mastitis Markers in Dairy Ruminants—A Systematic Review. Vet. Res. Commun. 2022, 46, 329. [Google Scholar] [CrossRef]
- Shimazaki, K.I.; Kawai, K. Advances in Lactoferrin Research Concerning Bovine Mastitis. Biochem. Cell Biol. 2017, 95, 69–75. [Google Scholar] [CrossRef]
- Åkerstedt, M.; Forsbck, L.; Larsen, T.; Svennersten-Sjaunja, K. Natural Variation in Biomarkers Indicating Mas-titis in Healthy Cows. J. Dairy Res. 2011, 78, 88–96. [Google Scholar] [CrossRef]
- Curone, G.; Filipe, J.; Cremonesi, P.; Trevisi, E.; Amadori, M.; Pollera, C.; Castiglioni, B.; Turin, L.; Tedde, V.; Vigo, D.; et al. What We Have Lost: Mastitis Resistance in Holstein Friesians and in a Local Cattle Breed. Res. Vet. Sci. 2018, 116, 88–98. [Google Scholar] [CrossRef]
- Cremonesi, P.; Ceccarani, C.; Curone, G.; Severgnini, M.; Pollera, C.; Bronzo, V.; Riva, F.; Addis, M.F.; Filipe, J.; Amadori, M.; et al. Milk Microbiome Diversity and Bacterial Group Prevalence in a Comparison between Healthy Holstein Friesian and Rendena Cows. PLoS ONE 2018, 13, e0205054. [Google Scholar] [CrossRef]
- Petrera, F.; Catillo, G.; Napolitano, F.; Malacarne, M.; Franceschi, P.; Summer, A.; Abeni, F. New Insights into the Quality Characteristics of Milk from Modenese Breed Compared with Italian Friesian. Ital. J. Anim. Sci. 2016, 15, 559–567. [Google Scholar] [CrossRef][Green Version]
- Nickerson, S.C.; Akers, R.M. Mammary Gland: Anatomy. Encycl. Dairy. Sci. Second. Ed. 2011, 328–337. [Google Scholar] [CrossRef]
- Sullivan, G.M.; Feinn, R. Using Effect Size-or Why the P Value Is Not Enough. J. Grad. Med. Educ. 2012, 4, 279–282. [Google Scholar] [CrossRef] [PubMed]
- Adkins, P.R.; Middleton, J.R.; Fox, L.K.; Pighetti, K.; Petersson-Wolfe, C. Laboratory Handbook on Bovine Mastitis; National Mastitis Council, Inc.: New Prague, MN, USA, 2017. [Google Scholar]
- Oikonomou, G.; Addis, M.F.; Chassard, C.; Nader-Macias, M.E.F.; Grant, I.; Delbès, C.; Bogni, C.I.; Le Loir, Y.; Even, S. Milk Microbiota: What Are We Exactly Talking About? Front. Microbiol. 2020, 11, 497338. [Google Scholar] [CrossRef] [PubMed]
- Kitchen, B.J.; Middleton, G.; Salmon, M. Bovine Milk N-Acetyl-Beta-D-Glucosaminidase and Its Significance in the Detection of Abnormal Udder Secretions. J. Dairy. Res. 1978, 45, 15–20. [Google Scholar] [CrossRef]
- Bertelloni, F.; Cilia, G.; Turchi, B.; Pinzauti, P.; Cerri, D.; Fratini, F. Epidemiology of Leptospirosis in North- Central Italy: Fifteen Years of Serological Data (2002–2016). Comp. Immunol. Microbiol. Infect. Dis. 2019, 65, 14–22. [Google Scholar] [CrossRef]
- Zecconi, A.; Piccinini, R.; Fiorina, S.; Cabrini, L.; Daprà, V.; Amadori, M. Evaluation of Interleukin-2 Treatment for Prevention of Intramammary Infections in Cows after Calving. Comp. Immunol. Microbiol. Infect. Dis. 2009, 32, 439–451. [Google Scholar] [CrossRef] [PubMed]
- Bannerman, D.D. Pathogen-Dependent Induction of Cytokines and Other Soluble Inflammatory Mediators dur-ing Intramammary Infection of Dairy Cows. J. Anim. Sci. 2009, 87, 10–25. [Google Scholar] [CrossRef]
- Petzl, W.; Zerbe, H.; Günther, J.; Yang, W.; Seyfert, H.M.; Nürnberg, G.; Schuberth, H.J. Escherichia Coli, but Not Staphylococcus aureus Triggers an Early Increased Expression of Factors Contributing to the Innate Immune Defense in the Udder of the Cow. Vet. Res. 2008, 39, 18–23. [Google Scholar] [CrossRef]
- Yang, Z.; Qiu, B.; Cheng, D.; Zhao, N.; Liu, Y.; Li, M.; Liu, Q. Virulent Staphylococcus aureus Colonizes Pediatric Nares by Resisting Killing of Human Antimicrobial Peptides. Int. J. Med. Microbiol. 2022, 312, 151550. [Google Scholar] [CrossRef]
- Metzger, S.A.; Hernandez, L.L.; Suen, G. Understanding the Milk Microbiota. Vet. Clin. N. Am. Food Anim. Pract. 2018, 34, 427–438. [Google Scholar] [CrossRef] [PubMed]
- Darbandi, A.; Asadi, A.; Mahdizade Ari, M.; Ohadi, E.; Talebi, M.; Halaj Zadeh, M.; Darb Emamie, A.; Ghanavati, R.; Kakanj, M. Bacteriocins: Properties and Potential Use as Antimicrobials. J. Clin. Lab. Anal. 2022, 36, e24093. [Google Scholar] [CrossRef] [PubMed]
- Ceotto-Vigoder, H.; Marques, S.L.S.; Santos, I.N.S.; Alves, M.D.B.; Barrias, E.S.; Potter, A.; Alviano, D.S.; Bastos, M.C.F. Nisin and Lysostaphin Activity against Preformed Biofilm of Staphylococcus aureus Involved in Bovine Mastitis. J. Appl. Microbiol. 2016, 121, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Rainard, P. The Complement in Milk and Defense of the Bovine Mammary Gland against Infections. Veter- Res. 2003, 34, 647–670. [Google Scholar] [CrossRef] [PubMed]
- Nagasawa, Y.; Kiku, Y.; Sugawara, K.; Hirose, A.; Kai, C.; Kitano, N.; Takahashi, T.; Nochi, T.; Aso, H.; Sawada, S.I.; et al. Staphylococcus aureus-Specific IgA Antibody in Milk Suppresses the Multiplication of S. Aureus in Infected Bovine Udder. BMC Veter. Res. 2019, 15, 286. [Google Scholar] [CrossRef] [PubMed]
- Mkaddem, S.B.; Christou, I.; Rossato, E.; Berthelot, L.; Lehuen, A.; Monteiro, R.C. IgA, IgA Receptors, and Their Anti-Inflammatory Properties. Curr. Top. Microbiol. Immunol. 2014, 382, 221–235. [Google Scholar] [CrossRef] [PubMed]
- Rani, R.S.; Pandita, S.; Ashutosh, M. Milk NAGase Activity as an Indicator of Subclinical and Clinical Mastitis in Sahiwal Cows. Int. J. Curr. Microbiol. Appl. Sci. 2020, 9, 4189–4194. [Google Scholar] [CrossRef]
- Chaneton, L.; Tirante, L.; Maito, J.; Chaves, J.; Bussmann, L.E. Relationship between Milk Lactoferrin and Etio-logical Agent in the Mastitic Bovine Mammary Gland. J. Dairy Sci. 2008, 91, 1865–1873. [Google Scholar] [CrossRef] [PubMed]
- Ploegaert, T.C.W.; Tijhaar, E.; Lam, T.J.G.M.; Taverne-Thiele, A.; van der Poel, J.J.; van Arendonk, J.A.M.; Savelkoul, H.F.J.; Parmentier, H.K. Natural Antibodies in Bovine Milk and Blood Plasma: Variability among Cows, Repeatability within Cows, and Relation between Milk and Plasma Titers. Vet. Immunol. Immunopathol. 2011, 144, 88–94. [Google Scholar] [CrossRef]
- Nakanjako, D.; Otiti-Sengeri, J.; Ssewanyana, I.; Nabatanzi, R.; Bayigga, L.; Kirimunda, S.; Joloba, M.; Manabe, Y.C.; Kambugu, A.; Colebunders, R.; et al. CD4 T-Cell Activation and Reduced Regulatory T-Cell Populations Are Associated with Early Development of Cataracts among HIV-Infected Adults in Uganda. Immunol. Lett. 2014, 161, 44–49. [Google Scholar] [CrossRef]
- Kathariya, R.; Jain, H.; Gujar, D.; Singh, A.; Ajwani, H.; Mandhyan, D. Pentraxins as Key Disease Markers for Periodontal Diagnosis. Dis. Markers 2013, 34, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Parente, R.; Sobacchi, C.; Bottazzi, B.; Mantovani, A.; Grčevic, D.; Inforzato, A. The Long Pentraxin PTX3 in Bone Homeostasis and Pathology. Front. Immunol. 2019, 10, 2628. [Google Scholar] [CrossRef] [PubMed]
- Parente, R.; Doni, A.; Bottazzi, B.; Garlanda, C.; Inforzato, A. The Complement System in Aspergillus Fumigatus Infections and Its Crosstalk with Pentraxins. FEBS Lett. 2020, 594, 2480–2501. [Google Scholar] [CrossRef] [PubMed]
- Huttunen, R.; Hurme, M.; Aittoniemi, J.; Huhtala, H.; Vuento, R.; Laine, J.; Jylhävä, J.; Syrjänen, J. High Plasma Level of Long Pentraxin 3 (PTX3) Is Associated with Fatal Disease in Bacteremic Patients: A Prospective Cohort Study. PLoS ONE 2011, 6, e17653. [Google Scholar] [CrossRef] [PubMed]
- Rainard, P.; Gitton, C.; Chaumeil, T.; Fassier, T.; Huau, C.; Riou, M.; Tosser-Klopp, G.; Krupova, Z.; Chaize, A.; Gilbert, F.B.; et al. Host Factors Determine the Evolution of Infection with Staphylococcus aureus to Gangrenous Mastitis in Goats. Vet. Res. 2018, 49, 1–17. [Google Scholar] [CrossRef]
- He, Y.; Song, M.; Zhang, Y.; Li, X.; Song, J.; Zhang, Y.; Yu, Y. Whole-Genome Regulation Analysis of Histone H3 Lysin 27 Trimethylation in Subclinical Mastitis Cows Infected by Staphylococcus aureus. BMC Genom. 2016, 17, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lutzow, Y.C.S.; Donaldson, L.; Gray, C.P.; Vuocolo, T.; Pearson, R.D.; Reverter, A.; Byrne, K.A.; Sheehy, P.A.; Windon, R.; Tellam, R.L. Identification of Immune Genes and Proteins Involved in the Response of Bovine Mam-mary Tissue to Staphylococcus aureus Infection. BMC Vet. Res. 2008, 4, 18. [Google Scholar] [CrossRef] [PubMed]
- Filipe, J.; Curone, G.; Bronzo, V.; Pisoni, G.; Cremonesi, P.; Pollera, C.; Turin, L.; Vigo, D.; Roccabianca, P.; Ca-niatti, M.; et al. Pentraxin 3 Is Up-Regulated in Epithelial Mammary Cells during Staphylococcus aureus Intra-Mammary Infection in Goat. Comp. Immunol. Microbiol. Infect. Dis. 2018, 59, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.L.; Li, Z.X.; Wang, L.J.; He, H.; Yang, J.; Chen, L.; Niu, F.B.; Liu, Y.; Guo, J.Z.; Liu, X.L. Polymorphism in PGLYRP-1 Gene by PCR-RFLP and Its Association with Somatic Cell Score in Chinese Holstein. Res. Vet. Sci. 2013, 95, 508–514. [Google Scholar] [CrossRef]
- Mudaliar, M.; Tassi, R.; Thomas, F.C.; McNeilly, T.N.; Weidt, S.K.; McLaughlin, M.; Wilson, D.; Burchmore, R.; Herzyk, P.; Eckersall, P.D.; et al. Mastitomics, the Integrated Omics of Bovine Milk in an Experimental Model of: Streptococcus Uberis Mastitis: 2. Label-Free Relative Quantitative Proteomics. Mol. Biosyst. 2016, 12, 2748–2761. [Google Scholar] [CrossRef]
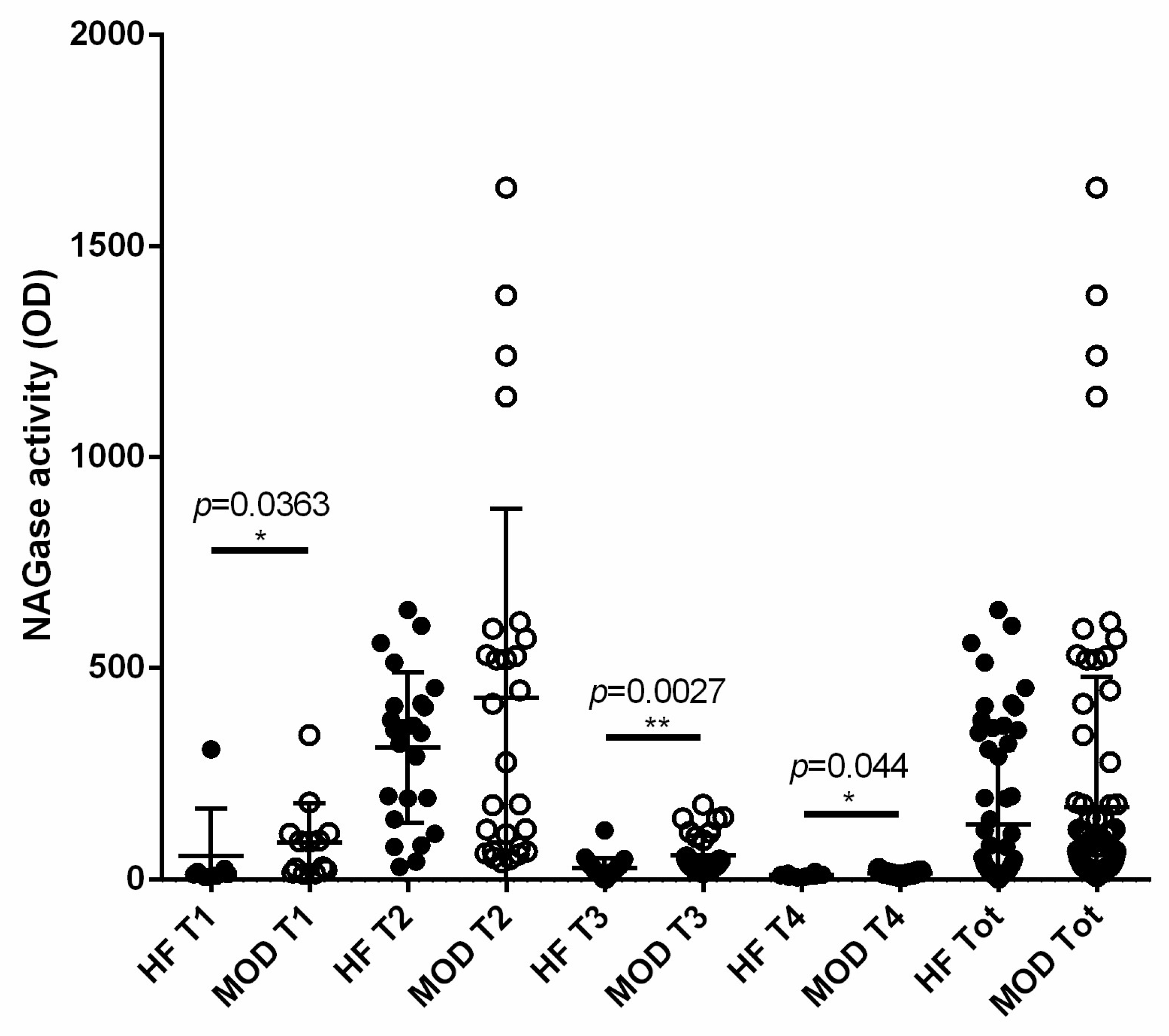
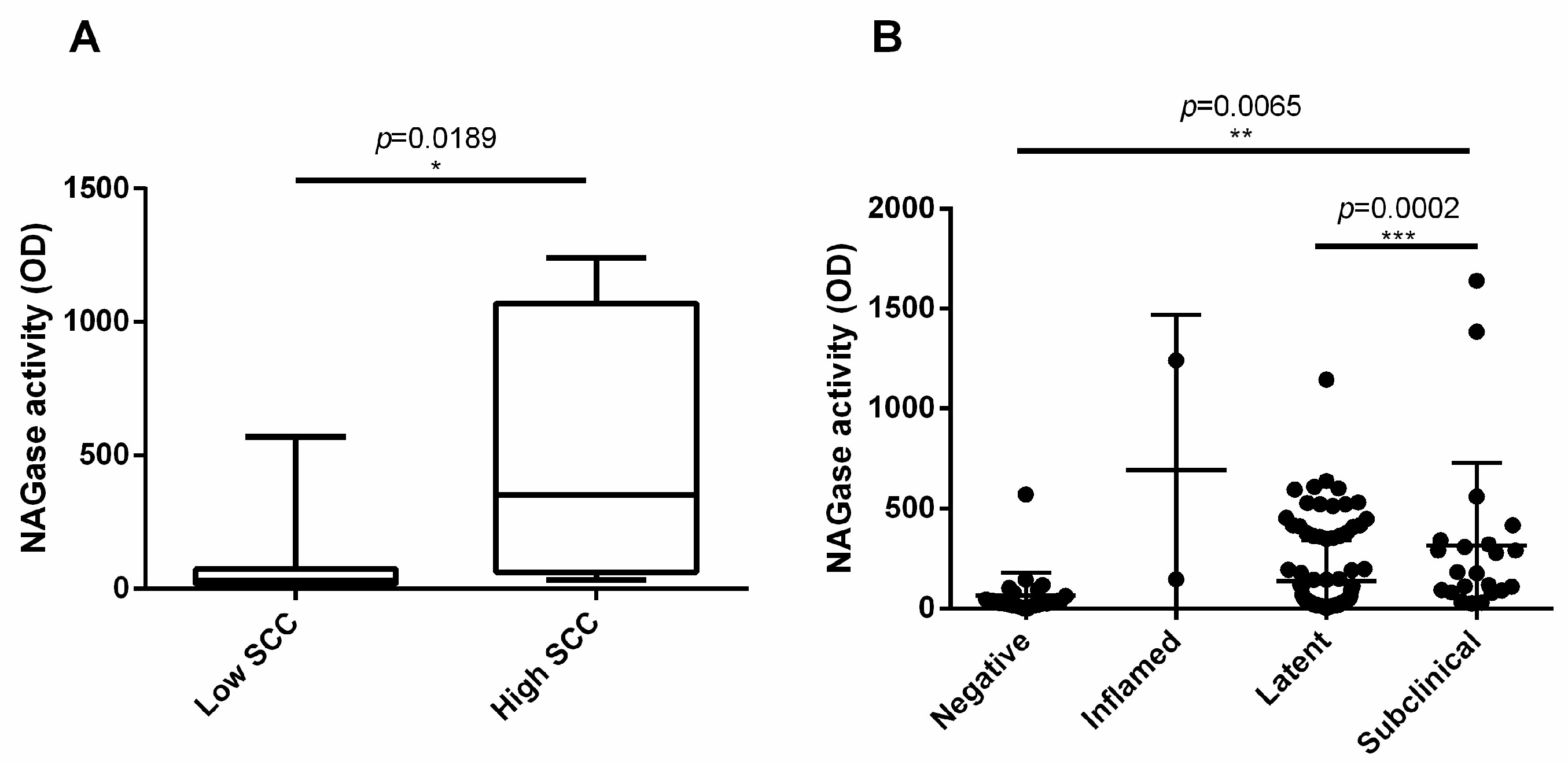
| Parameter | Correlation | p Value | R Spearman |
|---|---|---|---|
| E. coli killing activity | DIM | <0.0001 **** | −0.36 |
| Time point § | 0.0365 * | 0.15 | |
| Type of IMI §§ | 0.0015 ** | 0.24 | |
| NAGase activity | 0.018 * | 0.18 | |
| S. aureus killing activity | <0.0001 **** | 0.34 | |
| S. aureus killing activity | DIM | <0.0001 **** | −0.45 |
| E. coli killing activity | <0.0001 **** | 0.34 | |
| NAGase activity | <0.0001 **** | 0.44 | |
| Etiology of IMI | 0.0027 ** | 0.25 |
| Parameter | Correlation | p Value | R Spearman |
|---|---|---|---|
| NAGase activity | Time point § | <0.0001 **** | −0.54 |
| DIM | <0.0001 **** | −0.73 | |
| SCC | <0.0001 **** | 0.62 | |
| Type of IMI §§ | 0.0001 *** | 0.30 | |
| S. aureus killing activity | <0.0001 **** | 0.44 |
| Parameter | Correlation | p Value | R Spearman |
|---|---|---|---|
| NAGase activity | Time point § | <0.0001 **** | −0.68 |
| DIM | <0.0001 **** | −0.87 | |
| SCC | 0.0048 ** | 0.52 | |
| E. coli killing activity | 0.0111 * | 0.47 | |
| S. aureus killing activity | <0.0485 * | 0.40 |
| Parameter | Correlation | p Value | R Spearman |
|---|---|---|---|
| NAGase activity | Time point § | <0.0001 **** | −0.52 |
| DIM | <0.0001 **** | −0.85 | |
| SCC Type of IMI §§ | <0.0001 **** 0.0007 *** | 0.60 0.29 | |
| E. coli killing activity | 0.0177 * | 0.21 | |
| S. aureus killing activity | <0.0001 **** | 0.60 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Curone, G.; Filipe, J.; Inglesi, A.; Bronzo, V.; Pollera, C.; Comazzi, S.; Draghi, S.; Piccinini, R.; Ferlazzo, G.; Quattrone, A.; et al. Different Immune Control of Gram-Positive and Gram-Negative Mammary Infections in Dairy Cows. Vet. Sci. 2024, 11, 166. https://doi.org/10.3390/vetsci11040166
Curone G, Filipe J, Inglesi A, Bronzo V, Pollera C, Comazzi S, Draghi S, Piccinini R, Ferlazzo G, Quattrone A, et al. Different Immune Control of Gram-Positive and Gram-Negative Mammary Infections in Dairy Cows. Veterinary Sciences. 2024; 11(4):166. https://doi.org/10.3390/vetsci11040166
Chicago/Turabian StyleCurone, Giulio, Joel Filipe, Alessia Inglesi, Valerio Bronzo, Claudia Pollera, Stefano Comazzi, Susanna Draghi, Renata Piccinini, Gianluca Ferlazzo, Alda Quattrone, and et al. 2024. "Different Immune Control of Gram-Positive and Gram-Negative Mammary Infections in Dairy Cows" Veterinary Sciences 11, no. 4: 166. https://doi.org/10.3390/vetsci11040166
APA StyleCurone, G., Filipe, J., Inglesi, A., Bronzo, V., Pollera, C., Comazzi, S., Draghi, S., Piccinini, R., Ferlazzo, G., Quattrone, A., Vigo, D., Amadori, M., & Riva, F. (2024). Different Immune Control of Gram-Positive and Gram-Negative Mammary Infections in Dairy Cows. Veterinary Sciences, 11(4), 166. https://doi.org/10.3390/vetsci11040166










