A Preliminary Study Assessing a Transfer Learning Approach to Intestinal Image Analysis to Help Determine Treatment Response in Canine Protein-Losing Enteropathy
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
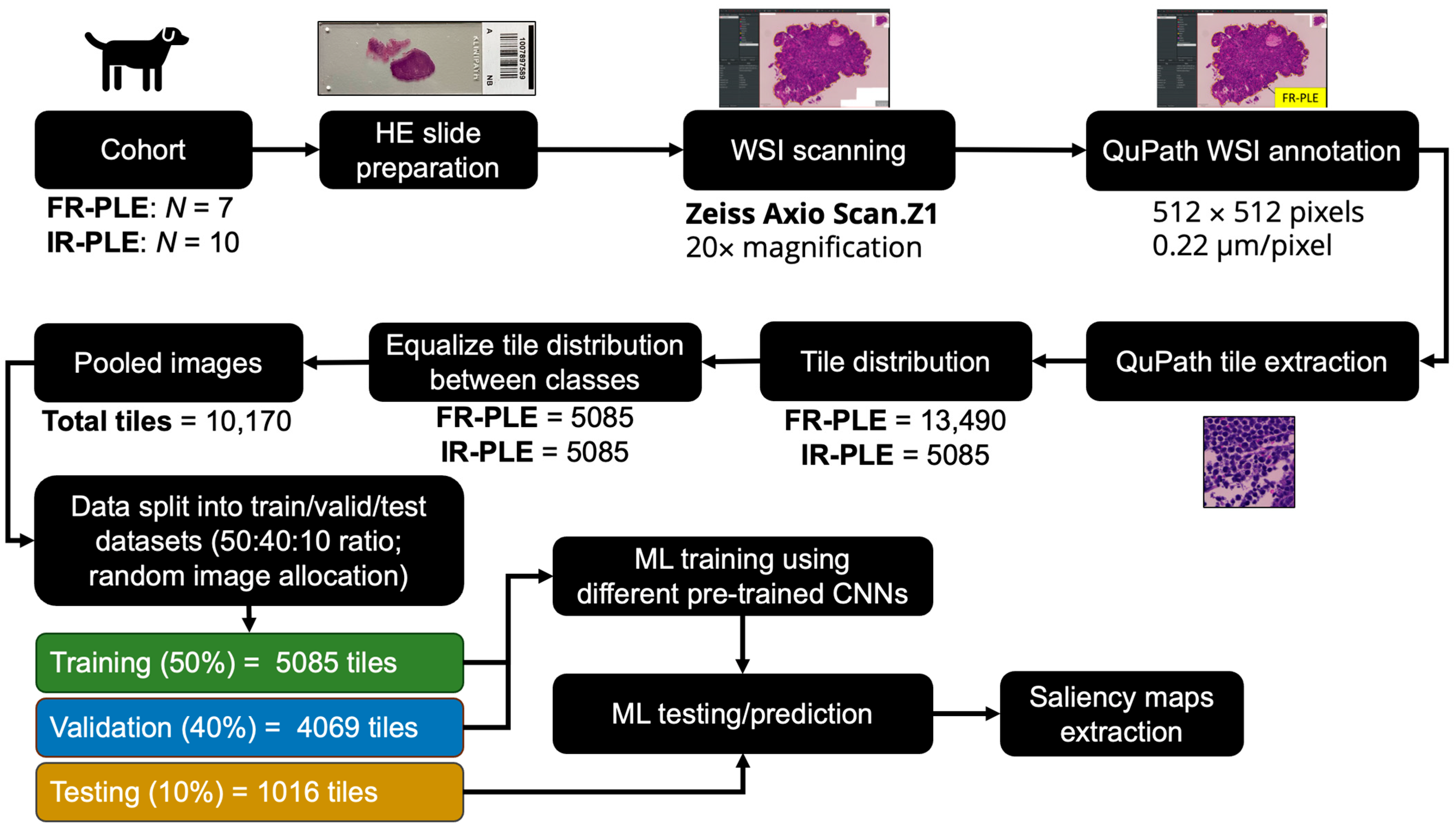
2.1. Study Design
2.2. Image Pre-Processing for Machine Learning
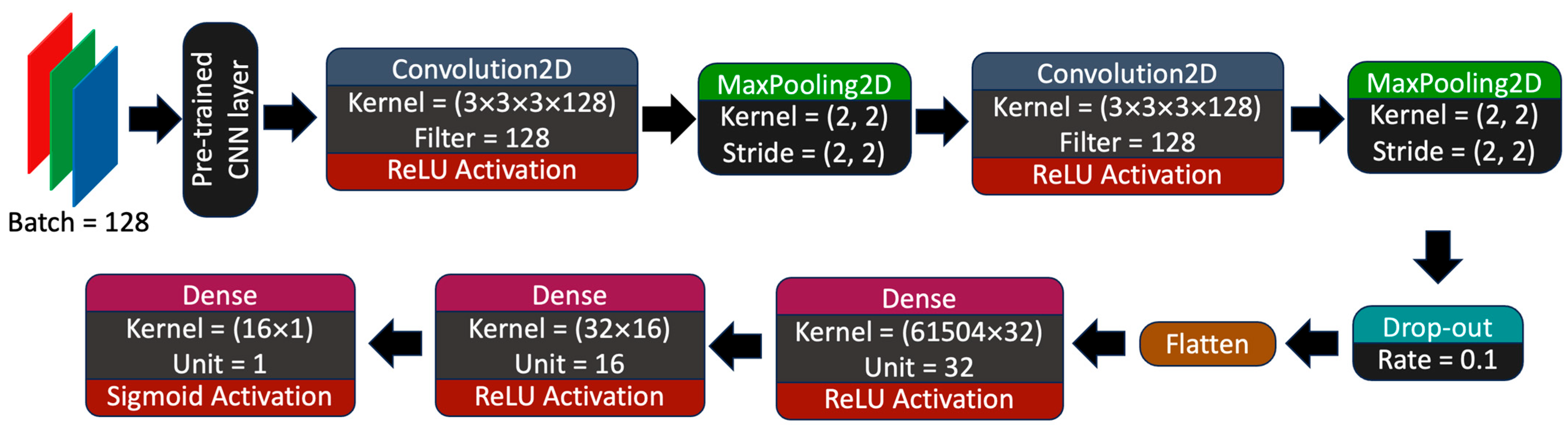
2.3. Machine Learning Model Training and Hyperparameter Optimization
2.4. Human Interpretable Machine Learning Vision
2.5. Statistical Analysis
2.6. Computational Hardware
2.7. Carbon Impact and Offsetting
3. Results
3.1. Study Population
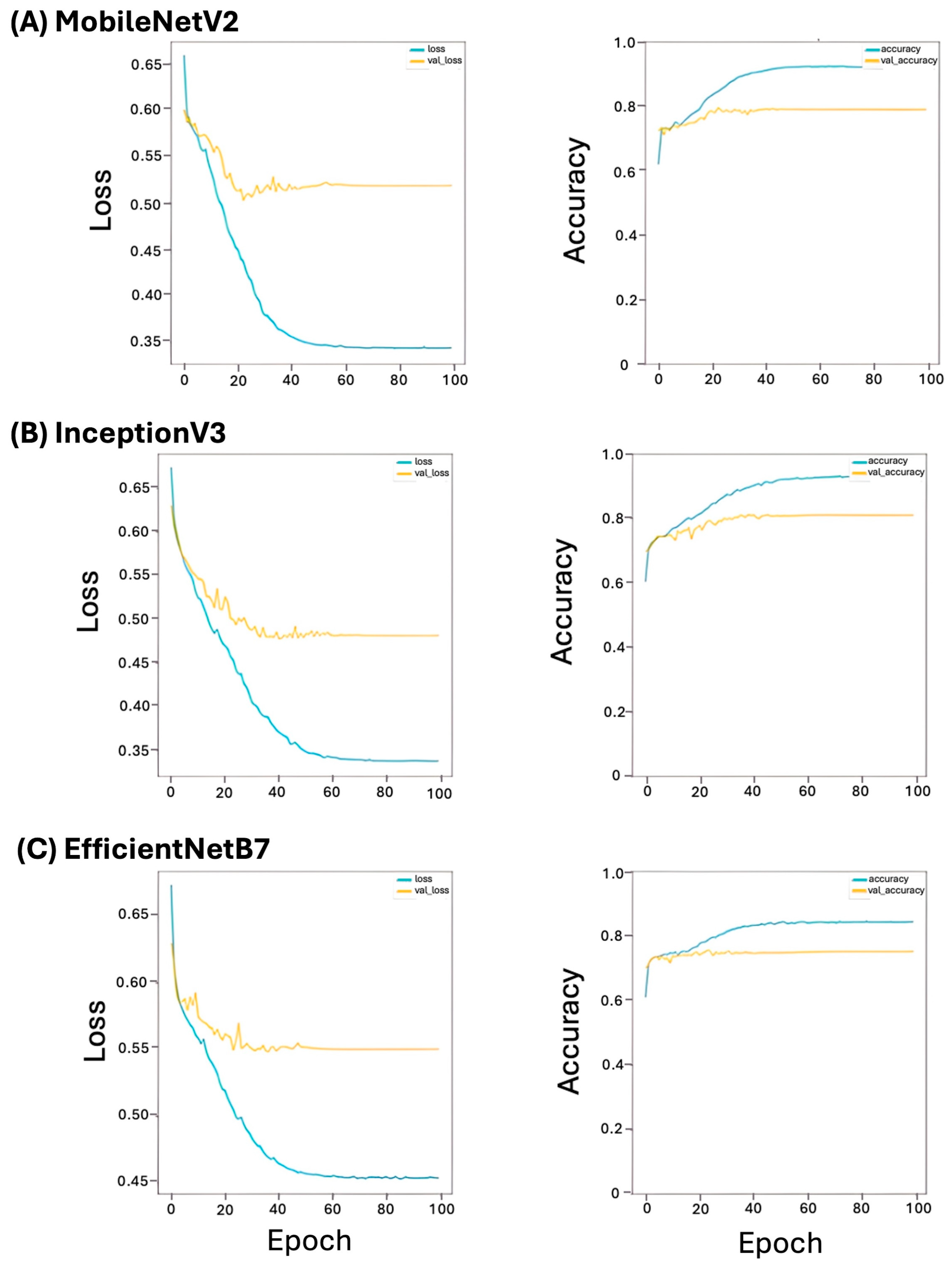
3.2. Tile Distribution and Training Performance
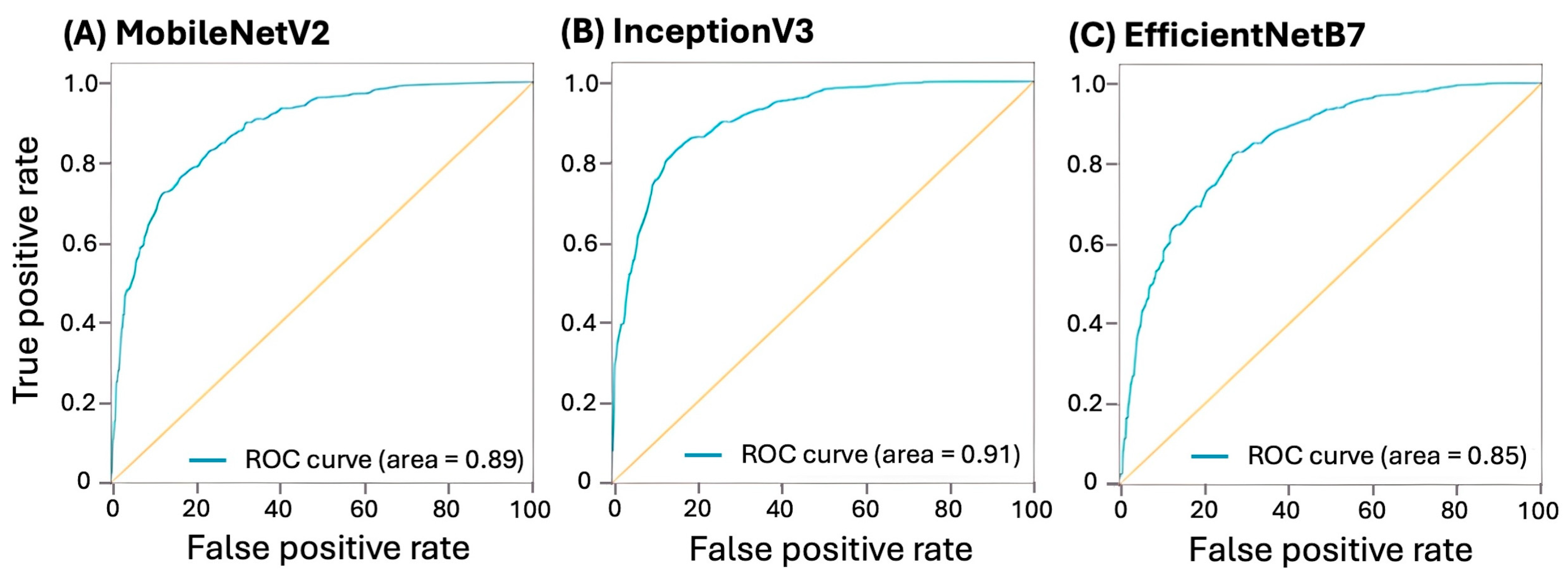
3.3. Model Prediction Performance
3.4. Human Explainable Machine Learning Vision
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stafford, I.S.; Kellermann, M.; Mossotto, E.; Beattie, R.M.; MacArthur, B.D.; Ennis, S. A systematic review of the applications of artificial intelligence and machine learning in autoimmune diseases. npj Digit. Med. 2020, 3, 30. [Google Scholar] [CrossRef]
- Abid, M.H.; Ashraf, R.; Mahmood, T.; Faisal, C.M.N. Multi-modal medical image classification using deep residual network and genetic algorithm. PLoS ONE 2023, 18, e0287786. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Kourou, K.; Exarchos, K.P.; Papaloukas, C.; Sakaloglou, P.; Exarchos, T.; Fotiadis, D.I. Applied machine learning in cancer research: A systematic review for patient diagnosis, classification and prognosis. Comput. Struct. Biotechnol. J. 2021, 19, 5546–5555. [Google Scholar] [CrossRef]
- Kraszewski, S.; Szczurek, W.; Szymczak, J.; Reguła, M.; Neubauer, K. Machine Learning Prediction Model for Inflammatory Bowel Disease Based on Laboratory Markers. Working Model in a Discovery Cohort Study. J. Clin. Med. 2021, 10, 4745. [Google Scholar] [CrossRef]
- Stafford, I.S.; Gosink, M.M.; Mossotto, E.; Ennis, S.; Hauben, M. A Systematic Review of Artificial Intelligence and Machine Learning Applications to Inflammatory Bowel Disease, with Practical Guidelines for Interpretation. Inflamm. Bowel Dis. 2022, 28, 1573–1583. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Picetti, D.; Dulai, P.S.; Jairath, V.; Sandborn, W.J.; Ohno-Machado, L.; Chen, P.L.; Singh, S. Machine Learning-based Prediction Models for Diagnosis and Prognosis in Inflammatory Bowel Diseases: A Systematic Review. J. Crohns Colitis 2022, 16, 398–413. [Google Scholar] [CrossRef] [PubMed]
- Iacucci, M.; Parigi, T.L.; Del Amor, R.; Meseguer, P.; Mandelli, G.; Bozzola, A.; Bazarova, A.; Bhandari, P.; Bisschops, R.; Danese, S.; et al. Artificial Intelligence Enabled Histological Prediction of Remission or Activity and Clinical Outcomes in Ulcerative Colitis. Gastroenterology 2023, 164, 1180–1188.e2. [Google Scholar] [CrossRef] [PubMed]
- Craven, M.D.; Washabau, R.J. Comparative pathophysiology and management of protein-losing enteropathy. J. Vet. Intern. Med. 2019, 33, 383–402. [Google Scholar] [CrossRef] [PubMed]
- Economu, L.; Chang, Y.M.; Priestnall, S.L.; Kathrani, A. The effect of assisted enteral feeding on treatment outcome in dogs with inflammatory protein-losing enteropathy. J. Vet. Intern. Med. 2021, 35, 1297–1305. [Google Scholar] [CrossRef] [PubMed]
- Allenspach, K.; Rizzo, J.; Jergens, A.E.; Chang, Y.M. Hypovitaminosis D is associated with negative outcome in dogs with protein losing enteropathy: A retrospective study of 43 cases. BMC Vet. Res. 2017, 13, 96. [Google Scholar] [CrossRef]
- Nagata, N.; Ohta, H.; Yokoyama, N.; Bin Teoh, Y.; Nisa, K.; Sasaki, N.; Osuga, T.; Morishita, K.; Takiguchi, M. Clinical characteristics of dogs with food-responsive protein-losing enteropathy. J. Vet. Intern. Med. 2020, 34, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Bovens, C.; Tennant, K.; Reeve, J.; Murphy, K. Basal serum cortisol concentration as a screening test for hypoadrenocorticism in dogs. J. Vet. Intern. Med. 2014, 28, 1541–1545. [Google Scholar] [CrossRef]
- Seraphin, T.P.; Luedde, M.; Roderburg, C.; van Treeck, M.; Scheider, P.; Buelow, R.D.; Boor, P.; Loosen, S.H.; Provaznik, Z.; Mendelsohn, D.; et al. Prediction of heart transplant rejection from routine pathology slides with self-supervised deep learning. Eur. Heart J. Digit. Health 2023, 4, 265–274. [Google Scholar] [CrossRef]
- Muti, H.S.; Heij, L.R.; Keller, G.; Kohlruss, M.; Langer, R.; Dislich, B.; Cheong, J.-H.; Kim, Y.-W.; Kim, H.; Kook, M.-C.; et al. Development and validation of deep learning classifiers to detect Epstein-Barr virus and microsatellite instability status in gastric cancer: A retrospective multicentre cohort study. Lancet Digit. Health 2021, 3, e654–e664. [Google Scholar] [CrossRef]
- Loeffler, C.M.; Gaisa, N.T.; Heij, L.R.; Grabsch, H.I.; Bruechle, N.O.; Kather, J.N. Predicting Mutational Status of Driver and Suppressor Genes Directly from Histopathology with Deep Learning: A Systematic Study Across 23 Solid Tumor Types. Front. Genet. 2021, 12, 806386. [Google Scholar] [CrossRef] [PubMed]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef] [PubMed]
- Lannelongue, L.; Grealey, J.; Inouye, M. Green Algorithms: Quantifying the Carbon Footprint of Computation. Adv. Sci. 2021, 8, 2100707. [Google Scholar] [CrossRef]
- Kallipolitis, A.R.K.; Maglogiannis, I. Ensembling EfficientNets for the Classification and Interpretation of Histopathology Images. Algorithms 2021, 14, 278. [Google Scholar] [CrossRef]
- Green, J.; Kathrani, A. Incidence of relapse of inflammatory protein-losing enteropathy in dogs and associated risk factors. J. Vet. Intern. Med. 2022, 36, 1981–1988. [Google Scholar] [CrossRef]
- Willard, M.D.; Jergens, A.E.; Duncan, R.B.; Leib, M.S.; McCracken, M.D.; DeNovo, R.C.; Helman, R.G.; Slater, M.R.; Harbison, J.L. Interobserver variation among histopathologic evaluations of intestinal tissues from dogs and cats. J. Am. Vet. Med. Assoc. 2002, 220, 1177–1182. [Google Scholar] [CrossRef] [PubMed]
- Day, M.J.; Bilzer, T.; Mansell, J.; Wilcock, B.; Hall, E.; Jergens, A.; Minami, T.; Willard, M.; Washabau, R. Histopathological standards for the diagnosis of gastrointestinal inflammation in endoscopic biopsy samples from the dog and cat: A report from the World Small Animal Veterinary Association Gastrointestinal Standardization Group. J. Comp. Pathol. 2008, 138 (Suppl. S1), S1–S43. [Google Scholar] [CrossRef] [PubMed]
- Allenspach, K.A.; Mochel, J.P.; Du, Y.; Priestnall, S.L.; Moore, F.; Slayter, M.; Rodrigues, A.; Ackermann, M.; Krockenberger, M.; Mansell, J.; et al. Correlating Gastrointestinal Histopathologic Changes to Clinical Disease Activity in Dogs with Idiopathic Inflammatory Bowel Disease. Vet. Pathol. 2019, 56, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Allenspach, K.; Wieland, B.; Gröne, A.; Gaschen, F. Chronic enteropathies in dogs: Evaluation of risk factors for negative outcome. J. Vet. Intern. Med. 2007, 21, 700–708. [Google Scholar] [CrossRef] [PubMed]
- Schreiner, N.M.; Gaschen, F.; Gröne, A.; Sauter, S.N.; Allenspach, K. Clinical signs, histology, and CD3-positive cells before and after treatment of dogs with chronic enteropathies. J. Vet. Intern. Med. 2008, 22, 1079–1083. [Google Scholar] [CrossRef] [PubMed]
- Procoli, F.; Mõtsküla, P.F.; Keyte, S.V.; Priestnall, S.; Allenspach, K. Comparison of histopathologic findings in duodenal and ileal endoscopic biopsies in dogs with chronic small intestinal enteropathies. J. Vet. Intern. Med. 2013, 27, 268–274. [Google Scholar] [CrossRef]
- Caulfield, S.; Priestnall, S.L.; Kathrani, A. Concordance of the Histopathologic Diagnosis of Concurrent Duodenal and Ileal Biopsy Specimens in Dogs. Animals 2021, 11, 2938. [Google Scholar] [CrossRef]
- Miotto, R.; Wang, F.; Wang, S.; Jiang, X.; Dudley, J.T. Deep learning for healthcare: Review, opportunities and challenges. Brief. Bioinform. 2018, 19, 1236–1246. [Google Scholar] [CrossRef]
- Choi, J.H.; Kim, H.A.; Kim, W.; Lim, I.; Lee, I.; Byun, B.H.; Noh, W.C.; Seong, M.-K.; Lee, S.-S.; Kim, B.I.; et al. Early prediction of neoadjuvant chemotherapy response for advanced breast cancer using PET/MRI image deep learning. Sci. Rep. 2020, 10, 21149. [Google Scholar] [CrossRef]

| FR-PLE | IR-PLE | ||
|---|---|---|---|
| Albumin (g/L) | Vitamin B12 (ng/L) | Albumin (g/L) | Vitamin B12 (ng/L) |
| 15.8 | <150 | 15.4 | <150 |
| 23.0 | <150 | 13.9 | 419 |
| 18.8 | <150 | 13.0 | 184 |
| 20.7 | 473 | 19.3 | <150 |
| 14.8 | 459 | 16.2 | <150 |
| 24.7 | 233 | 19.6 | 227 |
| 18.0 | WNL * | 22.3 | 158 |
| 25.7 | 298 | ||
| 12.9 | 174 | ||
| 13.7 | 308 | ||
| Architecture | Precision | Recall | Accuracy | F1 Score | AUROC | Confusion Matrix |
|---|---|---|---|---|---|---|
| MobileNetV2 | 83.0% | 79.1% | 80.4% | 0.79 | 0.88 | |
| InceptionV3 | 85.8% | 80.9% | 83.8% | 0.83 | 0.91 | |
| EfficientNetB7 | 76.7% | 74.6% | 76.0% | 0.76 | 0.85 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kathrani, A.; Trewin, I.; Ancheta, K.; Psifidi, A.; Le Calvez, S.; Williams, J. A Preliminary Study Assessing a Transfer Learning Approach to Intestinal Image Analysis to Help Determine Treatment Response in Canine Protein-Losing Enteropathy. Vet. Sci. 2024, 11, 129. https://doi.org/10.3390/vetsci11030129
Kathrani A, Trewin I, Ancheta K, Psifidi A, Le Calvez S, Williams J. A Preliminary Study Assessing a Transfer Learning Approach to Intestinal Image Analysis to Help Determine Treatment Response in Canine Protein-Losing Enteropathy. Veterinary Sciences. 2024; 11(3):129. https://doi.org/10.3390/vetsci11030129
Chicago/Turabian StyleKathrani, Aarti, Isla Trewin, Kenneth Ancheta, Androniki Psifidi, Sophie Le Calvez, and Jonathan Williams. 2024. "A Preliminary Study Assessing a Transfer Learning Approach to Intestinal Image Analysis to Help Determine Treatment Response in Canine Protein-Losing Enteropathy" Veterinary Sciences 11, no. 3: 129. https://doi.org/10.3390/vetsci11030129
APA StyleKathrani, A., Trewin, I., Ancheta, K., Psifidi, A., Le Calvez, S., & Williams, J. (2024). A Preliminary Study Assessing a Transfer Learning Approach to Intestinal Image Analysis to Help Determine Treatment Response in Canine Protein-Losing Enteropathy. Veterinary Sciences, 11(3), 129. https://doi.org/10.3390/vetsci11030129









