CircRNA-5335 Regulates the Differentiation and Proliferation of Sheep Preadipocyte via the miR-125a-3p/STAT3 Pathway
Abstract
Simple Summary
Abstract
1. Introduction
1.1. Intramuscular Fat Accumulation and Preadipocytes in Sheep
1.2. IMF Regulatory Factors and Pathways
1.3. CircRNA and IMF
2. Materials and Methods
2.1. Cell Separation and Culture
2.2. Dual-Luciferase Activity Assay
2.3. Identification of circRNA
2.4. Preadipocyte Differentiation
2.5. Oil Red O(ORO) Staining
2.6. Cell Proliferation Assay
2.7. Real-Time Quantitative PCR (RT-qPCR)
2.8. Western Blotting (WB)
2.9. Statistical Analysis
3. Results
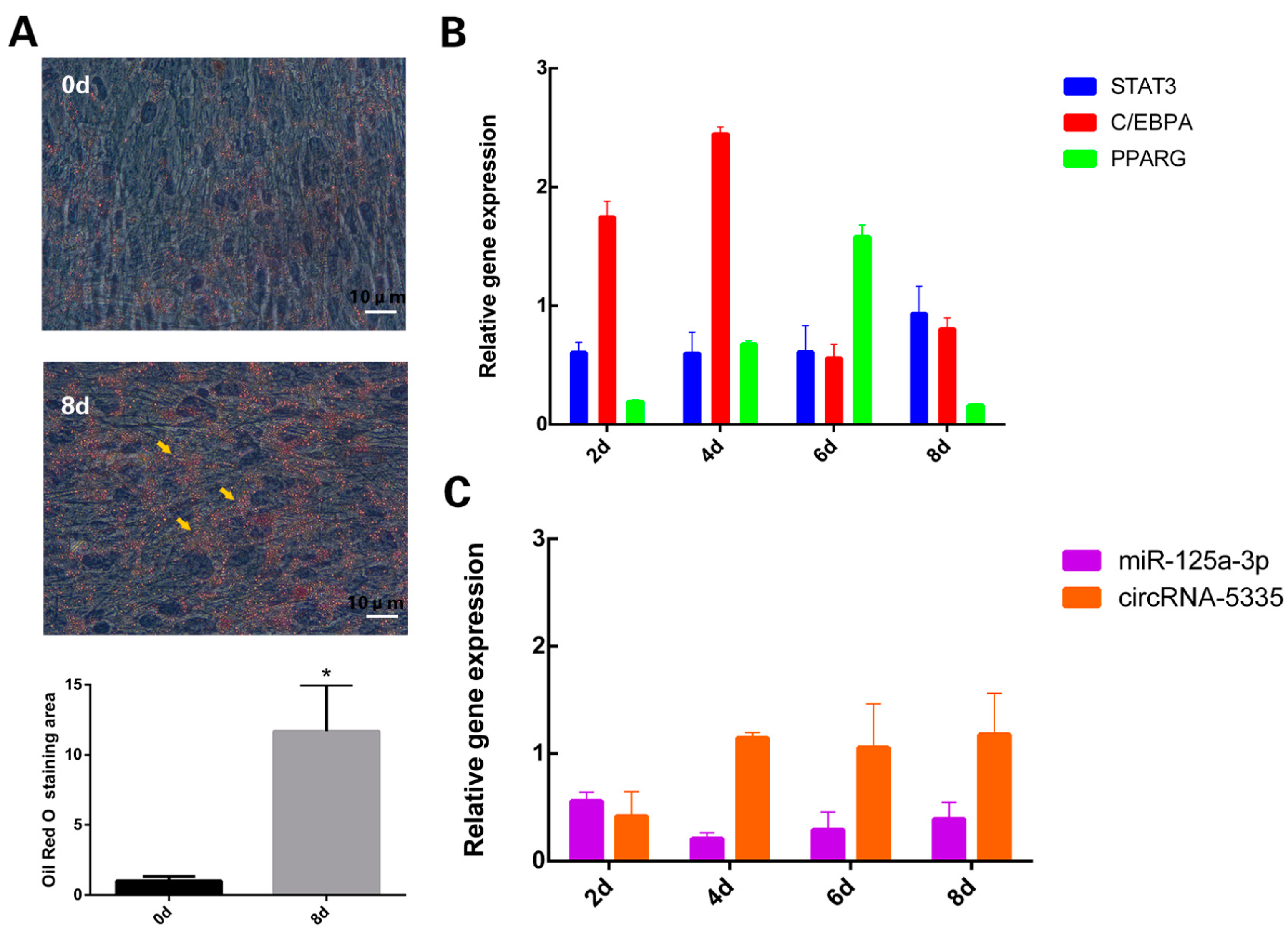
3.1. Expression Levels of circRNA-5335/miR-125a-3p/STAT3 during Sheep Preadipocyte Differentiation
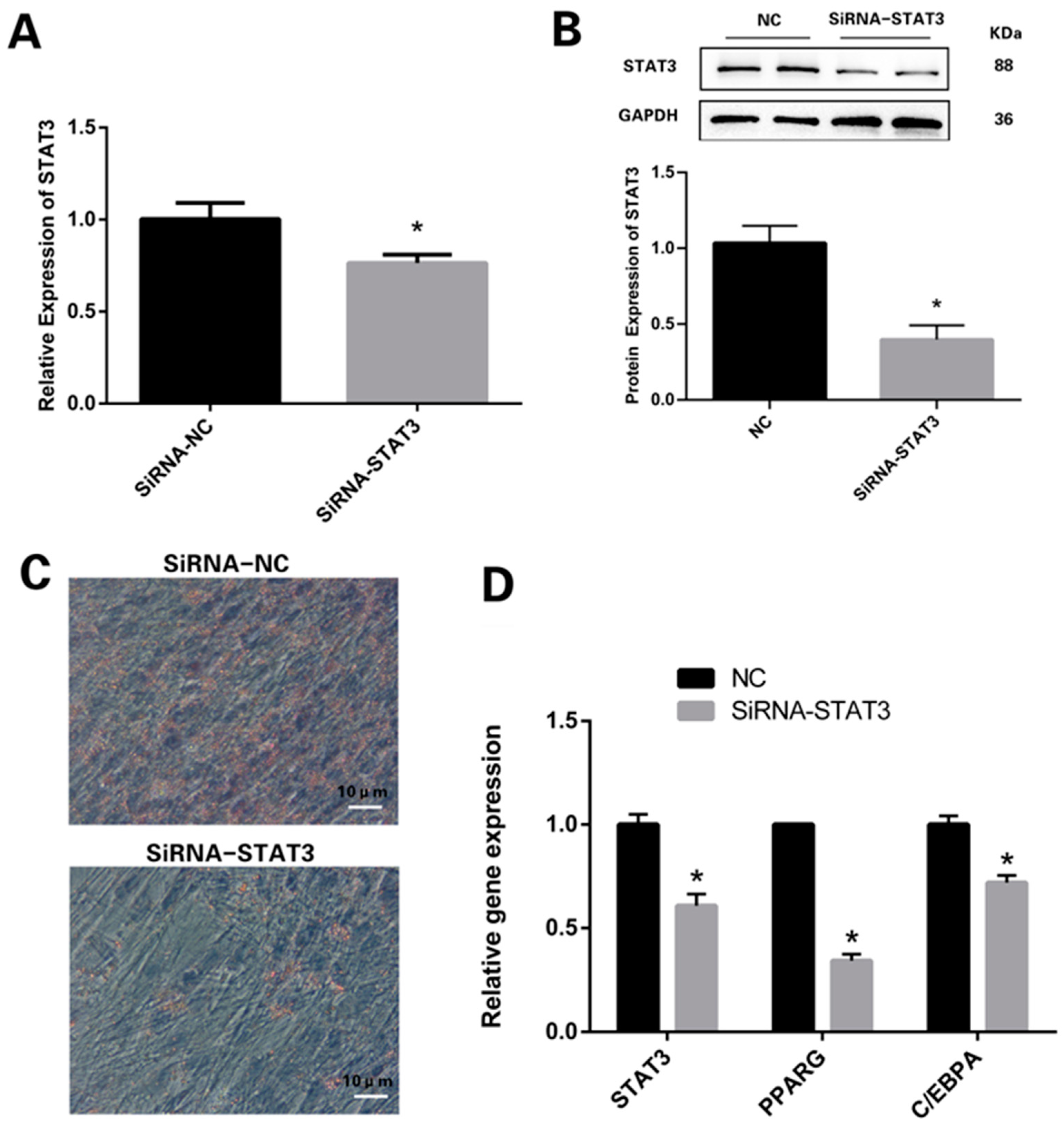
3.2. Knockdown of STAT3 Inhibits Differentiation Process of Preadipocytes
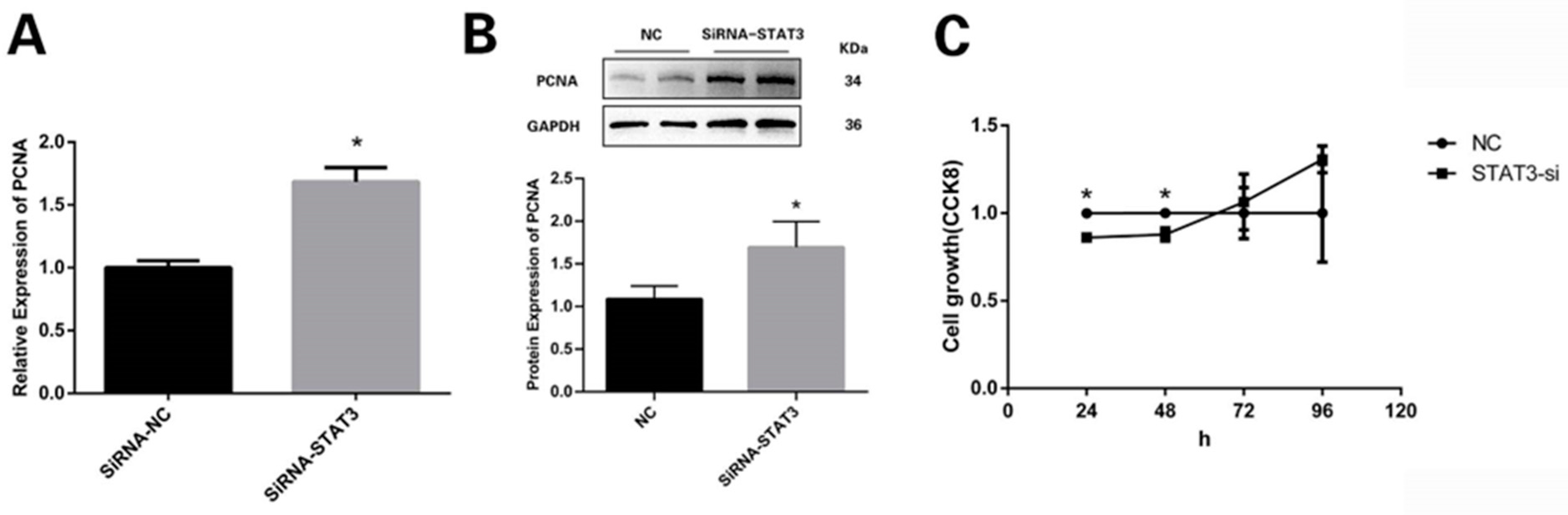
3.3. STAT3 Prevents Preadipocytes from Multiplying
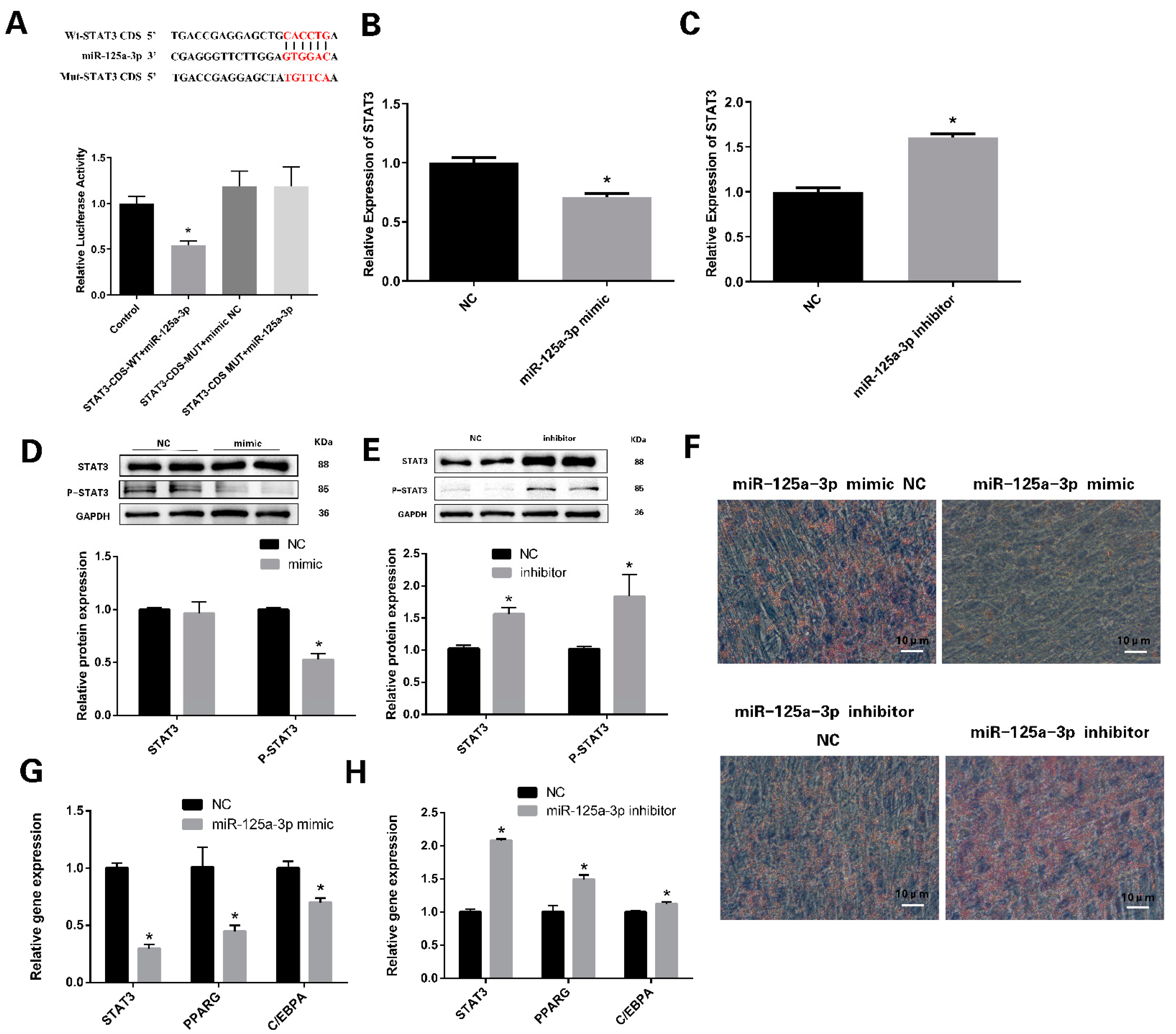
3.4. miR-125a-3p Regulates the Differentiation of Preadipocytes through STAT3 Signaling Pathway
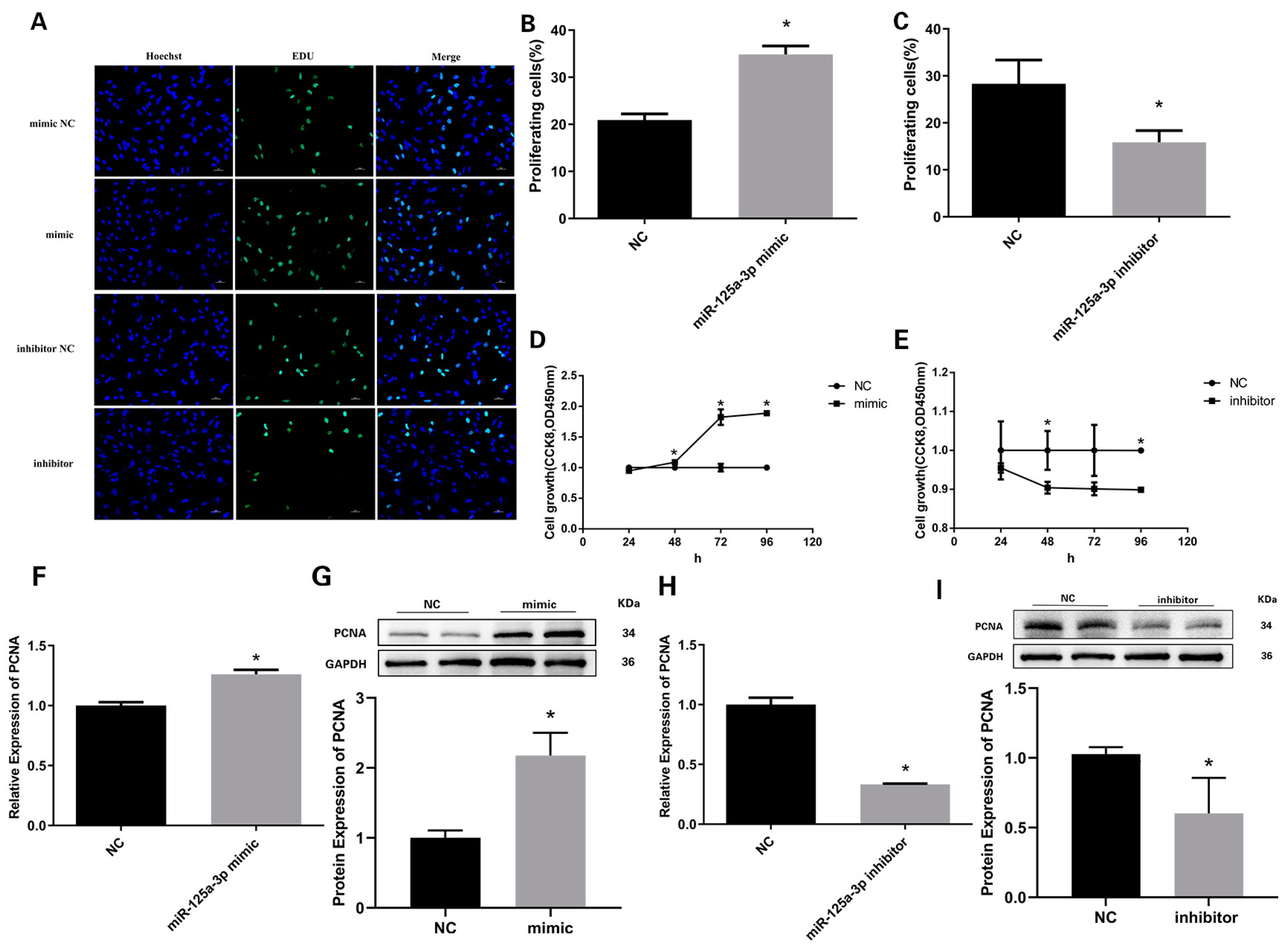
3.5. miR-125a-3p Inhibits the Proliferation of Preadipocytes in Sheep
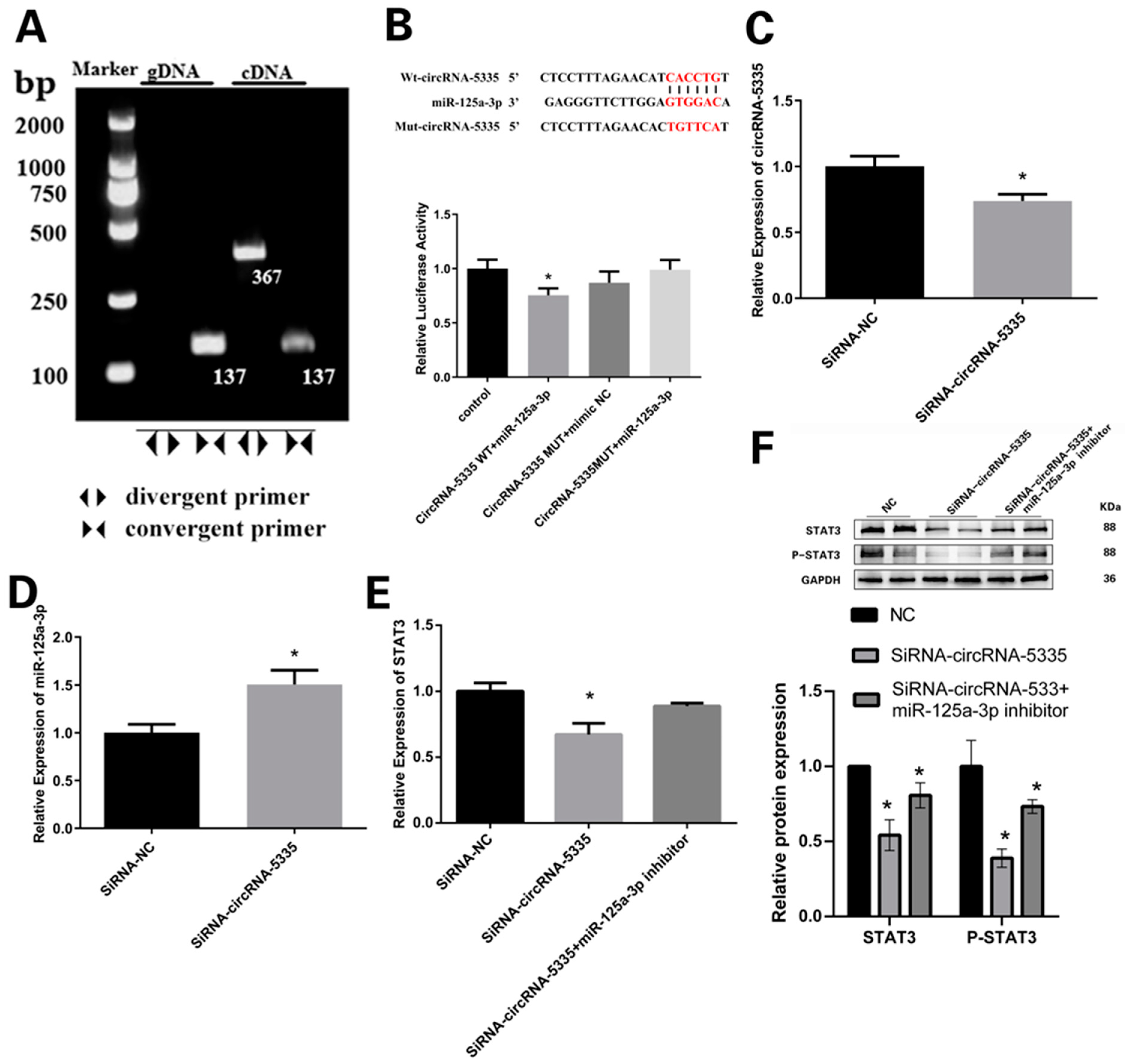
3.6. CircRNA-5335 Regulates the Expression of STAT3 by Binding to miR-125a-3p
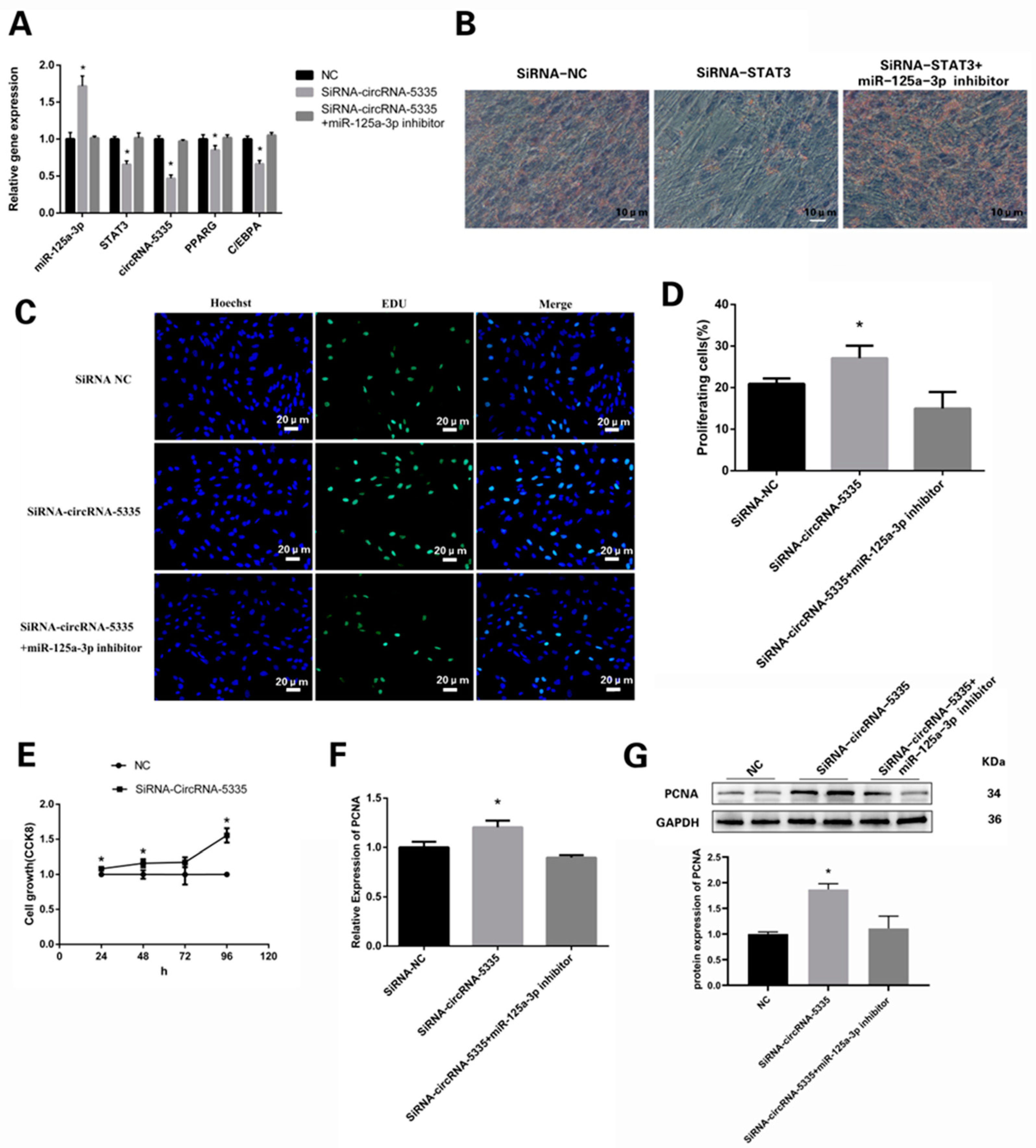
3.7. circRNA-5335/miR-125a-3p/STAT3 Regulates the Differentiation and Proliferation of Preadipocytes
4. Discussion
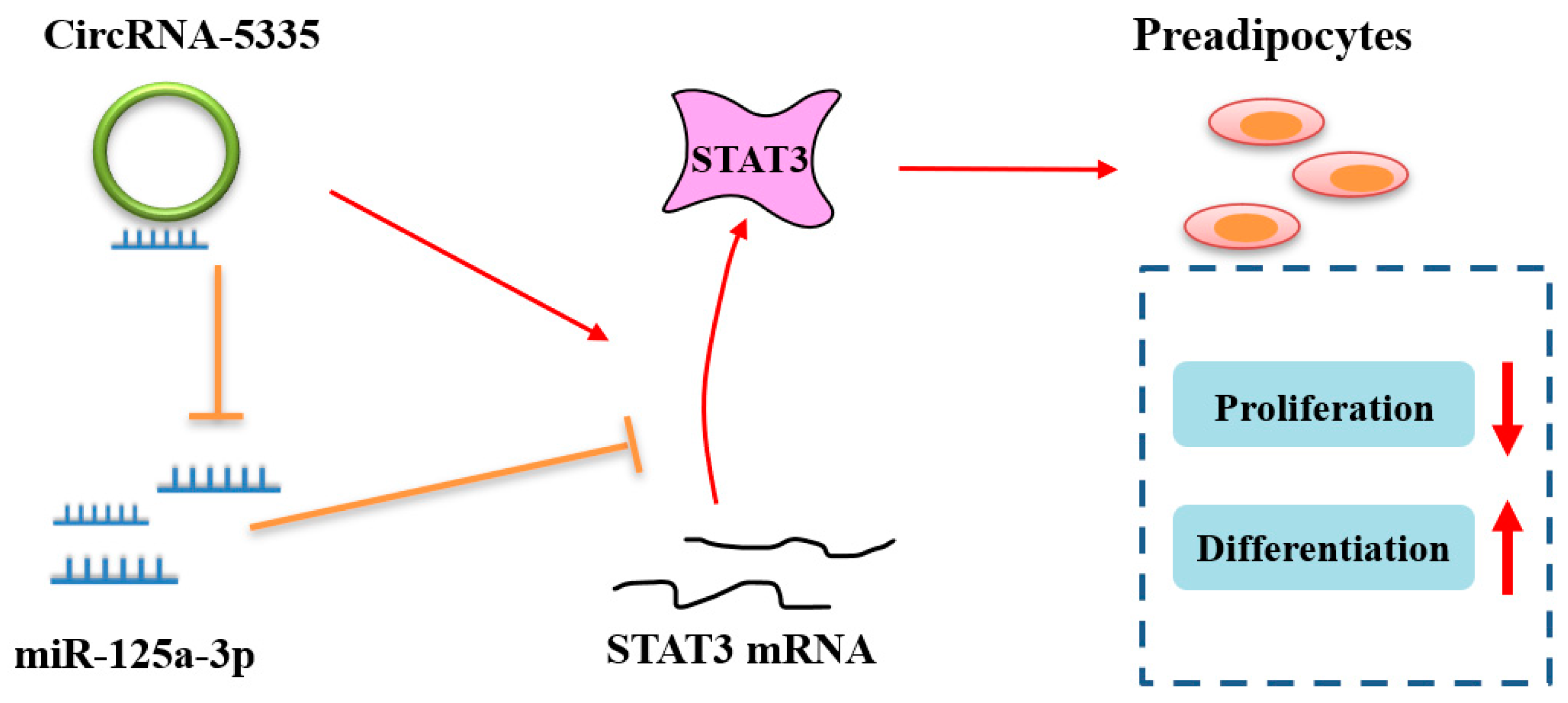
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xiao, C.; Wei, T.; Liu, L.X.; Liu, J.Q.; Wang, C.X.; Yuan, Z.Y.; Ma, H.H.; Jin, H.G.; Zhang, L.C.; Cao, Y. Whole-Transcriptome Analysis of Preadipocyte and Adipocyte and Construction of Regulatory Networks to Investigate Lipid Metabolism in Sheep. Front. Genet. 2021, 12, 662143. [Google Scholar] [CrossRef]
- Hausman, G.J.; Basu, U.; Du, M.; Fernyhough-Culver, M.; Dodson, M.V. Intermuscular and intramuscular adipose tissues: Bad vs. good adipose tissues. Adipocyte 2014, 3, 242–255. [Google Scholar] [CrossRef]
- Li, J.; Yang, Y.; Tang, C.; Yue, S.; Zhao, Q.; Li, F.; Zhang, J. Changes in lipids and aroma compounds in intramuscular fat from Hu sheep. Food Chem. 2022, 383, 132611. [Google Scholar] [CrossRef] [PubMed]
- Pannier, L.; Gardner, G.E.; Pearce, K.L.; McDonagh, M.; Ball, A.J.; Jacob, R.H.; Pethick, D.W. Associations of sire estimated breeding values and objective meat quality measurements with sensory scores in Australian lamb. Meat Sci. 2014, 96 Pt B, 1076–1087. [Google Scholar] [CrossRef]
- Realini, C.E.; Pavan, E.; Johnson, P.L.; Font-I-Furnols, M.; Jacob, N.; Agnew, M.; Craigie, C.R.; Moon, C.D. Consumer liking of M. longissimus lumborum from New Zealand pasture-finished lamb is influenced by intramuscular fat. Meat Sci. 2021, 173, 108380. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhou, Y.; Lei, W.; Zhang, K.; Shi, J.; Hu, Y.; Shu, G.; Song, J. Signal transducer and activator of transcription 3 (STAT3) regulates adipocyte differentiation via peroxisome-proliferator-activated receptor gamma (PPARgamma). Biol. Cell 2009, 102, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Prache, S.; Schreurs, N.; Guillier, L. Review: Factors affecting sheep carcass and meat quality attributes. Animal 2022, 16, 100330. [Google Scholar] [CrossRef] [PubMed]
- Mortimer, S.I.; van der Werf, J.H.; Jacob, R.H.; Hopkins, D.L.; Pannier, L.; Pearce, K.L.; Gardner, G.E.; Warner, R.D.; Geesink, G.H.; Edwards, J.E.; et al. Genetic parameters for meat quality traits of Australian lamb meat. Meat Sci. 2014, 96 Pt B, 1016–1024. [Google Scholar] [CrossRef]
- Luo, G.; Hong, T.; Yu, L.; Ren, Z. FTO Regulated Intramuscular Fat by Targeting APMAP Gene via an m6A-YTHDF2-dependent Manner in Rex Rabbits. Cells 2023, 12, 369. [Google Scholar] [CrossRef] [PubMed]
- Al Dow, M.; Silveira, M.A.D.; Poliquin, A.; Tribouillard, L.; Fournier, É.; Trébaol, E.; Secco, B.; Villot, R.; Tremblay, F.; Bilodeau, S.; et al. Control of adipogenic commitment by a STAT3-VSTM2A axis. Am. J. Physiol. Endocrinol. Metab. 2021, 320, 259–269. [Google Scholar] [CrossRef]
- Zhang, K.; Guo, W.; Yang, Y.; Wu, J. JAK2/STAT3 pathway is involved in the early stage of adipogenesis through regulating C/EBPβ transcription. Cell Biochem. 2011, 112, 488–497. [Google Scholar] [CrossRef] [PubMed]
- Ambele, M.A.; Dhanraj, P.; Giles, R.; Pepper, M.S. Adipogenesis: A Complex Interplay of Multiple Molecular Determinants and Pathways. Int. J. Mol. Sci. 2020, 21, 4283. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, P.R.; Gnudi, L.; Tozzo, E.; Yang, H.; Leach, F.; Kahn, B.B. Adipose cell hyperplasia and enhanced glucose disposal in transgenic mice overexpressing GLUT4 selectively in adipose tissue. J. Biol. Chem. 1993, 268, 22243–22246. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.; Zheng, H.; Wu, Z.; Chen, M.; Huang, Y. Circular RNA-protein interactions: Functions, mechanisms, and identification. Theranostics 2020, 10, 3503–3517. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Yang, Z.; Peng, T.; Lv, Y. Circular RNAs: Rising stars in lipid metabolism and lipid disorders. J. Cell Physiol. 2021, 236, 4797–4806. [Google Scholar] [CrossRef] [PubMed]
- Kang, Z.; Zhang, S.; Jiang, E.; Wang, X.; Wang, Z.; Chen, H.; Lan, X. circFLT1 and lncCCPG1 Sponges miR-93 to Regulate the Proliferation and Differentiation of Adipocytes by Promoting lncSLC30A9 Expression. Mol. Ther. Nucleic Acids 2020, 22, 484–499. [Google Scholar] [CrossRef]
- Du, J.; Xu, Y.; Zhang, P.; Zhao, X.; Gan, M.; Li, Q.; Ma, J.; Tang, G.; Jiang, Y.; Wang, J.; et al. MicroRNA-125a-5p Affects Adipocytes Proliferation, Differentiation and Fatty Acid Composition of Porcine Intramuscular Fat. Int. J. Mol. Sci. 2018, 19, 501. [Google Scholar] [CrossRef]
- Liu, H.; Wen, J.; Tian, X.; Li, T.; Zhao, J.; Cheng, J.; Huang, L.; Zhao, Y.; Cao, Q.; Jiang, J. miR-125a-3p regulates the expression of FSTL1, a pro-inflammatory factor, during adipogenic differentiation, and inhibits adipogenesis in mice. FASEB J. 2023, 37, e23146. [Google Scholar] [CrossRef]
- Li, M.; Li, J.; Ji, M.; An, J.; Zhao, T.; Yang, Y.; Cai, C.; Gao, P.; Cao, G.; Guo, X.; et al. CircHOMER1 inhibits porcine adipogenesis via the miR-23b/SIRT1 axis. FASEB J. 2023, 37, e22828. [Google Scholar] [CrossRef]
- Zhao, L.; Zhou, L.; Hao, X.; Wang, L.; Han, F.; Liu, L.; Duan, X.; Guo, F.; He, J.; Liu, N. Identification and Characterization of Circular RNAs in Association with the Deposition of Intramuscular Fat in Aohan Fine-Wool Sheep. Front. Genet. 2021, 12, 759747. [Google Scholar] [CrossRef]
- Han, F.; Zhou, L.; Zhao, L.; Wang, L.; Liu, L.; Li, H.; Qiu, J.; He, J.; Liu, N. Identification of miRNA in Sheep Intramuscular Fat and the Role of miR-193a-5p in Proliferation and Differentiation of 3T3-L1. Front. Genet. 2021, 12, 633295. [Google Scholar] [CrossRef]
- Shi, T.; Yan, X.; Qiao, L.; Li, B.; Cheng, L.; Pan, Y.; Jing, J.; Cao, N.; Liu, W. MiR-330-5p negatively regulates ovine preadipocyte differentiation by targeting branched-chain aminotransferase 2. Anim. Sci. J. 2018, 89, 858–867. [Google Scholar] [CrossRef]
- Wang, Y.; Li, X.; Cao, Y.; Xiao, C.; Liu, Y.; Jin, H.; Cao, Y. Effect of the ACAA1 Gene on Preadipocyte Differentiation in Sheep. Front. Genet. 2021, 12, 649140. [Google Scholar] [CrossRef] [PubMed]
- Abumrad, N.A.; el-Maghrabi, M.R.; Amri, E.Z.; Lopez, E.; Grimaldi, P.A. Cloning of a rat adipocyte membrane protein implicated in binding or transport of long-chain fatty acids that is induced during preadipocyte differentiation. Homology with human CD36. J. Biol. Chem. 1993, 268, 17665–17668. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Yao, X.; Wang, Z.; Li, X.; Li, X.; An, S.; Wei, Z.; Zhang, G.; Wang, F. Long non-coding RNA366.2 controls endometrial epithelial cell proliferation and migration by upregulating WNT6 as a ceRNA of miR-1576 in sheep uterus. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194606. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Fang, X.; Gao, M.; Mi, J.; Zhang, X.; Xia, L.; Zhao, Z.; Albrecht, E.; Maak, S.; Yang, R. Isolation and Identification of Bovine Preadipocytes and Screening of MicroRNAs Associated with Adipogenesis. Animals 2020, 10, 818. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.C.; Nadeau, K.; Abbasi, M.; Lachance, C.; Nguyen, M.; Fenrich, J. The Ultimate RT-qPCR Experiment: Producing Publication Quality, Reproducible Data the First Time. Trends Biotechnol. 2019, 37, 761–774. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Li, L.; Liu, M.; Wang, L.; Gao, X.; Zhou, L.; Liu, N.; He, J. miR-27a Targeting PIK3R3 Regulates the Proliferation and Apoptosis of Sheep Hair Follicle Stem Cells. Animals 2022, 13, 141. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.; Joo, S.T.; Warner, R. Consumer Acceptability of Intramuscular Fat. Korean J. Food Sci. Anim. Resour. 2016, 36, 699–708. [Google Scholar] [CrossRef]
- Kang, H.J.; Seo, H.A.; Go, Y.; Oh, C.J.; Jeoung, N.H.; Park, K.G.; Lee, I.K. Dimethylfumarate suppresses adipogenic differentiation in 3T3-L1 preadipocytes through inhibition of STAT3 activity. PLoS ONE 2013, 8, e61411. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Lin, F.; Yin, Y.; Shang, Y.; Xiao, Z.; Xu, W. Adipocyte-derived exosomal miR-30c-5p promotes ovarian angiogenesis in polycystic ovary syndrome via the SOCS3/STAT3/VEGFA pathway. J. Steroid Biochem. Mol. Biol. 2023, 230, 106278. [Google Scholar] [CrossRef]
- Chong, Y.; Liu, G.; Girmay, S.; Jiang, X. Novel mutations in the signal transducer and activator of transcription 3 gene are associated with sheep body weight and fatness traits. Mamm. Genome 2021, 32, 38–49. [Google Scholar] [CrossRef]
- Siao, A.C.; Lin, Y.Y.; Shih, L.J.; Tsuei, Y.W.; Chuu, C.P.; Kuo, Y.C.; Kao, Y.H. Endothelin-1 stimulates preadipocyte growth via the PKC, STAT3, AMPK, c-JUN, ERK, sphingosine kinase, and sphingomyelinase pathways. Am. J. Physiol. Cell Physiol. 2020, 319, 839–857. [Google Scholar] [CrossRef]
- Yuan, Y.; Xi, Y.; Chen, J.; Zhu, P.; Kang, J.; Zou, Z.; Wang, F.; Bu, S. STAT3 stimulates adipogenic stem cell proliferation and cooperates with HMGA2 during the early stage of differentiation to promote adipogenesis. Biochem. Biophys. Res. Commun. 2017, 482, 1360–1366. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, H.; Li, Y.; Mao, R.; Yang, H.; Zhang, Y.; Zhang, Y.; Guo, P.; Zhan, D.; Zhang, T. Circular RNA SAMD4A controls adipogenesis in obesity through the miR-138-5p/EZH2 axis. Theranostics 2020, 10, 4705–4719. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Du, Y.; Lu, W.; Gui, W.; Sun, S.; Zhu, Y.; Wang, G.; Eserberg, D.T.; Zheng, F.; Zhou, J.; et al. CircRNF111 Protects Against Insulin Resistance and Lipid Deposition via Regulating miR-143-3p/IGF2R Axis in Metabolic Syndrome. Front. Cell Dev. Biol. 2021, 9, 663148. [Google Scholar] [CrossRef] [PubMed]
- Qi, K.; Liu, Y.; Li, C.; Li, X.; Li, X.; Wang, K.; Qiao, R.; Han, X. Construction of circRNA-related ceRNA networks in longissimus dorsi muscle of Queshan Black and Large White pigs. Mol. Genet. Genom. 2022, 297, 101–112. [Google Scholar] [CrossRef]
- Patop, I.L.; Wüst, S.; Kadener, S. Past, present, and future of CircRNAs. EMBO J. 2019, 38, 100836. [Google Scholar] [CrossRef]
- Zheng, Z.; Zeng, X.; Zhu, Y.; Leng, M.; Zhang, Z.; Wang, Q.; Liu, X.; Zeng, S.; Xiao, Y.; Hu, C.; et al. CircPPAP2B controls metastasis of clear cell renal cell carcinoma via HNRNPC-dependent alternative splicing and targeting the miR-182-5p/CYP1B1 axis. Mol. Cancer 2024, 23, 4. [Google Scholar] [CrossRef]
- Shi, H.; Zhou, Y.; Jia, E.; Liu, Z.; Pan, M.; Bai, Y.; Zhao, X.; Ge, Q. Comparative analysis of circular RNA enrichment methods. RNA Biol. 2022, 19, 55–67. [Google Scholar] [CrossRef]
- Zaiou, M. The Emerging Role and Promise of Circular RNAs in Obesity and Related Metabolic Disorders. Cells 2020, 9, 1473. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; He, Y.; Wu, W.; Tan, X.; Wang, Z.; Irwin, D.M.; Wang, Z.; Zhan, S. Circular RNA Profiling Identifies Novel circPPARA that Promotes Intramuscular Fat Deposition in Pigs. J. Agric. Food Chem. 2022, 70, 4123–4137. [Google Scholar] [CrossRef]
- Arcinas, C.; Tan, W.; Fang, W.; Desai, T.P.; Teh DC, S.; Degirmenci, U.; Xu, D.; Foo, R.; Sun, L. Adipose circular RNAs exhibit dynamic regulation in obesity and functional role in adipogenesis. Nat. Metab. 2019, 1, 688–703. [Google Scholar] [CrossRef] [PubMed]
- Song, X.H.; He, N.; Xing, Y.T.; Jin, X.Q.; Li, Y.W.; Liu, S.S.; Gao, Z.Y.; Guo, C.; Wang, J.J.; Huang, Y.Y.; et al. A Novel Age-Related Circular RNA Circ-ATXN2 Inhibits Proliferation, Promotes Cell Death and Adipogenesis in Rat Adipose Tissue-Derived Stromal Cells. Front. Genet. 2021, 12, 761926. [Google Scholar] [CrossRef] [PubMed]
| Target Gene | Primer Sequence (5′-3′) | Product Size (bp) | Accession Number |
|---|---|---|---|
| STAT3 | F:5′-GGAACCTTACACCAAACAGCA-3′ R:5′-AGGGTAGAGATAGACCAGCGG-3′ | 115 | XM_042255967.1 |
| PCNA | F:5′-GTGGAGAACTTGGAAATGGAA-3′ R:5′-GAGACAGTGGAGTGGCTTATG-3′ | 153 | XM_004014340.5 |
| PPARG | F:5′-CATTTCTGCTCCGCACTAC-3′ R:5′-TGGAACCCTGACGCTTT-3′ | 234 | NM_001100921.1 |
| CEBPA | F:5′-GTGGAGACGCAACAGAAGGT-3′ R:5′-AGTTCGCGGCTCAGTTGTT-3′ | 83 | NM_001308574.1 |
| GAPDH | F:5′-AAGTTCAACGGCACAGTCA-3′ R:5′-ACCACATACTCAGCACCAGC-3′ | 125 | NM_001190390.1 |
| Target Gene | Primer Sequence (5′-3′) |
|---|---|
| CircRNA-5335 convergent | F: CGACAAAGAGGAAATAGCAAT R: ACAGGTGATGTTCTAAAGGAG |
| CircRNA-5335 divergent | F: CACACTCTTGGATTCAGCAGC R: CCTCTTTGATAGGACACTCGT |
| miRNA-125a-3p | F: CGCGACAGGTGAGGTTCTT R: AGTGCAGGGTCCGAGGTATT RT:GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCTCCC |
| U6 | F: GGAACGATACAGAGAAGATTAGC R: TGGAACGCTTCACGAATTTGCG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, W.; Ciwang, R.; Wang, L.; Zhang, S.; Liu, N.; Zhao, J.; Zhou, L.; Li, H.; Gao, X.; He, J. CircRNA-5335 Regulates the Differentiation and Proliferation of Sheep Preadipocyte via the miR-125a-3p/STAT3 Pathway. Vet. Sci. 2024, 11, 70. https://doi.org/10.3390/vetsci11020070
Guo W, Ciwang R, Wang L, Zhang S, Liu N, Zhao J, Zhou L, Li H, Gao X, He J. CircRNA-5335 Regulates the Differentiation and Proliferation of Sheep Preadipocyte via the miR-125a-3p/STAT3 Pathway. Veterinary Sciences. 2024; 11(2):70. https://doi.org/10.3390/vetsci11020070
Chicago/Turabian StyleGuo, Wei, Renzeng Ciwang, Lei Wang, Shuer Zhang, Nan Liu, Jinshan Zhao, Lisheng Zhou, Hegang Li, Xiaoxiao Gao, and Jianning He. 2024. "CircRNA-5335 Regulates the Differentiation and Proliferation of Sheep Preadipocyte via the miR-125a-3p/STAT3 Pathway" Veterinary Sciences 11, no. 2: 70. https://doi.org/10.3390/vetsci11020070
APA StyleGuo, W., Ciwang, R., Wang, L., Zhang, S., Liu, N., Zhao, J., Zhou, L., Li, H., Gao, X., & He, J. (2024). CircRNA-5335 Regulates the Differentiation and Proliferation of Sheep Preadipocyte via the miR-125a-3p/STAT3 Pathway. Veterinary Sciences, 11(2), 70. https://doi.org/10.3390/vetsci11020070




