The Evolution and Role of Molecular Tools in Measuring Diversity and Genomic Selection in Livestock Populations (Traditional and Up-to-Date Insights): A Comprehensive Exploration
Simple Summary
Abstract
1. Introduction
2. Materials and Methods (Methodology)
3. Animal Genetic Resources (AnGR) Biodiversity
4. Developments in Estimating Animal Genetic Values
5. The Potential of Genetic Diversity in Livestock Populations
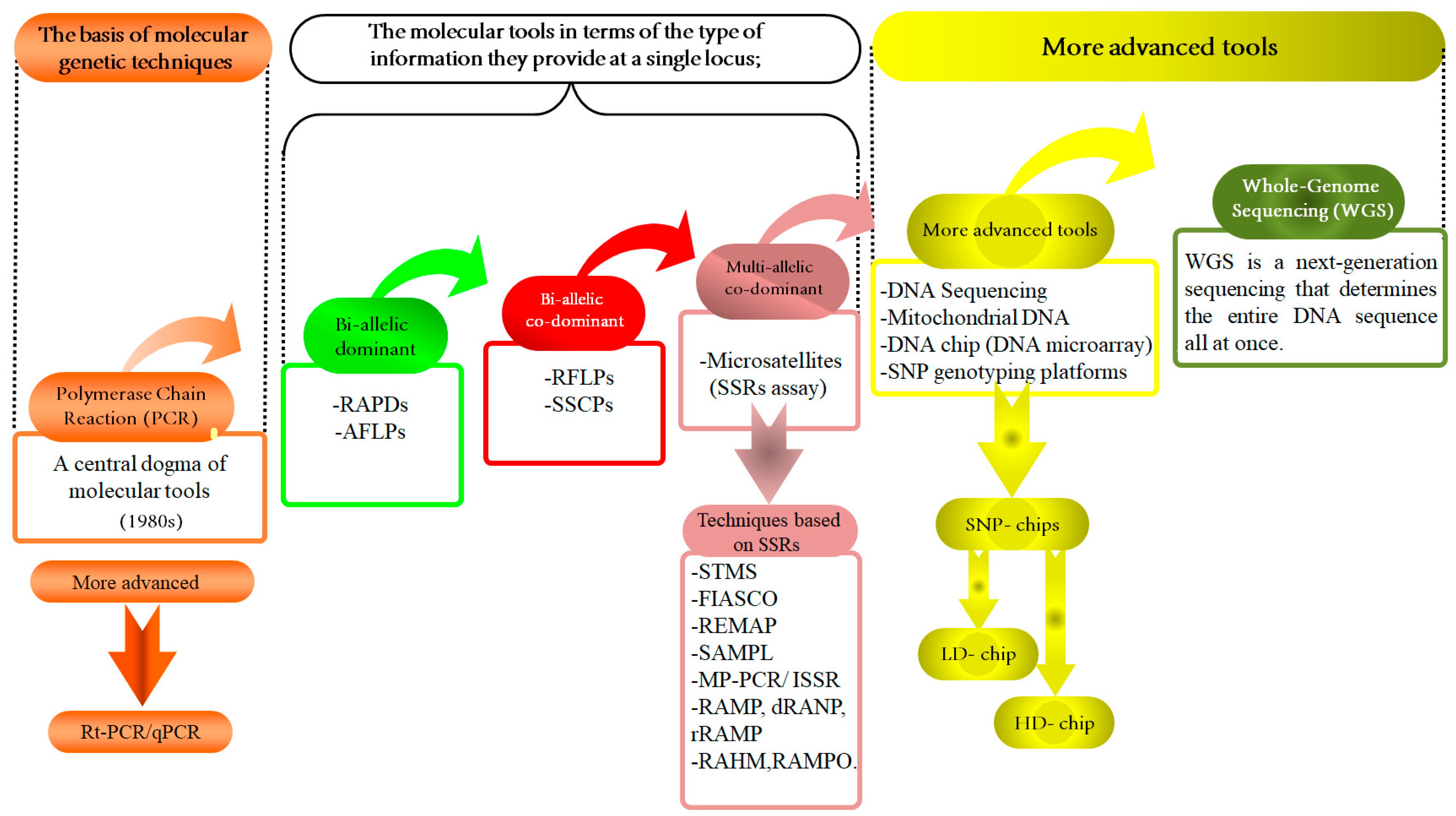
6. Advanced Molecular Techniques for Assessing Biodiversity
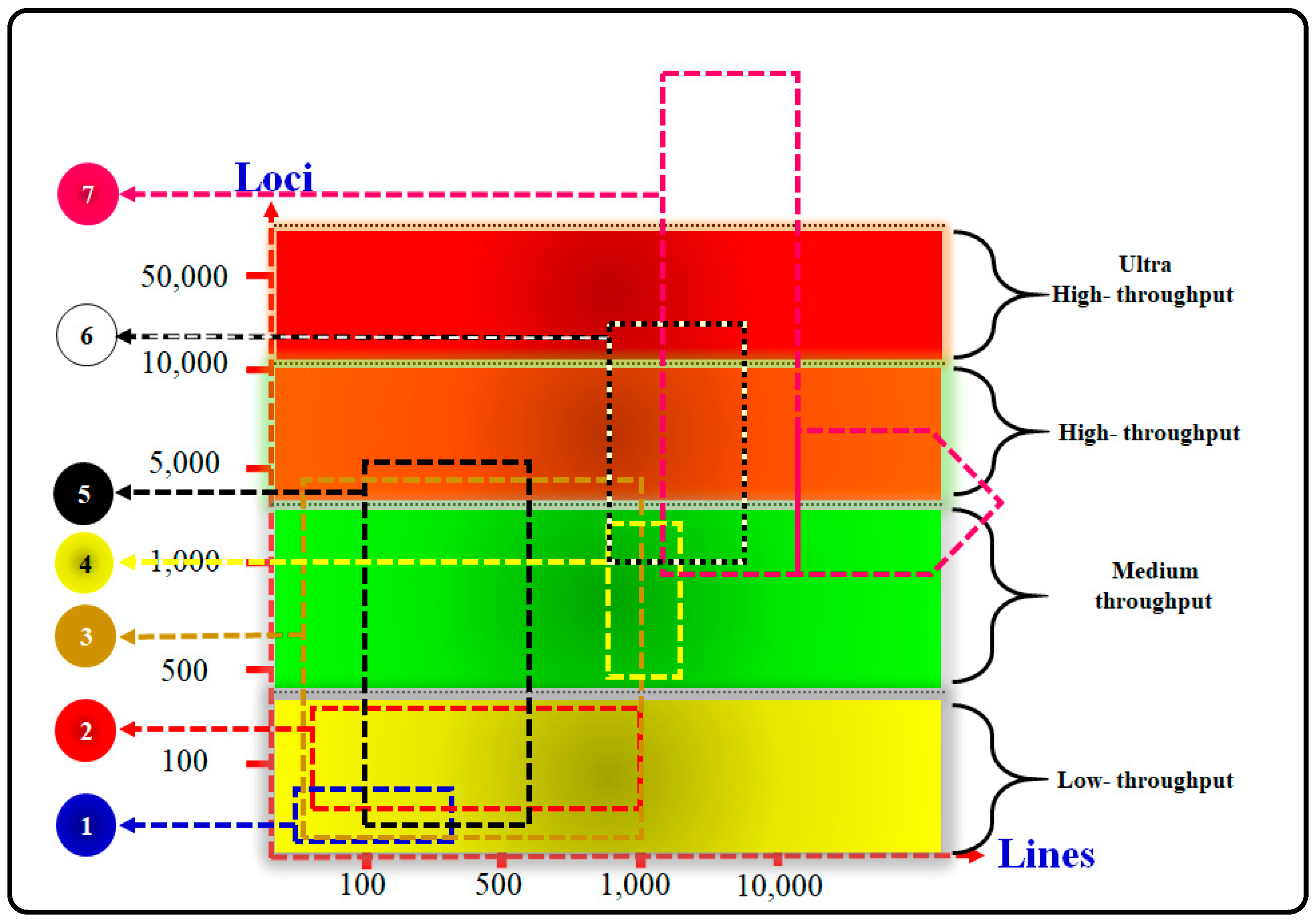
6.1. The High-Density of SNP Genotyping
6.1.1. Maxam and Gilbert Sequencing
6.1.2. Next-Generation Sequencing (NGS) “SNP Chips and Genotyping”
6.1.3. Third-Generation “Single-Molecule Real-Time (SMRT)”
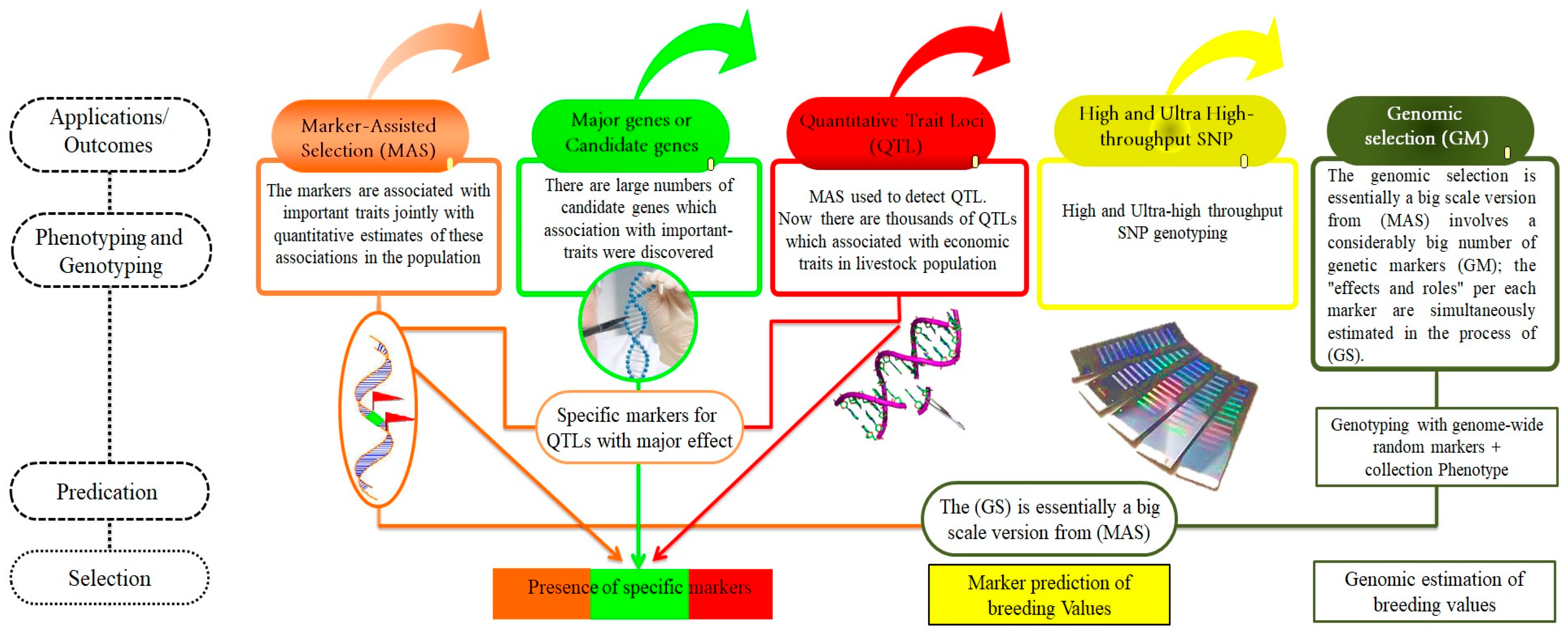
7. Outcomes of Utilizing Molecular Tools in Animal Breeding
7.1. Marker-Assisted Selection (MAS)
7.2. The Global Usefulness of High-Throughput SNP (SNP Panels)
7.3. Exploring the Mitochondrial DNA Sequence
7.4. Identification of Potential Genes Associated with Economic Traits in Livestock
7.5. Detection of QTLs
| No. | Species | No. QTLs | Publications No. | Traits | Ref. |
|---|---|---|---|---|---|
| 1 | Goat | 128 | 6 | Represent 25 different traits | [166,167,168,169,170] |
| 2 | Chicken | 16,656 | 376 | Represent 370 different traits | |
| 3 | Sheep | 4416 | 226 | Represent 266 different traits | |
| 4 | Cattle | 193,216 | 1111 | Represent 684 different traits | |
| 5 | Horse | 2636 | 106 | Represent 65 different traits | |
| 6 | Pig | 35,846 | 773 | Represent 693 different traits |
7.6. Obtaining the Whole-Genome Sequencing (WGS)
7.6.1. The Genome-Wide Studies (GWS)
7.6.2. Genome-Wide Association Studies (GWAS)
7.7. Detection of SS
7.8. The Contribution of Advanced Molecular Tools to the Detect of SS in Livestock Populations
7.9. Detection of Selective Sweeps
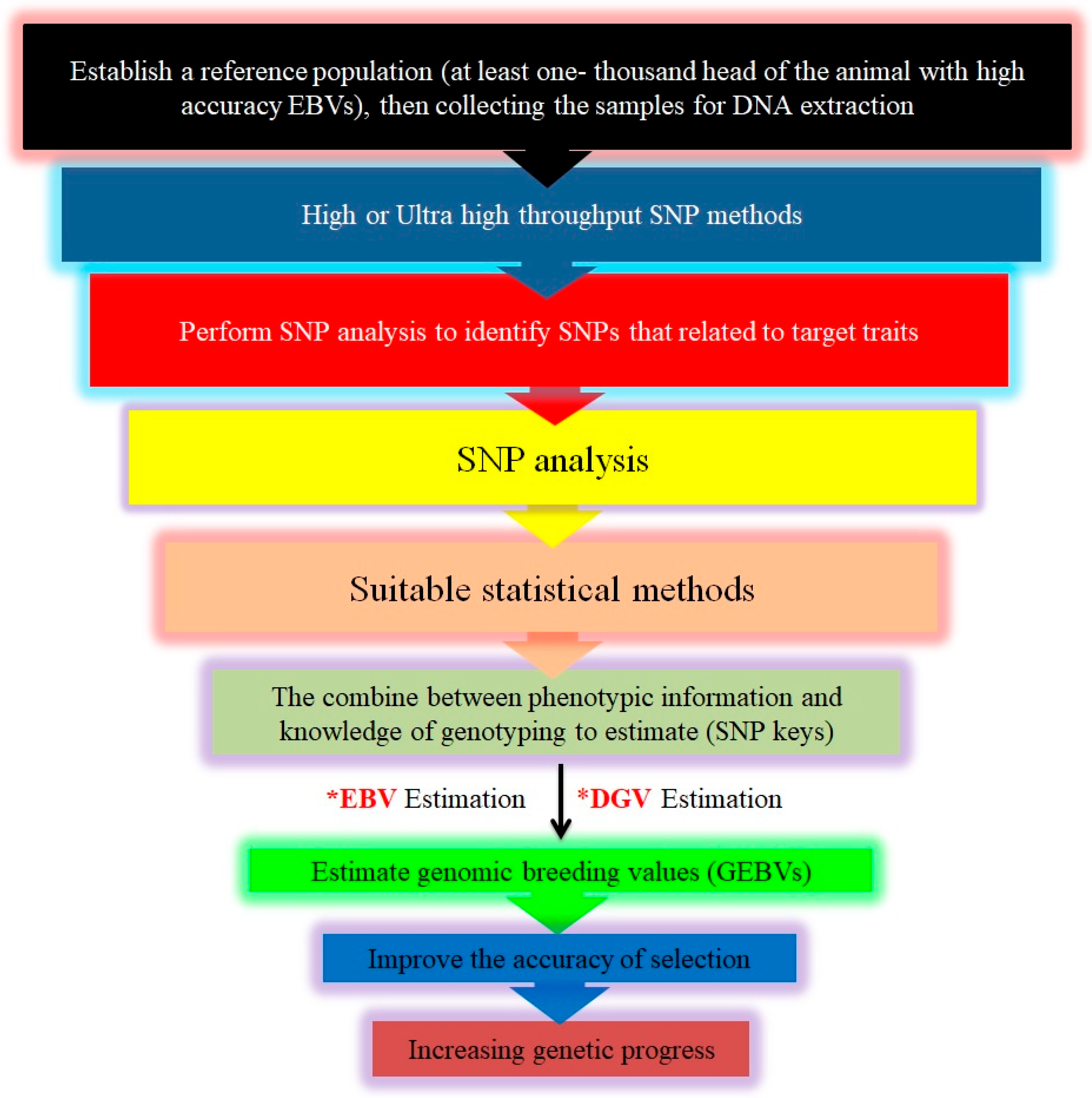
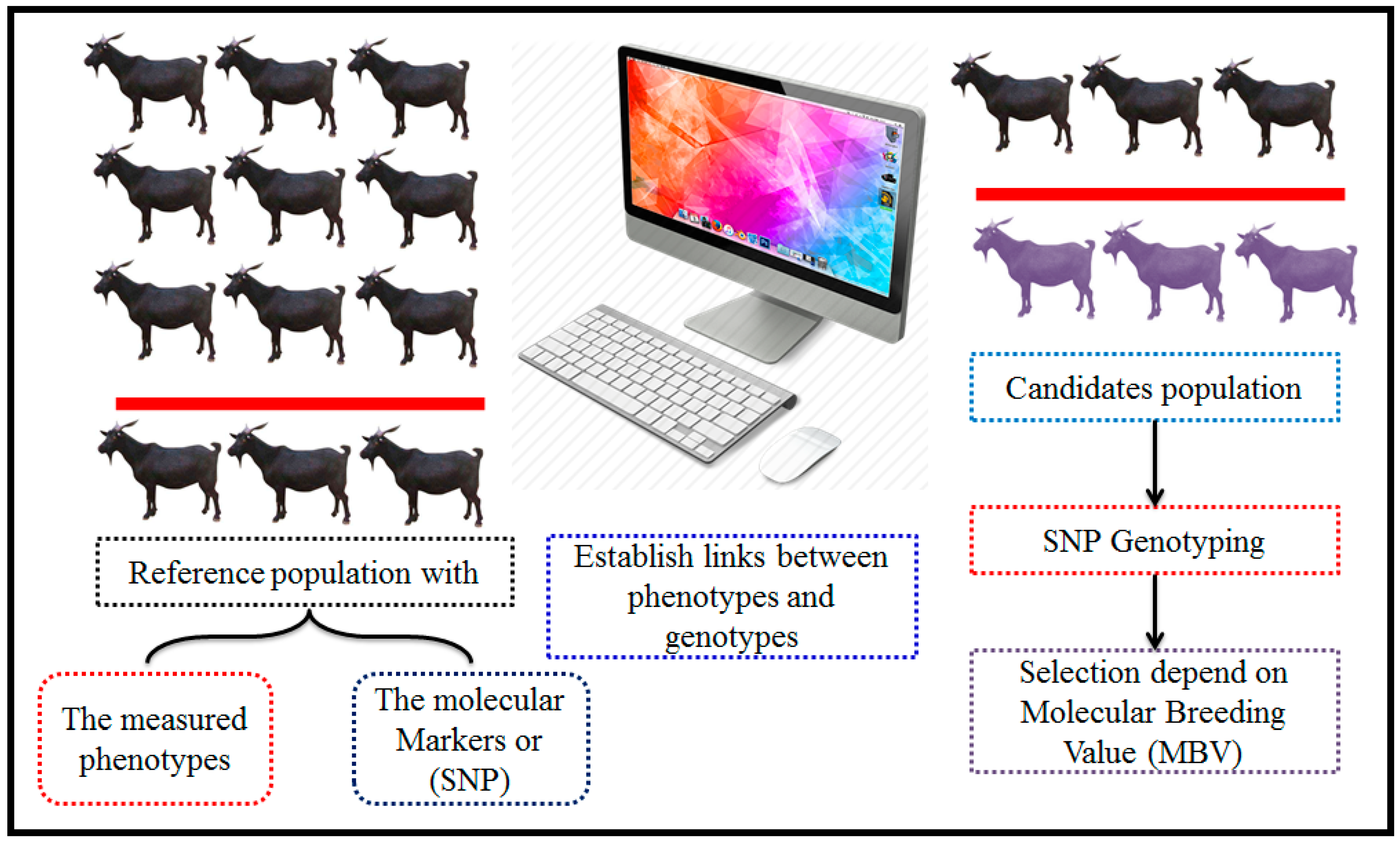
8. Genomic-Selection Applications
The Methodology of Genomic Selection
9. Future Perspectives
10. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Müller, A. The role of livestock in agroecology and sustainable food systems. In Feeding the People: Agroecology for Nourishing the World and Transforming the Agri-Food System; Chapter 6; IFOAM EU Group: Brussels, Belgium, 2015; pp. 30–33. Available online: http://www.ifoam-eu.org/sites/default/files/ifoameu_policy_ffe_feedingthepeople.pdf (accessed on 3 December 2024).
- Torpman, O.; Röös, E. Are Animals Needed for Food Supply, Efficient Resource Use, and Sustainable Cropping Systems? An Argumentation Analysis Regarding Livestock Farming. Food Ethics 2024, 9, 15. [Google Scholar] [CrossRef]
- Pacheco, A.; Banos, G.; Lambe, N.; McLaren, A.; McNeilly, T.N.; Conington, J. Genome-wide association studies of parasite resistance, productivity, and immunology traits in Scottish Blackface sheep. Animal 2024, 18, 101069. [Google Scholar] [CrossRef]
- Quigley, K.M.; Donelson, J.M. Selective breeding and promotion of naturally heat-tolerant coral reef species. In Oceanographic Processes of Coral Reefs; CRC Press: Boca Raton, FL, USA, 2024; pp. 341–357. [Google Scholar]
- Tapio, M.; Grigaliunaite, I.; Holm, L.-E.; Jeppson, S.; Kantanen, J.; Miceikiene, I.; Olsaker, I.; Viinalass, H.; Eythorsdottir, E. Mitochondrial differentiation in Northern European sheep. In Proceedings of the 7th World Congress on Genetics Applied to Livestock Production (WCGALP), Montpellier, France, 19–23 August 2002; p. 621. [Google Scholar]
- Sharma, P.; Doultani, S.; Hadiya, K.; George, L.; Highland, H. Overview of Marker-assisted Selection in Animal Breeding. J. Adv. Biol. Biotechnol. 2024, 27, 303–318. [Google Scholar] [CrossRef]
- Adebayo, O.M.; Popoola, M.A.; Kuusu, D.J.; Fanwo, R.R.; Shoyombo, A.J.; Ndiomu, E.P.; Egbeyan, J.A.; Moses, A.A. Application of Bioinformatics in Animal Breeding and Genetics: A Review. In Proceedings of the 2024 IEEE International Conference on Science, Engineering and Business for Driving Sustainable Development Goals (SEB4SDG), Omu-Aran, Nigeria, 2–4 April 2024; pp. 1–7. [Google Scholar]
- Thénard, V.; Quénon, J.; Arsenos, G.; Bailo, G.; Baptista, T.; Byrne, T.; De Barbieri, I.; Bruni, G.; Freire, F.; Theodoridis, A. Identifying selection strategies based on the practices and preferences of small ruminant farmers to improve the sustainability of their breeding systems. Animal 2024, 18, 101208. [Google Scholar] [CrossRef]
- Hossein-Zadeh, N.G. An overview of recent technological developments in bovine genomics. Vet. Anim. Sci. 2024, 25, 100382. [Google Scholar] [CrossRef]
- Abbott, K.A. Veterinary Services to Sheep Farms. In Sheep Veterinary Practice; CRC Press: Boca Raton, FL, USA, 2024; pp. 1–18. [Google Scholar]
- Ciani, E.; Burger, P.; Zappaterra, M.; Pastrana, C.I. How Early Domestication and Modern Genomics Contribute to Camel Welfare. In Dromedary Camel Behavior and Welfare: Camel Friendly Management Practices; Springer: New York, NY, USA, 2024; pp. 17–29. [Google Scholar]
- Selli, A.; Miller, S.P.; Ventura, R.V. The Use of Interactive Visualizations for Tracking Haplotypic Inheritance in Livestock. Ruminants 2024, 4, 90–111. [Google Scholar] [CrossRef]
- Boye, C.; Nirmalan, S.; Ranjbaran, A.; Luca, F. Genotype × environment interactions in gene regulation and complex traits. Nat. Genet. 2024, 56, 1057–1068. [Google Scholar] [CrossRef]
- Leimar, O. Environmental and genetic cues in the evolution of phenotypic polymorphism. Evol. Ecol. 2009, 23, 125–135. [Google Scholar] [CrossRef]
- Saleh, A.A.; Hassan, T.G.; El-Hedainy, D.K.; El-Barbary, A.S.; Sharaby, M.A.; Hafez, E.E.; Rashad, A.M. IGF-I and GH Genes polymorphism and their association with milk yields, composition and reproductive performance in Holstein–Friesian dairy cattle. BMC Vet. Res. 2024, 20, 341. [Google Scholar] [CrossRef] [PubMed]
- Saleh, A.A.; Rashad, A.M.; Hassanine, N.N.; Sharaby, M.A.; Sallam, S.M. Chapter: History of the Goat and Modern Versus Old Strategies to enhance the genetic performance. In Goat Science—From Keeping to Precision Production; IntechOpen: Rijeka, Croatia, 2023. [Google Scholar] [CrossRef]
- Safdar, M.; Kaleem, M.; Duarte, P.M.; Tazerji, S.S.; Ozaslan, M.; Hassanpour, S.; Rath, J.; Priyadarsini, S.; Rizwan, M.A. Review on optimizing dairy sector efficiency: Integrating of genetic markers with managemental techniques. Ecol. Genet. Genom. 2024, 32, 100259. [Google Scholar] [CrossRef]
- Gebreselase, H.B.; Nigussie, H.; Wang, C.; Luo, C. Genetic Diversity, Population Structure and Selection Signature in Begait Goats Revealed by Whole-Genome Sequencing. Animals 2024, 14, 307. [Google Scholar] [CrossRef] [PubMed]
- Kanthaswamy, S. Wildlife forensic genetics—Biological evidence, DNA markers, analytical approaches, and challenges. Anim. Genet. 2024, 55, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, R.Q.; del Ser, S.D.; Raschka, T.; Hofmann-Apitius, M.; Kodamullil, A.T.; Mubeen, S.; Domingo-Fernández, D. Elucidating gene expression patterns across multiple biological contexts through a large-scale investigation of transcriptomic datasets. BMC Bioinform. 2022, 23, 231. [Google Scholar] [CrossRef] [PubMed]
- Wojcik, G.L.; Graff, M.; Nishimura, K.K.; Tao, R.; Haessler, J.; Gignoux, C.R.; Highland, H.M.; Patel, Y.M.; Sorokin, E.P.; Avery, C.L.; et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature 2019, 570, 514–518. [Google Scholar] [CrossRef] [PubMed]
- Antonopoulos, A.; Gilleard, J.S.; Charlier, J. Next-generation sequencing technologies for helminth diagnostics and surveillance in ruminants: Shifting diagnostic barriers. Trends Parasitol. 2024, 40, 511–526. [Google Scholar] [CrossRef]
- Banerjee, P.; Diniz, W.J. Advancing Dairy and Beef Genetics Through Genomic Technologies. Vet. Clin. Food Anim. Pract. 2024, 40, 447–458. [Google Scholar] [CrossRef]
- Huang, J.; Lin, X. Advances in Animal Disease Resistance Research: Discoveries of Genetic Markers for Disease Resistance in Cattle through GWAS. Biol. Evid. 2024, 14, 24–31. [Google Scholar] [CrossRef]
- Plemyashov, K.; Krutikova, A.; Belikova, A.; Kuznetsova, T.; Semenov, B. Genome-Wide Association Study (GWAS) For Left Displaced Abomasum in Highly Productive Russian Holstein Cattle. Animals 2024, 14, 2795. [Google Scholar] [CrossRef]
- Hidalgo, J.; Tsuruta, S.; Gonzalez, D.; de Oliveira, G.; Sanchez, M.; Kulkarni, A.; Przybyla, C.; Vargas, G.; Vukasinovic, N.; Misztal, I. Converting estimated breeding values from the observed to probability scale for health traits. J. Dairy Sci. 2024, 107, 9628–9637. [Google Scholar] [CrossRef]
- Akdemir, D.; Beavis, W.; Fritsche-Neto, R.; Singh, A.K.; Isidro-Sánchez, J. Multi-objective optimized genomic breeding strategies for sustainable food improvement. Heredity 2019, 122, 672–683. [Google Scholar] [CrossRef]
- Cole, J.B.; Dürr, J.W.; Nicolazzi, E.L. Invited review: The future of selection decisions and breeding programs: What are we breeding for, and who decides? J. Dairy Sci. 2021, 104, 5111–5124. [Google Scholar] [CrossRef] [PubMed]
- Brito, L.F.; Bedere, N.; Douhard, F.; Oliveira, H.R.; Arnal, M.; Peñagaricano, F.; Schinckel, A.P.; Baes, C.F.; Miglior, F. Review: Genetic selection of high-yielding dairy cattle toward sustainable farming systems in a rapidly changing world. Animal 2021, 15, 100292. [Google Scholar] [CrossRef] [PubMed]
- Jones, H.E.; Wilson, P.B. Progress and opportunities through use of genomics in animal production. Trends Genet. 2022, 38, 1228–1252. [Google Scholar] [CrossRef]
- Sanchez, M.-P.; Tribout, T.; Kadri, N.K.; Chitneedi, P.K.; Maak, S.; Hozé, C.; Boussaha, M.; Croiseau, P.; Philippe, R.; Spengeler, M.; et al. Sequence-based GWAS meta-analyses for beef production traits. Genet. Sel. Evol. 2023, 55, 70. [Google Scholar] [CrossRef]
- Mukhopadhyay, C.S.; Kaur, B. NGS-Based Biomarkers in Livestock. In Biotechnological Interventions Augmenting Livestock Health and Production; Mukhopadhyay, C.S., Choudhary, R.K., Panwar, H., Malik, Y.S., Eds.; Springer Nature: Singapore, 2023; pp. 107–148. [Google Scholar]
- Khan, R.; Li, A.; Raza, S.H.A. Editorial: Genetic Regulation of Meat Quality Traits in Livestock Species. Front. Genet. 2023, 13, 1092562. [Google Scholar] [CrossRef]
- Saleh, A.A.; Rashad, A.M.A.; Hassanine, N.; Sharaby, M.A.; Zhao, Y. Assessment of hair and cashmere properties and their genetic background of several goat breeds in Southwest China. Sci. Rep. 2022, 12, 11135. [Google Scholar] [CrossRef] [PubMed]
- Saleh, A.A.; Xue, L.; Zhao, Y. Screening Indels from the whole genome to identify the candidates and their association with economic traits in several goat breeds. Funct. Integr. Genom. 2023, 23, 58. [Google Scholar] [CrossRef] [PubMed]
- Saleh, A.A.; Hassanine, N.N.A.M. Egyptian Sheep Breeds and Genetic Tools to Improve; LAMBERT Academic Publishing: Heidelberg, Germany, 2019. [Google Scholar] [CrossRef]
- Sonstegard, T.S.; Flórez, J.M.; Garcia, J.F. Commercial perspectives: Genome editing as a breeding tool for health and well-being in dairy cattle. JDS Commun. 2024, 5, 767–771. [Google Scholar] [CrossRef]
- Tuggle, C.K.; Clarke, J.L.; Murdoch, B.M.; Lyons, E.; Scott, N.M.; Beneš, B.; Campbell, J.D.; Chung, H.; Daigle, C.L.; Das Choudhury, S. Current challenges and future of agricultural genomes to phenomes in the USA. Genome Biol. 2024, 25, 8. [Google Scholar] [CrossRef]
- Martinell, D.P. New technologies as a means to achieve sustainability. In An Introduction to Sustainable Aquaculture; Routledge: London, UK, 2024; pp. 286–314. [Google Scholar]
- Saleh, A.A.; Easa, A.A.; El-Hedainy, D.K.; Rashad, A.M.A. Prediction of some milk production traits using udder and teat measurements with a spotlight on their genetic background in Friesian cows. Sci. Rep. 2023, 13, 16193. [Google Scholar] [CrossRef]
- Rehman, S.U.; Zhen, Y.; Ding, L.; Saleh, A.A.; Zhang, Y.; Zhang, J.; He, F.; Husien, H.M.; Zhou, P.; Wang, M. Integrative Meta-Analysis: Unveiling Genetic Factors in Meat Sheep Growth and Muscular Development through QTL and Transcriptome Studies. Animals 2024, 14, 1679. [Google Scholar] [CrossRef]
- Saleh, A.A.; Rashad, A.; Hassanine, N.A.M.; Sharaby, M.A. Modern Strategies to Enhance Goat Genetic Performance; LAP LAMBERT Acad. Publishing: Saarbrücken, Germany, 2019; ISBN 978-620-0-47020-1. EAN: 9786200470201; Available online: https://www.morebooks.de/shop-ui/shop/product/978-620-0-47020-1 (accessed on 3 December 2024). [CrossRef]
- Hall, S.J.; Bradley, D.G. Conserving livestock breed biodiversity. Trends Ecol. Evol. 1995, 10, 267–270. [Google Scholar] [CrossRef]
- Moazami-Goudarzi, K.; Furet, J.; Grosclaude, F.; Laloë, D. Analysis of genetic relationships between 10 cattle breeds with 17 microsatellites. Anim. Genet. 1997, 28, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Pearson, W.R.; Lipman, D.J. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 1988, 85, 2444–2448. [Google Scholar] [CrossRef] [PubMed]
- Bruford, M.W.; Ginja, C.; Hoffmann, I.; Joost, S.; Orozco-terWengel, P.; Alberto, F.J.; Amaral, A.J.; Barbato, M.; Biscarini, F.; Colli, L. Prospects and challenges for the conservation of farm animal genomic resources, 2015–2025. Front. Genet. 2015, 6, 314. [Google Scholar] [CrossRef] [PubMed]
- Thornton, P.K. Livestock production: Recent trends, future prospects. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 2853–2867. [Google Scholar] [CrossRef]
- Rupp, R.; Mucha, S.; Larroque, H.; McEwan, J.; Conington, J. Genomic application in sheep and goat breeding. Anim. Front. 2016, 6, 39–44. [Google Scholar] [CrossRef]
- Dong, Y.; Xie, M.; Jiang, Y.; Xiao, N.; Du, X.; Zhang, W.; Tosser-Klopp, G.; Wang, J.; Yang, S.; Liang, J. Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus). Nat. Biotechnol. 2013, 31, 135. [Google Scholar] [CrossRef]
- Jiang, Y.; Xie, M.; Chen, W.; Talbot, R.; Maddox, J.F.; Faraut, T.; Wu, C.; Muzny, D.M.; Li, Y.; Zhang, W. The sheep genome illuminates biology of the rumen and lipid metabolism. Science 2014, 344, 1168–1173. [Google Scholar] [CrossRef]
- Sharma, A.; Park, J.-E.; Chai, H.-H.; Jang, G.-W.; Lee, S.-H.; Lim, D. Next generation sequencing in livestock species: A review. J. Anim. Breed. Genom. JABG 2017, 1, 23–30. [Google Scholar]
- Patel, S.M.; Koringa, P.G.; Nathani, N.M.; Patel, N.V.; Shah, T.M.; Joshi, C.G. Exploring genetic polymorphism in innate immune genes in Indian cattle (Bos indicus) and buffalo (Bubalus bubalis) using next generation sequencing technology. Meta Gene 2015, 3, 50–58. [Google Scholar] [CrossRef]
- Taylor, J.F.; McKay, S.D.; Rolf, M.M.; Ramey, H.R.; Decker, J.E.; Schnabel, R.D. Genomic selection in beef cattle. Bov. Genom. 2012, 2012, 211–233. [Google Scholar]
- Adams, M.D.; Celniker, S.E.; Holt, R.A.; Evans, C.A.; Gocayne, J.D.; Amanatides, P.G.; Scherer, S.E.; Li, P.W.; Hoskins, R.A.; Galle, R.F. The genome sequence of Drosophila melanogaster. Science 2000, 287, 2185–2195. [Google Scholar] [CrossRef]
- Guénet, J.L. The mouse genome. Genom. Res. 2005, 15, 1729–1740. [Google Scholar] [CrossRef] [PubMed]
- Ostrander, E.A.; Lindblad-Toh, K.; Lander, E.S. Sequencing the Genome of the Domestic Dog Canis Familiaris. Available online: http://www.genome.gov/Pages/Research/Sequencing/SeqProposals/CanineSEQedited.pdf (accessed on 16 June 2024).
- Schmid, M.; Nanda, I.; Burt, D.W. Second report on chicken genes and chromosomes 2005. Cytogenet. Genome Res. 2005, 109, 415–479. [Google Scholar]
- Rat Genome Sequencing Consortium; de Jong, P.J.; Osoegawa, K.; Zhu, B.; Marra, M.; Schein, J.; Bosdet, I.; Fjell, C. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 2004, 428, 493–521. [Google Scholar]
- Sodergren, E.; Weinstock, G.M.; Davidson, E.H.; Cameron, R.A.; Gibbs, R.A.; Angerer, R.C.; Angerer, L.M.; Arnone, M.I.; Burgess, D.R.; Burke, R.D. The genome of the sea urchin Strongylocentrotus purpuratus. Science 2006, 314, 941–952. [Google Scholar] [PubMed]
- Venkatesh, B.; Kirkness, E.F.; Loh, Y.H.; Halpern, A.L.; Lee, A.P.; Johnson, J.; Dandona, N.; Viswanathan, L.D.; Tay, A.; Venter, J.C. Survey sequencing and comparative analysis of the elephant shark (Callorhinchus milii) genome. PLoS Biol. 2007, 5, e101. [Google Scholar] [CrossRef]
- Rhesus Macaque Genome Sequencing and Analysis Consortium. The Rhesus macaque genome sequence informs biomedical and evolutionary analyses. Science 2007, 316, 222–234. [Google Scholar] [CrossRef]
- Pontius, J.U.; Mullikin, J.C.; Smith, D.R.; Lindblad-Toh, K.; Gnerre, S.; Clamp, M.; Chang, J.; Stephens, R.; Neelam, B.; Volfovsky, N. Initial sequence and comparative analysis of the cat genome. Genome Res. 2007, 17, 1675–1689. [Google Scholar] [CrossRef] [PubMed]
- Warren, W.C.; Gruetzner, F. The enigma of the platypus genome. Aust. J. Zool. 2009, 57, 157–165. [Google Scholar] [CrossRef]
- Groenen, M.A.; Archibald, A.L.; Uenishi, H.; Tuggle, C.K.; Takeuchi, Y.; Rothschild, M.F.; Rogel-Gaillard, C.; Park, C.; Milan, D.; Megens, H.J. Analyses of pig genomes provide insight into porcine demography and evolution. Nature 2012, 491, 393–398. [Google Scholar] [CrossRef]
- International Sheep Genomics Consortium; Archibald, A.; Cockett, N.; Dalrymple, B.; Faraut, T.; Kijas, J.; Maddox, J.; McEwan, J.; Hutton Oddy, V.; Raadsma, H. The sheep genome reference sequence: A work in progress. Anim. Genet. 2010, 41, 449–453. [Google Scholar] [CrossRef] [PubMed]
- Heaton, M.P.; Smith, T.P.; Bickhart, D.M.; Vander Ley, B.L.; Kuehn, L.A.; Oppenheimer, J.; Shafer, W.R.; Schuetze, F.T.; Stroud, B.; McClure, J.C. A reference genome assembly of Simmental cattle, Bos taurus taurus. J. Hered. 2021, 112, 184–191. [Google Scholar] [CrossRef]
- Wade, C.; Giulotto, E.; Sigurdsson, S.; Zoli, M.; Gnerre, S.; Imsland, F.; Lear, T.; Adelson, D.; Bailey, E.; Bellone, R. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 2009, 326, 865–867. [Google Scholar] [CrossRef]
- Hellsten, U.; Harland, R.M.; Gilchrist, M.J.; Hendrix, D.; Jurka, J.; Kapitonov, V.; Ovcharenko, I.; Putnam, N.H.; Shu, S.; Taher, L.; et al. The genome of the Western clawed frog Xenopus tropicalis. Science 2010, 328, 633–636. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Fan, W.; Tian, G.; Zhu, H.; He, L.; Cai, J.; Huang, Q.; Cai, Q.; Li, B.; Bai, Y.; et al. The sequence and de novo assembly of the giant panda genome. Nature 2010, 463, 311–317. [Google Scholar] [CrossRef]
- Backström, N.; Forstmeier, W.; Schielzeth, H.; Mellenius, H.; Nam, K.; Bolund, E.; Webster, M.T.; Ost, T.; Schneider, M.; Kempenaers, B.; et al. The recombination landscape of the zebra finch Taeniopygia guttata genome. Genome Res. 2010, 20, 485–495. [Google Scholar] [CrossRef]
- Dalloul, R.A.; Long, J.A.; Zimin, A.V.; Aslam, L.; Beal, K.; Blomberg, L.A.; Bouffard, P.; Burt, D.W.; Crasta, O.; Crooijmans, R.P.; et al. Multi-Platform Next-Generation Sequencing of the Domestic Turkey (Meleagris gallopavo): Genome Assembly and Analysis. PLoS Biol. 2010, 8, e1000475. [Google Scholar] [CrossRef]
- The Bactrian Camels Genome Sequencing and Analysis Consortium. Genome sequences of wild and domestic bactrian camels. Nat. Commun. 2012, 3, 1202. [Google Scholar] [CrossRef]
- Wang, C.; Webley, L.; Wei, K.-j.; Wakefield, M.J.; Patel, H.R.; Deakin, J.E.; Alsop, A.; Marshall Graves, J.A.; Cooper, D.W.; Nicholas, F.W. A second-generation anchored genetic linkage map of the tammar wallaby (Macropus eugenii). BMC Genet. 2011, 12, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Alföldi, J.; Di Palma, F.; Grabherr, M.; Williams, C.; Kong, L.; Mauceli, E.; Russell, P.; Lowe, C.B.; Glor, R.E.; Jaffe, J.D.; et al. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 2011, 477, 587–591. [Google Scholar] [CrossRef] [PubMed]
- Star, B.; Nederbragt, A.J.; Jentoft, S.; Grimholt, U.; Malmström, M.; Gregers, T.F.; Rounge, T.B.; Paulsen, J.; Solbakken, M.H.; Sharma, A.; et al. The genome sequence of Atlantic cod reveals a unique immune system. Nature 2011, 477, 207–210. [Google Scholar] [CrossRef]
- Kim, E.B.; Fang, X.; Fushan, A.A.; Huang, Z.; Lobanov, A.V.; Han, L.; Marino, S.M.; Sun, X.; Turanov, A.A.; Yang, P.; et al. Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature 2011, 479, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Marsh, H.L. Apes in the Information Age: An Investigation of Information Management by Orangutans (Pongo abelii). Ph.D. Thesis, York University, Toronto, ON, Canada, 2012. [Google Scholar]
- Wu, H.; Luo, L.-Y.; Zhang, Y.-H.; Zhang, C.-Y.; Huang, J.-H.; Mo, D.-X.; Zhao, L.-M.; Wang, Z.-X.; Wang, Y.-C.; He-Hua, E.E.; et al. Telomere-to-telomere genome assembly of a male goat reveals variants associated with cashmere traits. Nat. Commun. 2024, 15, 10041. [Google Scholar] [CrossRef] [PubMed]
- Burger, P.A. The history of Old World camelids in the light of molecular genetics. Trop. Anim. Health Prod. 2016, 48, 905–913. [Google Scholar] [CrossRef]
- Batra, S.S.; Levy-Sakin, M.; Robinson, J.; Guillory, J.; Durinck, S.; Vilgalys, T.P.; Kwok, P.-Y.; Cox, L.A.; Seshagiri, S.; Song, Y.S.; et al. Accurate assembly of the olive baboon (Papio anubis) genome using long-read and Hi-C data. GigaScience 2020, 9, 1–13. [Google Scholar] [CrossRef]
- Yin, Z.; Zhang, F.; Smith, J.; Kuo, R.; Hou, Z.-C. Full-length transcriptome sequencing from multiple tissues of duck, Anas platyrhynchos. Sci. Data 2019, 6, 275. [Google Scholar] [CrossRef]
- Amemiya, C.T.; Alföldi, J.; Lee, A.P.; Fan, S.; Philippe, H.; MacCallum, I.; Braasch, I.; Manousaki, T.; Schneider, I.; Rohner, N.; et al. The African coelacanth genome provides insights into tetrapod evolution. Nature 2013, 496, 311–316. [Google Scholar] [CrossRef]
- Smith, J.J.; Kuraku, S.; Holt, C.; Sauka-Spengler, T.; Jiang, N.; Campbell, M.S.; Yandell, M.D.; Manousaki, T.; Meyer, A.; Bloom, O.E.; et al. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat. Genet. 2013, 45, 415–421. [Google Scholar] [CrossRef]
- Vonk, F.J.; Casewell, N.R.; Henkel, C.V.; Heimberg, A.M.; Jansen, H.J.; McCleary, R.J.; Kerkkamp, H.M.; Vos, R.A.; Guerreiro, I.; Calvete, J.J.; et al. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc. Natl. Acad. Sci. USA 2013, 110, 20651–20656. [Google Scholar] [CrossRef]
- Williamson, N.G.; Walsh, C.M.; Kijimoto, T. Comparative metabolomic analysis of polyphenic horn development in the dung beetle Onthophagus taurus. PLoS ONE 2022, 17, e0265222. [Google Scholar] [CrossRef]
- Ho, C.W.; Chen, J.J.-W.; Lee, T.H.; Lin, H.J. Complete mitochondrial genome of Oncorhynchus masou formosanus (Jordan & Oshima, 1919) (Pisces, Salmonidae). Mitochondrial DNA Part B 2016, 1, 295–296. [Google Scholar]
- Saleh, A.A.; Hammoud, M.; Dabour, N.A.; Hafez, E.; Sharaby, M.A. BMPR-1B, BMP-15 and GDF-9 genes structure and their relationship with litter size in six sheep breeds reared in Egypt. BMC Res. Notes 2020, 13, 119–126. [Google Scholar] [CrossRef]
- Saleh, A.A.; Rashad, A.M.; Hassanine, N.N.; Sharaby, M.A.; Zhao, Y. Comparative analysis of IGFBP-3 gene sequence in Egyptian sheep, cattle, and buffalo. BMC Res. Notes 2019, 12, 623. [Google Scholar] [CrossRef] [PubMed]
- Benjelloun, B.; Alberto, F.J.; Streeter, I.; Boyer, F.; Coissac, E.; Stucki, S.; Ben Bati, M.; Ibnelbachyr, M.; Chentouf, M.; Bechchari, A.; et al. Characterizing neutral genomic diversity and selection signatures in indigenous populations of Moroccan goats (Capra hircus) using WGS data. Front. Genet. 2015, 6, 107. [Google Scholar] [CrossRef] [PubMed]
- Snyder, M.; Du, J.; Gerstein, M. Personal genome sequencing: Current approaches and challenges. Genes Dev. 2010, 24, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Schaeffer, L. Strategy for applying genome-wide selection in dairy cattle. J. Anim. Breed. Genet. 2006, 123, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Dekkers, C.M.J. Application of genomics tools to animal breeding. Curr. Genom. 2012, 13, 207–212. [Google Scholar] [CrossRef]
- Saleh, A.A. Utilization of Molecular Markers to Detect some Genes and Mutations Affecting Economic Traits in Egyptian Sheep Breeds. Figshare 2016, 1, 24–55. [Google Scholar] [CrossRef]
- Saleh, A.A.; Rashad, A.; Hassanine, N.N.A.M.; Sharaby, M.A.; Zhao, Y. Traditional Versus Modern Methods for Fertility Evaluation. Figshare 2020, 7, 37–69. [Google Scholar] [CrossRef]
- Tosser-Klopp, G.; Bardou, P.; Bouchez, O.; Cabau, C.; Crooijmans, R.; Dong, Y.; Donnadieu-Tonon, C.; Eggen, A.; Heuven, H.C.; Jamli, S.; et al. Design and characterization of a 52K SNP chip for goats. PLoS ONE 2014, 9, e86227. [Google Scholar] [CrossRef] [PubMed]
- Munoz, P.R.; Resende, M.F.R.; Huber, D.A.; Quesada, T.; Resende, M.D.V.; Neale, D.B.; Wegrzyn, J.L.; Kirst, M.; Peter, G.F. Genomic relationship matrix for correcting pedigree errors in breeding populations: Impact on genetic parameters and genomic selection accuracy. Crop Sci. 2014, 54, 1115–1123. [Google Scholar] [CrossRef]
- Michelmore, R.; Hulbert, S. Molecular markers for genetic analysis of phytopathogenic fungi. Annu. Rev. Phytopathol. 1987, 25, 383–404. [Google Scholar] [CrossRef]
- Kantanen, J.; Vilkki, J.; Elo, K.; Mäki-Tanila, A. Random amplified polymorphic DNA in cattle and sheep: Application for detecting genetic variation. Anim. Genet. 1995, 26, 315–320. [Google Scholar] [CrossRef]
- Ogedengbe, M.; Barta, J.; Ogedengbe, J.; Akanbi, B.; Ogo, I. Use of a Sequence Characterized Amplified Regions (SCARS)-Multiplex PCR method to identify Eimeria species of chickens from widely distributed geographic areas. Trop. Vet. 2009, 27, 36–44. [Google Scholar]
- Fukuoka, S.; Inoue, T.; Miyao, A.; Monna, L.; Zhong, H.S.; Sasaki, T.; Minobe, Y. Mapping of sequence-tagged sites in rice by single strand conformation polymorphism. DNA Res. 1994, 1, 271–277. [Google Scholar] [CrossRef]
- Kemp, S.J.; Teale, A. Randomly primed PCR amplification of pooled DNA reveals polymorphism in a ruminant repetitive DNA sequence which differentiates Bos indicus and B. taurus. Anim. Genet. 1994, 25, 83–88. [Google Scholar] [CrossRef]
- Ganai, T.; Singh, R.; Butchiah, G. DNA amplification fingerprinting of cattle and buffalo genome by RAPD-PCR utilizing arbitrary oligonucleotide primers. Buff. J. 2000, 16, 331–339. [Google Scholar]
- Leroux, C.; Martin, P.; MAHÉ, M.F.; Levéziel, H.; MERCIER, J.C. Restriction fragment length polymorphism identification of goat αs1-casein alleles: A potential tool in selection of individuals carrying alleles associated with a high-level protein synthesis. Anim. Genet. 1990, 21, 341–351. [Google Scholar] [CrossRef]
- Fearnley, C.; On, S.L.; Kokotovic, B.; Manning, G.; Cheasty, T.; Newell, D.G. Application of fluorescent amplified fragment length polymorphism for comparison of human and animal isolates of Yersinia enterocolitica. Appl. Environ. Microbiol. 2005, 71, 4960–4965. [Google Scholar] [CrossRef]
- Knapik, E.W.; Goodman, A.; Atkinson, O.S.; Roberts, C.T.; Shiozawa, M.; Sim, C.U.; Weksler-Zangen, S.; Trolliet, M.R.; Futrell, C.; Innes, B.A. A reference cross DNA panel for zebrafish (Danio rerio) anchored with simple sequence length polymorphisms. Development 1996, 123, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Zamani, P.; Akhondi, M.; Mohammadabadi, M. Associations of inter-simple sequence repeat loci with predicted breeding values of body weight in sheep. Small Rumin. Res. 2015, 132, 123–127. [Google Scholar] [CrossRef]
- Je, S.; Ku, B.K.; Jeon, B.-Y.; Kim, J.-M.; Jung, S.-C.; Cho, S.-N. Extent of Mycobacterium bovis transmission among animals of dairy and beef cattle and deer farms in South Korea determined by variable-number tandem repeats typing. Vet. Microbiol. 2015, 176, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Morgan, U.M.; Constantine, C.C.; O’Donoghue, P.; Meloni, B.P.; O’Brien, P.A.; Thompson, R.A. Molecular characterization of Cryptosporidium isolates from humans and other animals using random amplified polymorphic DNA analysis. Am. J. Trop. Med. Hyg. 1995, 52, 559–564. [Google Scholar] [CrossRef][Green Version]
- Brandeis, M.; Kafri, T.; Ariel, M.; Chaillet, J.; McCarrey, J.; Razin, A.; Cedar, H. The ontogeny of allele-specific methylation associated with imprinted genes in the mouse. EMBO J. 1993, 12, 3669–3677. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, L.; Zhan, Q.; Chen, Y. Establishment of Sorghum bicolor expressed sequence tag-simple sequence repeat (EST-SSR) marker and its preliminary application to Sorghum sudanense. Pratacult. Sci. 2010, 27, 112–117. [Google Scholar]
- Talenti, A.; Nicolazzi, E.; Chessa, S.; Frattini, S.; Moretti, R.; Coizet, B.; Nicoloso, L.; Colli, L.; Pagnacco, G.; Stella, A. A method for single nucleotide polymorphism selection for parentage assessment in goats. J. Dairy Sci. 2016, 99, 3646–3653. [Google Scholar] [CrossRef]
- Chen, J.; Hu, Q.; Zhang, Y.; Lu, C.; Kuang, H. P-MITE: A database for plant miniature inverted-repeat transposable elements. Nucleic Acids Res. 2014, 42, D1176–D1181. [Google Scholar] [CrossRef]
- Qureshi, M.I.; Sabir, J.; Mutawakil, M.; El Hanafy, A.; Ashmaoui, H.; Ramadan, H.; Anwar, Y.; Sadek, A.; Alsoud, M.; Saini, K.S. Review of modern strategies to enhance livestock genetic performance: From molecular markers to next-generation sequencing technologies in goats. J. Food Agric. Environ. 2014, 12, 5. [Google Scholar]
- Vignal, A.; Milan, D.; SanCristobal, M.; Eggen, A. A review on SNP and other types of molecular markers and their use in animal genetics. Genet. Sel. Evol. 2002, 34, 275. [Google Scholar] [CrossRef] [PubMed]
- Garafutdinov, R.R.; Sakhabutdinova, A.R.; Slominsky, P.A.; Aminev, F.G.; Chemeris, A.V. A new digital approach to SNP encoding for DNA identification. Forensic Sci. Int. 2020, 317, 110520. [Google Scholar] [CrossRef] [PubMed]
- Maxam, A.; Gilbert, W. A new method for sequencing DNA. Proc. Natl. Acad. Sci. USA 1977, 74, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Pariset, L.; Cuteri, A.; Ligda, C.; Ajmone-Marsan, P.; Valentini, A. Geographical patterning of sixteen goat breeds from Italy, Albania and Greece assessed by single nucleotide polymorphisms. BMC Ecol. 2009, 9, 20. [Google Scholar] [CrossRef] [PubMed]
- Talenti, A.; Palhière, I.; Tortereau, F.; Pagnacco, G.; Stella, A.; Nicolazzi, E.L.; Crepaldi, P.; Tosser-Klopp, G. Functional SNP panel for parentage assessment and assignment in worldwide goat breeds. Genet. Sel. Evol. 2018, 50, 55. [Google Scholar] [CrossRef] [PubMed]
- International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature 2007, 449, 851. [Google Scholar] [CrossRef]
- International Chicken Polymorphism Map Consortium. A genetic variation map for chicken with 2.8 million single-nucleotide polymorphisms. Nature 2004, 432, 717. [Google Scholar] [CrossRef]
- Lindblad-Toh, K.; Wade, C.M.; Mikkelsen, T.S.; Karlsson, E.K.; Jaffe, D.B.; Kamal, M.; Clamp, M.; Chang, J.L.; Kulbokas III, E.J.; Zody, M.C. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 2005, 438, 803. [Google Scholar]
- Frazer, K.A.; Eskin, E.; Kang, H.M.; Bogue, M.A.; Hinds, D.A.; Beilharz, E.J.; Gupta, R.V.; Montgomery, J.; Morenzoni, M.M.; Nilsen, G.B. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature 2007, 448, 1050. [Google Scholar] [CrossRef]
- Van Tassell, C.P.; Smith, T.P.; Matukumalli, L.K.; Taylor, J.F.; Schnabel, R.D.; Lawley, C.T.; Haudenschild, C.D.; Moore, S.S.; Warren, W.C.; Sonstegard, T.S. SNP discovery and allele frequency estimation by deep sequencing of reduced representation libraries. Nat. Methods 2008, 5, 247. [Google Scholar] [CrossRef]
- Charlier, C.; Coppieters, W.; Rollin, F.; Desmecht, D.; Agerholm, J.S.; Cambisano, N.; Carta, E.; Dardano, S.; Dive, M.; Fasquelle, C. Highly effective SNP-based association mapping and management of recessive defects in livestock. Nat. Genet. 2008, 40, 449. [Google Scholar] [CrossRef] [PubMed]
- Castañeda-Rico, S.; León-Paniagua, L.; Edwards, C.W.; Maldonado, J.E. Ancient DNA from museum specimens and next generation sequencing help resolve the controversial evolutionary history of the critically endangered Puebla deer mouse. Front. Ecol. Evol. 2020, 8, 94. [Google Scholar] [CrossRef]
- Wu, F.; Sun, H.; Lu, S.; Gou, X.; Yan, D.; Xu, Z.; Zhang, Z.; Qadri, Q.R.; Zhang, Z.; Wang, Z. Genetic diversity and selection signatures within Diannan Small-Ear pigs revealed by next-generation sequencing. Front. Genet. 2020, 11, 733. [Google Scholar] [CrossRef]
- Gu, J.; Li, S. Next-generation sequencing of the complete mitochondrial genome of the Nixi chicken (Gallus gallus). Mitochondrial DNA Part B 2020, 5, 3271–3272. [Google Scholar] [CrossRef]
- Lee, J.J.; Wedow, R.; Okbay, A.; Kong, E.; Maghzian, O.; Zacher, M.; Nguyen-Viet, T.A.; Bowers, P.; Sidorenko, J.; Linnér, R.K. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 2018, 50, 1112. [Google Scholar] [CrossRef]
- Stella, A.; Nicolazzi, E.L.; Van Tassell, C.P.; Rothschild, M.F.; Colli, L.; Rosen, B.D.; Sonstegard, T.S.; Crepaldi, P.; Tosser-Klopp, G.; Joost, S. AdaptMap: Exploring goat diversity and adaptation. BMC Genom. 2018, 19, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Michelizzi, V.N.; DM, Z.; Pan, Z.; Amaral, M.E.J.; Michal, J.J.; McLean, D.J.; Womack, J.E.; Jiang, Z. Water buffalo genome science comes of age. Int. J. Biol. Sci. 2010, 6, 333–349. [Google Scholar] [CrossRef]
- Groenen, M.A.; Megens, H.J.; Zare, Y.; Warren, W.C.; Hillier, L.W.; Crooijmans, R.P.; Vereijken, A.; Okimoto, R.; Muir, W.M.; Cheng, H.H. The development and characterization of a 60K SNP chip for chicken. BMC Genom. 2011, 12, 274. [Google Scholar] [CrossRef]
- Matukumalli, L.K.; Lawley, C.T.; Schnabel, R.D.; Taylor, J.F.; Allan, M.F.; Heaton, M.P.; O’connell, J.; Moore, S.S.; Smith, T.P.; Sonstegard, T.S. Development and characterization of a high-density SNP genotyping assay for cattle. PLoS ONE 2009, 4, e5350. [Google Scholar] [CrossRef]
- Bickhart, D.M.; Rosen, B.D.; Koren, S.; Sayre, B.L.; Hastie, A.R.; Chan, S.; Lee, J.; Lam, E.T.; Liachko, I.; Sullivan, S.T. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat. Genet. 2017, 49, 643. [Google Scholar] [CrossRef]
- Bertolini, F.; Servin, B.; Talenti, A.; Rochat, E.; Kim, E.S.; Oget, C.; Palhière, I.; Crisà, A.; Catillo, G.; Steri, R. Signatures of selection and environmental adaptation across the goat genome post-domestication. Genet. Sel. Evol. 2018, 50, 57. [Google Scholar] [CrossRef] [PubMed]
- Bleidorn, C. Third generation sequencing: Technology and its potential impact on evolutionary biodiversity research. Syst. Biodivers. 2016, 14, 1–8. [Google Scholar] [CrossRef]
- Qiu, B.; Fang, S.; Ikhwanuddin, M.; Wong, L.; Ma, H. Genome survey and development of polymorphic microsatellite loci for Sillago sihama based on Illumina sequencing technology. Mol. Biol. Rep. 2020, 47, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Chou, H.-J. An Algorithm for Structural Variant Detection with Third-Generation Sequencing. Ph.D. Thesis, Graduate School Camden Rutgers, The State University of New Jersey, Camden, NJ, USA, 2017. Available online: https://rucore.libraries.rutgers.edu/rutgers-lib/52726/PDF/1/play (accessed on 3 December 2024). [CrossRef]
- Arias, J.A.; Keehan, M.; Fisher, P.; Coppieters, W.; Spelman, R. A high density linkage map of the bovine genome. BMC Genet. 2009, 10, 18. [Google Scholar] [CrossRef]
- Bidanel, J.P. Biology and genetics of reproduction. In The Genetics of the Pig; CABI: Wallingford, UK, 2011; Volume 1, pp. 218–241. [Google Scholar] [CrossRef]
- Yadav, V.; Singh, N.; Sharma, S.; Lakhani, N.; Bhimte, A.; Khare, A.; Yousuf, S. Genomic selection and its application in livestock improvement. J. Ento. Zoo. Stud. 2018, 6, 1838–1844. Available online: https://www.entomoljournal.com/archives/2018/vol6issue3/PartY/6-3-307-471.pdf (accessed on 3 December 2024).
- Lashmar, S.; Visser, C.; van Marle-Köster, E. Validation of the 50k Illumina goat SNP chip in the South African Angora goat. S. Afr. J. Anim. Sci. 2015, 45, 56–59. [Google Scholar] [CrossRef]
- Brito, L.F.; Kijas, J.W.; Ventura, R.V.; Sargolzaei, M.; Porto-Neto, L.R.; Cánovas, A.; Feng, Z.; Jafarikia, M.; Schenkel, F.S. Genetic diversity and signatures of selection in various goat breeds revealed by genome-wide SNP markers. BMC Genom. 2017, 18, 229. [Google Scholar] [CrossRef]
- Ren, X.; Yang, G.-L.; Peng, W.-F.; Zhao, Y.-X.; Zhang, M.; Chen, Z.-H.; Wu, F.-A.; Kantanen, J.; Shen, M.; Li, M.-H. A genome-wide association study identifies a genomic region for the polycerate phenotype in sheep (Ovis aries). Sci. Rep. 2016, 6, 1–8. [Google Scholar]
- Colli, L.; Milanesi, M.; Talenti, A.; Bertolini, F.; Chen, M.; Crisà, A.; Daly, K.G.; Del Corvo, M.; Guldbrandtsen, B.; Lenstra, J.A. Genome-wide SNP profiling of worldwide goat populations reveals strong partitioning of diversity and highlights post-domestication migration routes. Genet. Sel. Evol. 2018, 50, 1–20. [Google Scholar] [CrossRef]
- Qiao, X.; Su, R.; Wang, Y.; Wang, R.; Yang, T.; Li, X.; Chen, W.; He, S.; Jiang, Y.; Xu, Q. Genome-wide target enrichment-aided chip design: A 66K SNP chip for cashmere goat. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Goudenège, D.; Bris, C.; Hoffmann, V.; Desquiret-Dumas, V.; Jardel, C.; Rucheton, B.; Bannwarth, S.; Paquis-Flucklinger, V.; Lebre, A.S.; Colin, E. eKLIPse: A sensitive tool for the detection and quantification of mitochondrial DNA deletions from next-generation sequencing data. Genet. Med. 2019, 21, 1407–1416. [Google Scholar] [CrossRef]
- Ruo-Yu, L.; Gong-She, Y.; Chu-Zhao, L. The genetic diversity of mtDNA D-loop and the origin of Chinese goats. Acta Genet. Sin. 2006, 33, 420–428. [Google Scholar]
- Ladoukakis, E.D.; Zouros, E. Evolution and inheritance of animal mitochondrial DNA: Rules and exceptions. J. Biol. Res.-Thessalon. 2017, 24, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.; Turbill, C.; Suchentrunk, F. Introducing mother’s curse: Low male fertility associated with an imported mtDNA haplotype in a captive colony of brown hares. Mol. Ecol. 2010, 19, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Mannen, H.; Kojima, T.; Oyama, K.; Mukai, F.; Ishida, T.; Tsuji, S. Effect of mitochondrial DNA variation on carcass traits of Japanese Black cattle. J. Anim. Sci. 1998, 76, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Tsai, T.-S.; Rajasekar, S.; John, J.C.S. The relationship between mitochondrial DNA haplotype and the reproductive capacity of domestic pigs (Sus scrofa domesticus). BMC Genet. 2016, 17, 67. [Google Scholar] [CrossRef]
- Brown, D.; Koehler, C.; Lindberg, G.; Freeman, A.; Mayfield, J.; Myers, A.; Schutz, M.M.; Beitz, D. Molecular analysis of cytoplasmic genetic variation in Holstein cows. J. Anim. Sci. 1989, 67, 1926–1932. [Google Scholar] [CrossRef]
- Tanaka, M.; Gong, J.-S.; Zhang, J.; Yoneda, M.; Yagi, K. Mitochondrial genotype associated with longevity. Lancet 1998, 351, 185–186. [Google Scholar] [CrossRef]
- Ruiz-Pesini, E.; Mishmar, D.; Brandon, M.; Procaccio, V.; Wallace, D.C. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science 2004, 303, 223–226. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, J.; Zhao, F.; Ren, H.; Xu, L.; Lu, J.; Zhang, S.; Zhang, X.; Wei, C.; Lu, G. Genome-wide association studies for growth and meat production traits in sheep. PLoS ONE 2013, 8, e66569. [Google Scholar] [CrossRef]
- Gowane, G.; Akram, N.; Misra, S.; Prakash, V.; Kumar, A. Genetic diversity of Cahi DRB and DQB genes of caprine MHC class II in Sirohi goat. J. Genet. 2018, 97, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Våge, D.I.; Boman, I.A. A nonsense mutation in the beta-carotene oxygenase 2 (BCO2) gene is tightly associated with accumulation of carotenoids in adipose tissue in sheep (Ovis aries). BMC Genet. 2010, 11, 10. [Google Scholar] [CrossRef] [PubMed]
- Rupp, R.; Senin, P.; Sarry, J.; Allain, C.; Tasca, C.; Ligat, L.; Portes, D.; Woloszyn, F.; Bouchez, O.; Tabouret, G. A point mutation in suppressor of cytokine signalling 2 (Socs2) increases the susceptibility to inflammation of the mammary gland while associated with higher body weight and size and higher milk production in a sheep model. PLoS Genet. 2015, 11, e1005629. [Google Scholar] [CrossRef]
- Martin, P.; Raoul, J.; Bodin, L. Effects of the FecL major gene in the Lacaune meat sheep population. Genet. Sel. Evol. 2014, 46, 48. [Google Scholar] [CrossRef]
- Davis, G. Fecundity genes in sheep. Anim. Reprod. Sci. 2004, 82, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Dekkers, J.C. Commercial application of marker-and gene-assisted selection in livestock: Strategies and lessons. J. Anim. Sci. 2004, 82, E313–E328. [Google Scholar]
- Robinson, J.; Dombrowski, D.; Harpestad, G.; Shanks, R. Detection and prevalence of UMP synthase deficiency among dairy cattle. J. Hered. 1984, 75, 277–280. [Google Scholar] [CrossRef]
- Shuster, D.E.; Kehrli, M.E.; Ackermann, M.R.; Gilbert, R.O. Identification and prevalence of a genetic defect that causes leukocyte adhesion deficiency in Holstein cattle. Proc. Natl. Acad. Sci. USA 1992, 89, 9225–9229. [Google Scholar] [CrossRef]
- Kashi, Y.; Hallerman, E.; Soller, M. Marker-assisted selection of candidate bulls for progeny testing programmes. Anim. Sci. 1990, 51, 63–74. [Google Scholar] [CrossRef]
- Teneva, A. Molecular markers in animal genome analysis. Biotechnol. Anim. Husb. 2009, 25, 1267–1284. [Google Scholar]
- Database, A.Q. Animal QTLdb. 2022. Available online: https://www.animalgenome.org/cgi-bin/QTLdb/index (accessed on 24 August 2022).
- Hu, Z.-L.; Park, C.A.; Wu, X.-L.; Reecy, J.M. Animal QTLdb: An improved database tool for livestock animal QTL/association data dissemination in the post-genome era. Nucleic Acids Res. 2013, 41, D871–D879. [Google Scholar] [CrossRef] [PubMed]
- 2017, L.B. Week 5. 2017. Available online: https://xmlpipedb.cs.lmu.edu/biodb/fall2017/index.php/Week_5 (accessed on 28 September 2017).
- Bioinformatics U.S.N. Genome Informatics Resources. 2017. Available online: https://www.animalgenome.org/bioinfo/ (accessed on 1 October 2017).
- Hu, Z.-L.; Park, C.A.; Reecy, J.M. Bringing the Animal QTLdb and CorrDB into the future: Meeting new challenges and providing updated services. Nucleic Acids Res. 2022, 50, D956–D961. [Google Scholar] [CrossRef] [PubMed]
- Wang, X. Genomics and Epigenomics Analysis of Animal Production and Welfare in Denmark. Ph.D. Thesis, Department of Applied Mathematics and Computer Science, Statistics and Data Analysis. Technical University of Denmark., Lyngby, Denmark, 2020. Available online: https://backend.orbit.dtu.dk/ws/portalfiles/portal/221765870/Xiao_Wang_Thesis_Final_XiaoWang2020_updated.pdf (accessed on 3 December 2024).
- Fariello, M.-I.; Servin, B.; Tosser-Klopp, G.; Rupp, R.; Moreno, C.; San Cristobal, M.; Boitard, S.; Consortium, I.S.G. Selection signatures in worldwide sheep populations. PLoS ONE 2014, 9, e103813. [Google Scholar] [CrossRef]
- Kominakis, A.; Hager-Theodorides, A.L.; Zoidis, E.; Saridaki, A.; Antonakos, G.; Tsiamis, G. Combined GWAS and ‘guilt by association’-based prioritization analysis identifies functional candidate genes for body size in sheep. Genet. Sel. Evol. 2017, 49, 41. [Google Scholar] [CrossRef]
- Khanzadeh, H.; Ghavi Hossein-Zadeh, N.; Ghovvati, S. Genome Wide Association Studies, Next Generation Sequencing and Their Application in Animal Breeding and Genetics: A Review. Iran. J. Appl. Anim. Sci. 2020, 10, 395–404. [Google Scholar]
- Goddard, M.E.; Hayes, B.J. Mapping genes for complex traits in domestic animals and their use in breeding programmes. Nat. Rev. Genet. 2009, 10, 381. [Google Scholar] [CrossRef]
- Slatkin, M. Linkage disequilibrium—Understanding the evolutionary past and mapping the medical future. Nat. Rev. Genet. 2008, 9, 477. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.B.; Wiggans, G.R.; Ma, L.; Sonstegard, T.S.; Lawlor, T.J.; Crooker, B.A.; Van Tassell, C.P.; Yang, J.; Wang, S.; Matukumalli, L.K. Genome-wide association analysis of thirty-one production, health, reproduction, and body conformation traits in contemporary US Holstein cows. BMC Genom. 2011, 12, 408. [Google Scholar] [CrossRef]
- Kennedy, B.; Quinton, M.; Van Arendonk, J. Estimation of effects of single genes on quantitative traits. J. Anim. Sci. 1992, 70, 2000–2012. [Google Scholar] [CrossRef]
- Lai, F.-N.; Zhai, H.-L.; Cheng, M.; Ma, J.-Y.; Cheng, S.-F.; Ge, W.; Zhang, G.-L.; Wang, J.-J.; Zhang, R.-Q.; Wang, X. Whole-genome scanning for the litter size trait associated genes and SNPs under selection in dairy goat (Capra hircus). Sci. Rep. 2016, 6, 38096. [Google Scholar] [CrossRef]
- Chen, Q.; Huang, B.; Zhan, J.; Wang, J.; Qu, K.; Zhang, F.; Shen, J.; Jia, P.; Ning, Q.; Zhang, J. Whole-genome analyses identify loci and selective signals associated with body size in cattle. J. Anim. Sci. 2020, 98, skaa068. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Niu, Z.; Zeng, Z.; Jiang, Y.; Jiang, Y.; Ding, Y.; Tang, S.; Shi, H.; Ding, X. Using High-Density SNP Array to Reveal Selection Signatures Related to Prolificacy in Chinese and Kazakhstan Sheep Breeds. Animals 2020, 10, 1633. [Google Scholar] [CrossRef] [PubMed]
- Qanbari, S.; Simianer, H. Mapping signatures of positive selection in the genome of livestock. Livest. Sci. 2014, 166, 133–143. [Google Scholar] [CrossRef]
- Zhao, F.-P.; Wei, C.-H.; Zhang, L.; Liu, J.-S.; Wang, G.-k.; Tao, Z.; Du, L.-X. A genome scan of recent positive selection signatures in three sheep populations. J. Integr. Agric. 2016, 15, 162–174. [Google Scholar] [CrossRef]
- Li, X.; Su, R.; Wan, W.; Zhang, W.; Jiang, H.; Qiao, X.; Fan, Y.; Zhang, Y.; Wang, R.; Liu, Z. Identification of selection signals by large-scale whole-genome resequencing of cashmere goats. Sci. Rep. 2017, 7, 15142. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.-S.; Elbeltagy, A.; Aboul-Naga, A.; Rischkowsky, B.; Sayre, B.; Mwacharo, J.; Rothschild, M. Multiple genomic signatures of selection in goats and sheep indigenous to a hot arid environment. Heredity 2016, 116, 255. [Google Scholar] [CrossRef]
- Talenti, A.; Bertolini, F.; Pagnacco, G.; Pilla, F.; Ajmone-Marsan, P.; Rothschild, M.F.; Crepaldi, P.; Consortium, I.G. The Valdostana goat: A genome-wide investigation of the distinctiveness of its selective sweep regions. Mamm. Genome 2017, 28, 114–128. [Google Scholar] [CrossRef]
- Johnston, S.E.; McEwan, J.C.; Pickering, N.K.; Kijas, J.W.; Beraldi, D.; Pilkington, J.G.; Pemberton, J.M.; Slate, J. Genome-wide association mapping identifies the genetic basis of discrete and quantitative variation in sexual weaponry in a wild sheep population. Mol. Ecol. 2011, 20, 2555–2566. [Google Scholar] [CrossRef] [PubMed]
- Cheruiyot, E.K.; Bett, R.C.; Amimo, J.O.; Zhang, Y.; Mrode, R.; Mujibi, F.D. Signatures of selection in admixed dairy cattle in Tanzania. Front. Genet. 2018, 9, 607. [Google Scholar] [CrossRef]
- Weikard, R.; Widmann, P.; Buitkamp, J.; Emmerling, R.; Kuehn, C. Revisiting the quantitative trait loci for milk production traits on BTA6. Anim. Genet. 2012, 43, 318–323. [Google Scholar] [CrossRef]
- Yurchenko, A.A.; Daetwyler, H.D.; Yudin, N.; Schnabel, R.D.; Vander Jagt, C.J.; Soloshenko, V.; Lhasaranov, B.; Popov, R.; Taylor, J.F.; Larkin, D.M. Scans for signatures of selection in Russian cattle breed genomes reveal new candidate genes for environmental adaptation and acclimation. Sci. Rep. 2018, 8, 1–16. [Google Scholar] [CrossRef]
- Peripolli, E.; Reimer, C.; Ha, N.-T.; Geibel, J.; Machado, M.A.; do Carmo Panetto, J.C.; do Egito, A.A.; Baldi, F.; Simianer, H.; da Silva, M.V.G.B. Genome-wide detection of signatures of selection in indicine and Brazilian locally adapted taurine cattle breeds using whole-genome re-sequencing data. BMC Genom. 2020, 21, 624. [Google Scholar] [CrossRef] [PubMed]
- Gurgul, A.; Jasielczuk, I.; Semik-Gurgul, E.; Pawlina-Tyszko, K.; Stefaniuk-Szmukier, M.; Szmatoła, T.; Polak, G.; Tomczyk-Wrona, I.; Bugno-Poniewierska, M. A Genome-wide Scan for Diversifying Selection Signatures in Selected Horse Breeds. PLoS ONE 2019, 14, e0210751. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, R.; Williamson, S.; Kim, Y.; Hubisz, M.J.; Clark, A.G.; Bustamante, C. Genomic Scans for Selective Sweeps Using SNP Data. Genome Res. 2005, 15, 1566–1575. [Google Scholar] [CrossRef]
- Boitard, S.; Schlötterer, C.; Nolte, V.; Pandey, R.V.; Futschik, A. Detecting Selective Sweeps from Pooled Next-generation Sequencing Samples. Mol. Biol. Evol. 2012, 29, 2177–2186. [Google Scholar] [CrossRef]
- McRae, K.M.; McEwan, J.C.; Dodds, K.G.; Gemmell, N.J. Signatures of Selection in Sheep Bred for Resistance or Susceptibility to Gastrointestinal Nematodes. BMC Genom. 2014, 15, 637. [Google Scholar] [CrossRef]
- Ramey, H.R.; Decker, J.E.; McKay, S.D.; Rolf, M.M.; Schnabel, R.D.; Taylor, J.F. Detection of Selective Sweeps in Cattle Using Genome-wide SNP Data. BMC Genom. 2013, 14, 382. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, I.; Fernández, I.; Traoré, A.; Pérez-Pardal, L.; Menéndez-Arias, N.A.; Goyache, F. Genomic Scan of Selective Sweeps in Djallonké (West African Dwarf) Sheep Shed Light on Adaptation to Harsh Environments. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Guo, J.; Tao, H.; Li, P.; Li, L.; Zhong, T.; Wang, L.; Ma, J.; Chen, X.; Song, T.; Zhang, H. Whole-genome Sequencing Reveals Selection Signatures Associated with Important Traits in Six Goat Breeds. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Mariadassou, M.; Ramayo-Caldas, Y.; Charles, M.; Féménia, M.; Renand, G.; Rocha, D. Detection of Selection Signatures in Limousin Cattle Using Whole-genome Resequencing. Anim. Genet. 2020, 51, 815–819. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, K.; Peng, X.; Zhan, H.; Lu, J.; Xie, S.; Zhao, S.; Li, X.; Ma, Y. Selective Sweep Analysis Reveals Extensive Parallel Selection Traits between Large White and Duroc Pigs. Evolut. Appl. 2020, 13, 2807–2820. [Google Scholar] [CrossRef]
- Meuwissen, T.; Goddard, M. Accurate Prediction of Genetic Values for Complex Traits by Whole-genome Resequencing. Genetics 2010, 185, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Seidel, G. Brief Introduction to Whole-genome Selection in Cattle Using Single Nucleotide Polymorphisms. Reprod. Fertil. Dev. 2009, 22, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Brito, L.F.; Jafarikia, M.; Grossi, D.A.; Kijas, J.W.; Porto-Neto, L.R.; Ventura, R.V.; Salgorzaei, M.; Schenkel, F.S. Characterization of Linkage Disequilibrium, Consistency of Gametic Phase, and Admixture in Australian and Canadian Goats. BMC Genet. 2015, 16, 67. [Google Scholar] [CrossRef] [PubMed]
- Phua, S.; Hyndman, D.; Baird, H.; Auvray, B.; McEwan, J.; Lee, M.; Dodds, K. Towards Genomic Selection for Facial Eczema Disease Tolerance in the New Zealand Sheep Industry. Anim. Genet. 2014, 45, 559–564. [Google Scholar] [CrossRef]
- Pickering, N.K.; Auvray, B.; Dodds, K.G.; McEwan, J.C. Genomic Prediction and Genome-wide Association Study for Dagginess and Host Internal Parasite Resistance in New Zealand Sheep. BMC Genom. 2015, 16, 958. [Google Scholar] [CrossRef]
- Daetwyler, H.; Hickey, J.; Henshall, J.; Dominik, S.; Gredler, B.; Van Der Werf, J.; Hayes, B. Accuracy of Estimated Genomic Breeding Values for Wool and Meat Traits in a Multi-breed Sheep Population. Anim. Prod. Sci. 2010, 50, 1004–1010. [Google Scholar] [CrossRef]
- Larroque, H.; Robert-Granié, C. Comparison of Joint Versus Purebred Genomic Evaluation in the French Multi-breed Dairy Goat Population. Genet. Sel. Evol. 2014, 46, 67. [Google Scholar]
- Habier, D.; Fernando, R.L.; Kizilkaya, K.; Garrick, D.J. Extension of the Bayesian Alphabet for Genomic Selection. BMC Bioinform. 2011, 12, 186. [Google Scholar] [CrossRef]
- Van Marle-Köster, E.; Visser, C.; Berry, D. A Review of Genomic Selection-Implications for the South African Beef and Dairy Cattle Industries. S. Afr. J. Anim. Sci. 2013, 43, 1–17. [Google Scholar] [CrossRef][Green Version]

| Type of Marker | Description | Techniques Used | Applications | Advantages | Limitations |
|---|---|---|---|---|---|
| Molecular Markers | DNA-based markers used to identify genetic differences between individuals. |
| Gene mapping | High specificity | Can be expensive |
| Genetic diversity studies | Detects minute genetic variations | Requires technical expertise | ||
| Marker-assisted selection | ||||
| Parentage analysis | ||||
| Evolutionary studies | ||||
| |||||
| Biochemical Markers | Proteins or enzymes linked to biochemical pathways, reflecting phenotypic traits. | Isozyme analysis | Assessment of functional variations | Less expensive than DNA markers | Limited resolution |
| Protein electrophoresis | Genetic mapping of certain traits | Direct link to gene expression | Environmentally influenced | ||
| Identifying metabolic pathways | |||||
| Morphological Markers | Observable phenotypic traits such as seed shape, flower colour, or plant height. | Traditional breeding methods | Early genetic studies | Easy and inexpensive | Influenced by environment |
| Visual identification | Plant breeding | No need for complex technology | Often controlled by multiple genes | ||
| Taxonomy Conservation programs |
| Species | Genome Size (Gb/Mb) | Year | Reference |
|---|---|---|---|
| Fruit Fly (Drosophila melanogaster) | 0.18 | 2000 | [54] |
| Mouse (Mus musculus) | 2.6 | 2002 | [55] |
| Dog (Canis familiaris) | 2.4 | 2003 | [56] |
| Chicken (Gallus gallus) | 1.05 | 2004 | [57] |
| Norway Rat (Rattus norvegicus) | 2.75 | 2004 | [58] |
| Sea Urchin (Strongylocentrotus purpuratus) | 0.81 | 2006 | [59] |
| Elephant Shark (C. milii) | 910-Mb | 2007 | [60] |
| Monkeys (M. mulatta) | 3.09 | 2007 | [61] |
| Cat (Felis silvestris catus) | 2.7 | 2007 | [62] |
| Platypus (Ornithorhynchus anatinus) | 1.9 | 2008 | [63] |
| Pig (Sus scrofa) | 2.2 | 2008 | [64] |
| Sheep (Ovis aries) | 2.78 | 2009 | [65] |
| Cattle (Bos taurus) | 2.91 | 2009 | [66] |
| Horse (Equus caballus) | 2.47 | 2009 | [67] |
| Amphibians (Xenopus tropicalis) | 1.7 | 2010 | [68] |
| Giant Panda (Ailuropoda melanoleuca) | 2.4 | 2010 | [69] |
| Zebra Finch (Taeniopygia guttata) | 1.2 | 2010 | [70] |
| Turkey (Meleagris gallopavo) | 1.1 | 2010 | [71] |
| Camel (Camelus dromedarius) | 2.2 | 2011 | [72] |
| Tammar Wallaby (Macropus eugenii) | 3.1 | 2011 | [73] |
| Green Anole Lizard (Anolis carolinensis) | 1.78 | 2011 | [74] |
| Atlantic Cod (Gadus morhua) | 0.83 | 2011 | [75] |
| Naked Mole Rat (Heterocephalus glaber) | 2.7 | 2011 | [76] |
| Orangutan (Pongo abelii) | 3.08 | 2011 | [77] |
| Goat (Capra hircus) | 2.66 | 2012 | [78] |
| Bactrian Camel (Camelus bactrianus) | 2.38 | 2012 | [79] |
| Olive Baboon (Papio anubis) | 3.1 | 2012 | [80] |
| Wild Duck (Anas platyrhynchos) | 1.07 | 2013 | [81] |
| Coelacanth (Latimeria chalumnae) | 2.91 | 2013 | [82] |
| Sea Lamprey (Petromyzon marinus) | 0.81 | 2013 | [83] |
| King Cobra (Ophiophagus hannah) | 1.44 | 2013 | [84] |
| Dung Beetle (Onthophagus taurus) | 0.38 | 2022 | [85] |
| Cherry Salmon (Oncorhynchus masou) | 2.4 | 2024 | [86] |
| Type | Method | Most Applications | Ref. | |
|---|---|---|---|---|
| Biochemical markers | Isozymes | Allozymes | Animals | [97] |
| First Generation Markers | RAPD | Random Amplified Polymorphic DNA | Cattle and Sheep | [98] |
| SCAR | Sequence Characterized Amplified Regions | Chickens | [99] | |
| STS | Sequence Tagged Sites | Plants | [100] | |
| AP-PCR | Arbitrary Primed PCR | Cattle | [101] | |
| DAF | DNA Amplification Fingerprinting | Cattle and buffalo | [102] | |
| RFLP | Restriction Fragment Length Polymorphism | Sheep and goats | [88,103] | |
| Second Generation Markers | AFLP | Amplified Fragment Length Polymorphism | Humans and animals | [104] |
| SSLP | Simple Sequence Length Polymorphism | Fish | [105] | |
| ISSR | Inter Simple Sequence Repeat | Sheep | [106] | |
| VNTRs | Variable Number of Tandem Repeats | Cattle | [107] | |
| RAMPO | Random Amplified Micro satellite Polymorphism | Humans and animals | [108] | |
| CAPs | Cleaved Amplified Polymorphic Products | |||
| ASAP | Allele-Specific Associated Primers | Mouse | [109] | |
| Third Generation Markers | EST | Expressed Sequence Tag markers | Pigs | [110] |
| SNP | Single Nucleotide Polymorphism | Goats | [111] | |
| MITE | Miniature Inverted Repeat Transposable Elements | Plants | [112] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Husien, H.M.; Saleh, A.A.; Hassanine, N.N.A.M.; Rashad, A.M.A.; Sharaby, M.A.; Mohamed, A.Z.; Abdelhalim, H.; Hafez, E.E.; Essa, M.O.A.; Adam, S.Y.; et al. The Evolution and Role of Molecular Tools in Measuring Diversity and Genomic Selection in Livestock Populations (Traditional and Up-to-Date Insights): A Comprehensive Exploration. Vet. Sci. 2024, 11, 627. https://doi.org/10.3390/vetsci11120627
Husien HM, Saleh AA, Hassanine NNAM, Rashad AMA, Sharaby MA, Mohamed AZ, Abdelhalim H, Hafez EE, Essa MOA, Adam SY, et al. The Evolution and Role of Molecular Tools in Measuring Diversity and Genomic Selection in Livestock Populations (Traditional and Up-to-Date Insights): A Comprehensive Exploration. Veterinary Sciences. 2024; 11(12):627. https://doi.org/10.3390/vetsci11120627
Chicago/Turabian StyleHusien, Hosameldeen Mohamed, Ahmed A. Saleh, Nada N. A. M. Hassanine, Amr M. A. Rashad, Mahmoud A. Sharaby, Asmaa Z. Mohamed, Heba Abdelhalim, Elsayed E. Hafez, Mohamed Osman Abdalrahem Essa, Saber Y. Adam, and et al. 2024. "The Evolution and Role of Molecular Tools in Measuring Diversity and Genomic Selection in Livestock Populations (Traditional and Up-to-Date Insights): A Comprehensive Exploration" Veterinary Sciences 11, no. 12: 627. https://doi.org/10.3390/vetsci11120627
APA StyleHusien, H. M., Saleh, A. A., Hassanine, N. N. A. M., Rashad, A. M. A., Sharaby, M. A., Mohamed, A. Z., Abdelhalim, H., Hafez, E. E., Essa, M. O. A., Adam, S. Y., Chen, N., & Wang, M. (2024). The Evolution and Role of Molecular Tools in Measuring Diversity and Genomic Selection in Livestock Populations (Traditional and Up-to-Date Insights): A Comprehensive Exploration. Veterinary Sciences, 11(12), 627. https://doi.org/10.3390/vetsci11120627








