Epidemiology and Molecular Characterisation of Multidrug-Resistant Escherichia coli Isolated from Cow Milk
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
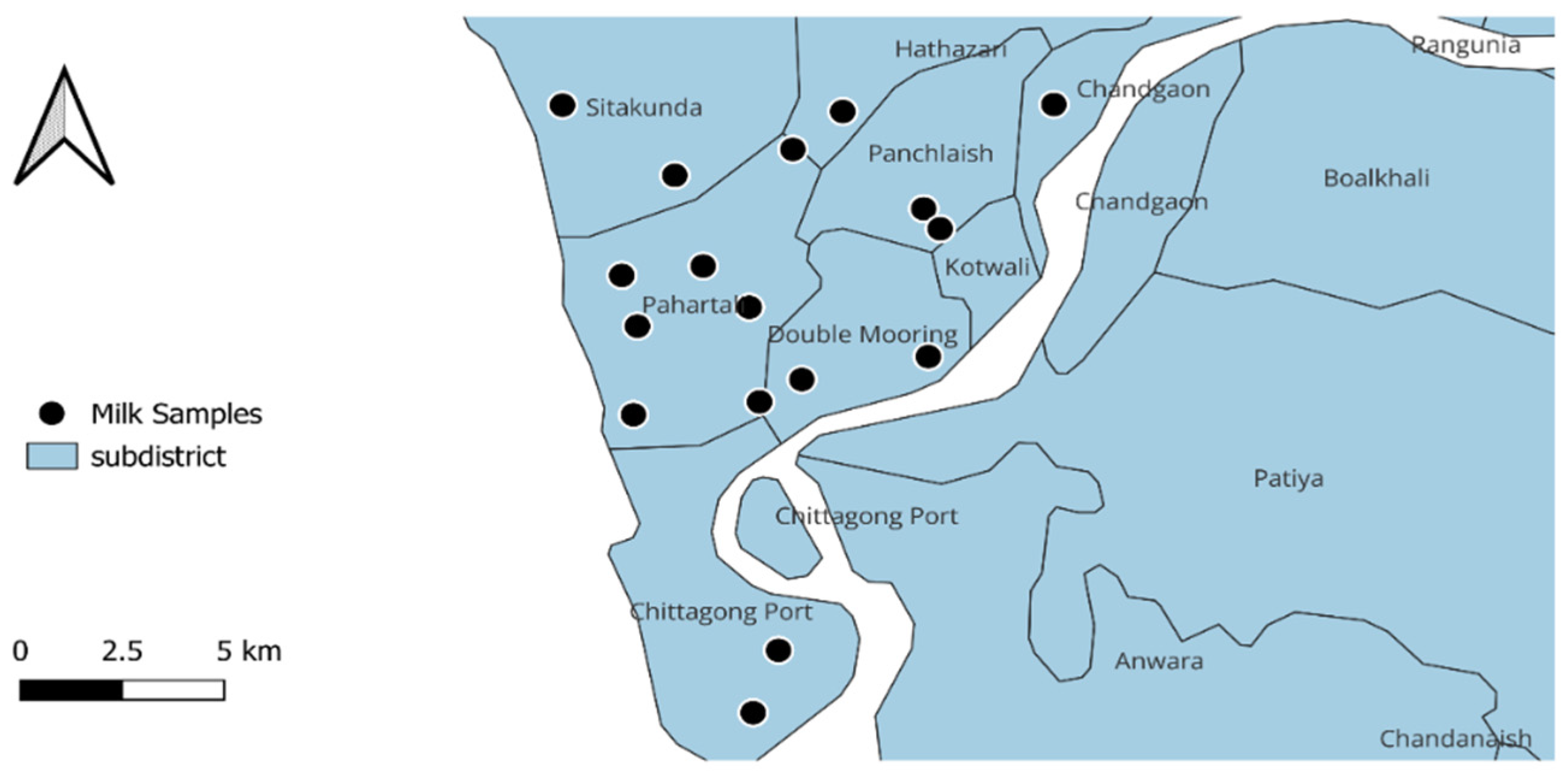
2.1. Study Design and Sample Collection
2.2. Sample Preparation
2.3. Phenotypic Isolation and Identification of E. coli
2.4. Molecular Confirmation of E. coli
2.5. Phenotypic Antimicrobial Resistance Profiles
2.6. Detection of AMR Genes
2.7. Statistical Analysis
3. Results
3.1. Prevalence of E. coli
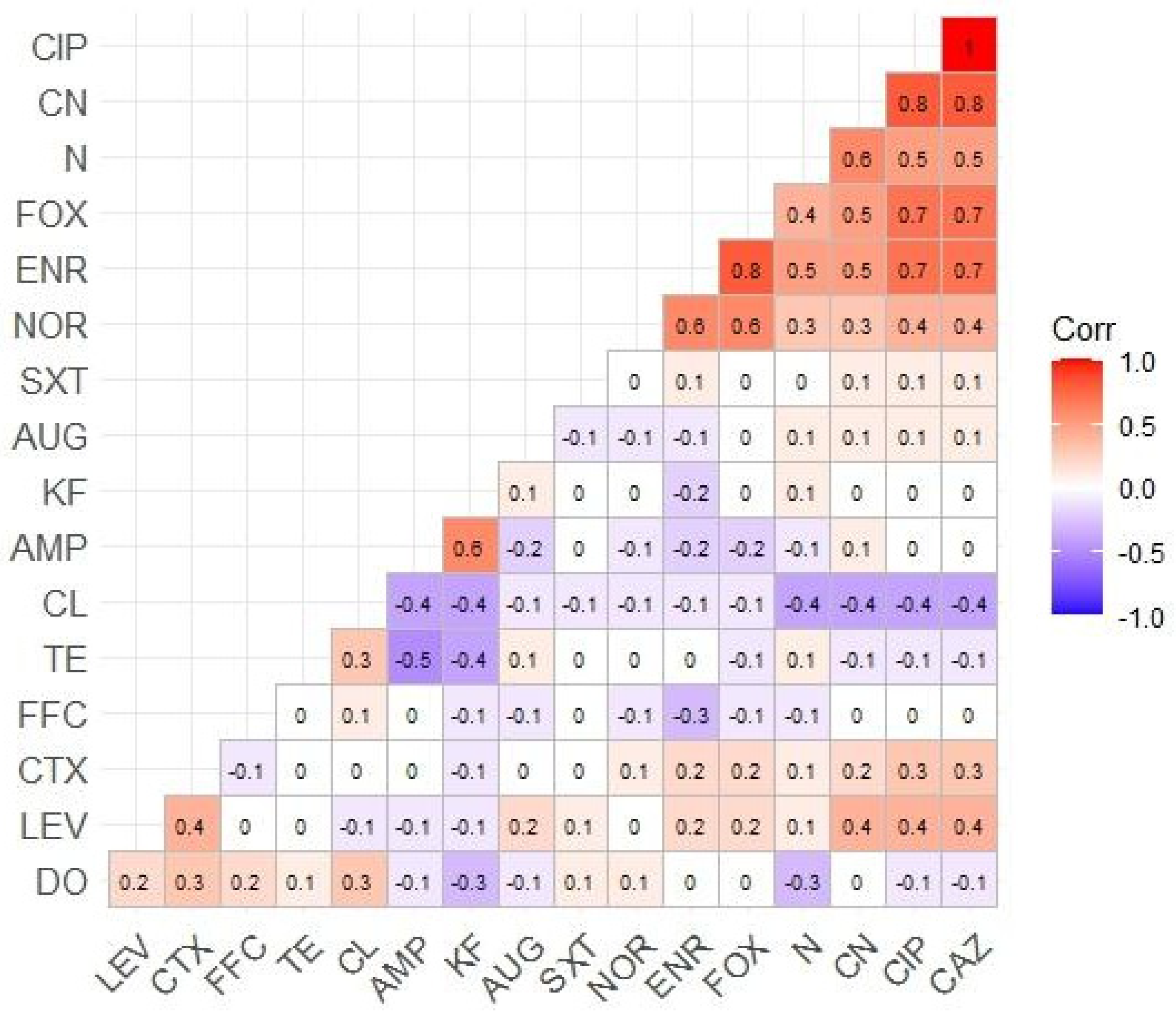
3.2. AMR Profiles of Isolated E. coli
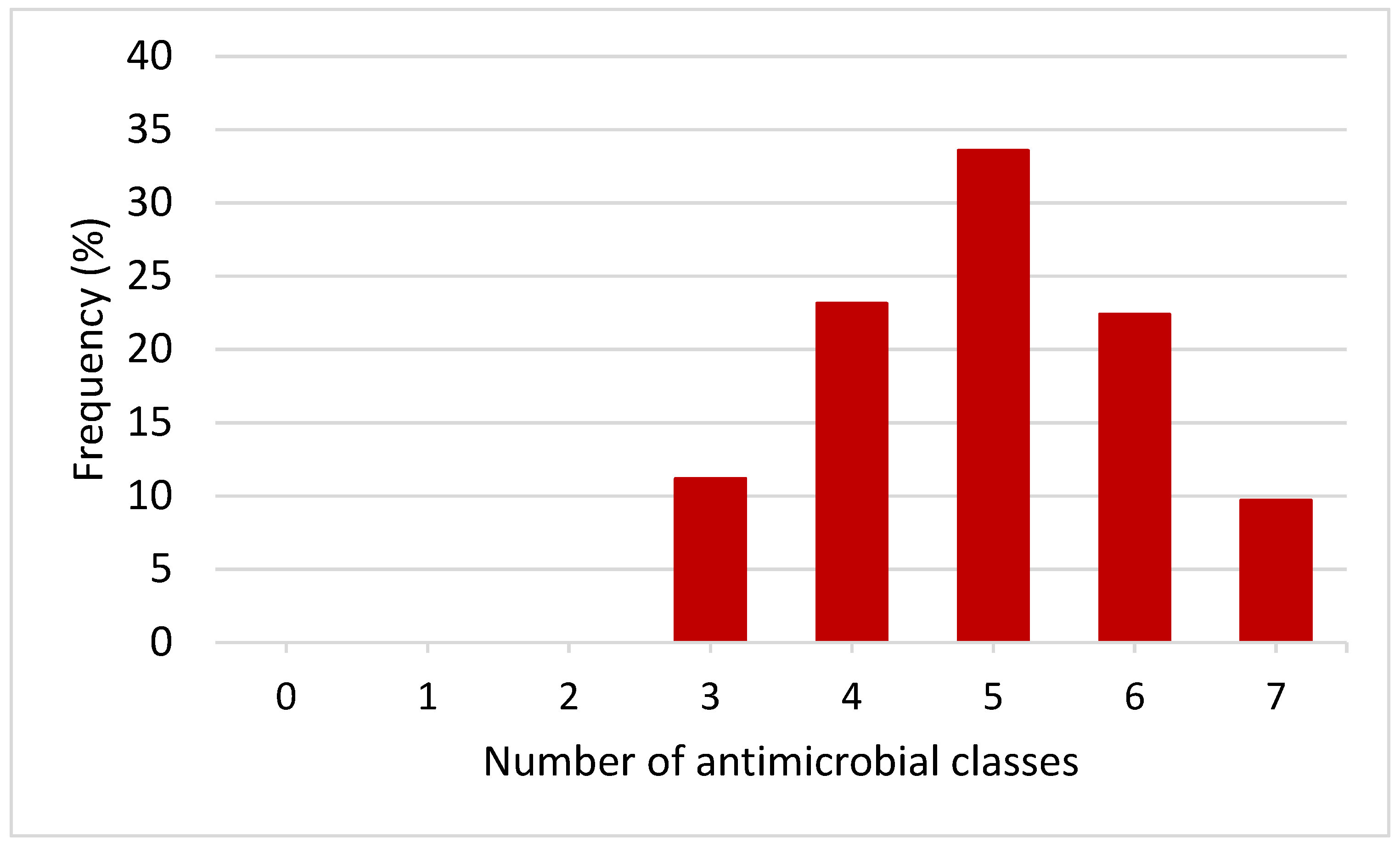
3.3. Phenotypic MDR Patterns of E. coli Isolates
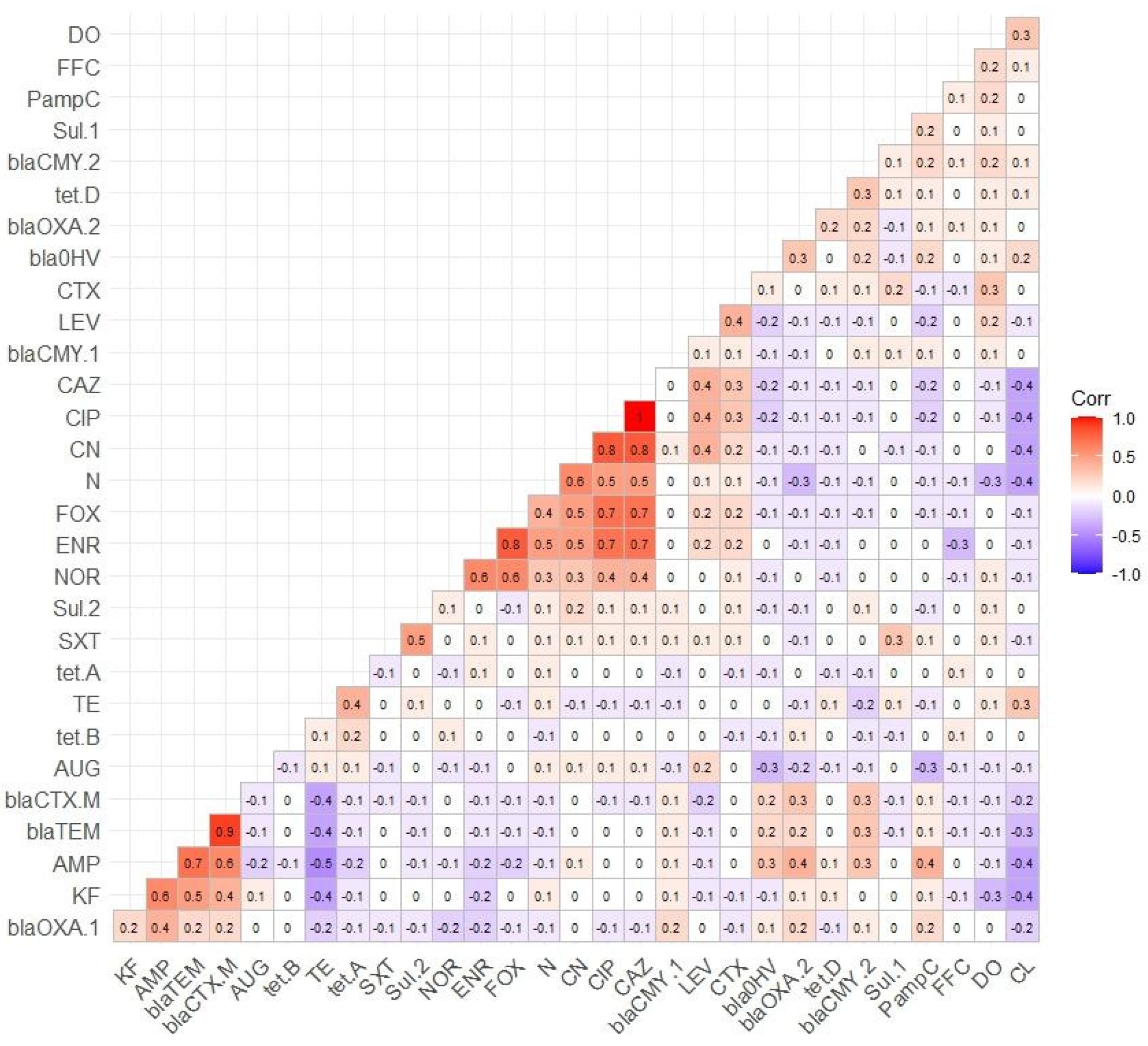
3.4. Distribution of AMR Genes
4. Discussion
5. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; Government of the United Kingdom: London, UK, 2016.
- Sakalauskienė, G.V.; Radzevičienė, A. Antimicrobial Resistance: What Lies Beneath This Complex Phenomenon? Diagnostics 2024, 14, 2319. [Google Scholar] [CrossRef] [PubMed]
- de Brito, F.A.; de Freitas, A.P.; Nascimento, M.S. Multidrug-resistant biofilms (MDR): Main mechanisms of tolerance and resistance in the food supply chain. Pathogens 2022, 11, 1416. [Google Scholar] [CrossRef] [PubMed]
- Sharma, C.; Rokana, N.; Chandra, M.; Singh, B.P.; Gulhane, R.D.; Gill, J.P.S.; Ray, P.; Puniya, A.K.; Panwar, H. Antimicrobial resistance: Its surveillance, impact, and alternative management strategies in dairy animals. Front. Vet. Sci. 2018, 4, 237. [Google Scholar] [CrossRef] [PubMed]
- Annunziato, G. Strategies to overcome antimicrobial resistance (AMR) making use of non-essential target inhibitors: A review. Int. J. Mol. Sci. 2019, 20, 5844. [Google Scholar] [CrossRef]
- Murugaiyan, J.; Kumar, P.A.; Rao, G.S.; Iskandar, K.; Hawser, S.; Hays, J.P.; Mohsen, Y.; Adukkadukkam, S.; Awuah, W.A.; Jose, R.A.M. Progress in alternative strategies to combat antimicrobial resistance: Focus on antibiotics. Antibiotics 2022, 11, 200. [Google Scholar] [CrossRef]
- Abebe, R.; Hatiya, H.; Abera, M.; Megersa, B.; Asmare, K. Bovine mastitis: Prevalence, risk factors and isolation of Staphylococcus aureus in dairy herds at Hawassa milk shed, South Ethiopia. BMC Vet. Res. 2016, 12, 270. [Google Scholar] [CrossRef]
- Mulchandani, R.; Wang, Y.; Gilbert, M.; Van Boeckel, T.P. Global trends in antimicrobial use in food-producing animals: 2020 to 2030. PLoS Glob. Public Health 2023, 3, e0001305. [Google Scholar] [CrossRef]
- Loo, E.; Lai, K.S.; Mansor, R. Antimicrobial usage and resistance in dairy cattle production. Vet. Med. Pharm. 2019, 7, 7–82. [Google Scholar]
- Lhermie, G.; Gröhn, Y.T.; Raboisson, D. Addressing antimicrobial resistance: An overview of priority actions to prevent suboptimal antimicrobial use in food-animal production. Front. Microbiol. 2017, 7, 2114. [Google Scholar] [CrossRef]
- Ayukekbong, J.A.; Ntemgwa, M.; Atabe, A.N. The threat of antimicrobial resistance in developing countries: Causes and control strategies. Antimicrob. Resist. Infect. Control 2017, 6, 47. [Google Scholar] [CrossRef]
- Batabyal, K.; Banerjee, A.; Pal, S.; Dey, S.; Joardar, S.N.; Samanta, I.; Isore, D.P.; Singh, A.D. Detection, characterization, and antibiogram of extended-spectrum beta-lactamase Escherichia coli isolated from bovine milk samples in West Bengal, India. Vet. World 2018, 11, 1423. [Google Scholar] [CrossRef] [PubMed]
- Almansour, A.M.; Alhadlaq, M.A.; Alzahrani, K.O.; Mukhtar, L.E.; Alharbi, A.L.; Alajel, S.M. The Silent Threat: Antimicrobial-Resistant Pathogens in Food-Producing Animals and Their Impact on Public Health. Microorganisms 2023, 11, 2127. [Google Scholar] [CrossRef]
- Skocková, A.; Cupáková, S.; Karpísková, R.; Janstová, B. Detection of tetracycline resistance genes in Escherichia coli from raw cow’s milk. J. Microbiol. Biotechnol. Food Sci. 2012, 1, 777. [Google Scholar]
- Metzger, S.; Hogan, J. Antimicrobial susceptibility and frequency of resistance genes in Escherichia coli isolated from bovine mastitis. J. Dairy Sci. 2013, 96, 3044–3049. [Google Scholar] [CrossRef] [PubMed]
- Kamaruzzaman, E.A.; Abdul Aziz, S.; Bitrus, A.A.; Zakaria, Z.; Hassan, L. Occurrence and characteristics of extended-spectrum β-lactamase-producing Escherichia coli from dairy cattle, milk, and farm environments in Peninsular Malaysia. Pathogens 2020, 9, 1007. [Google Scholar] [CrossRef]
- Bajaj, P.; Singh, N.S.; Virdi, J.S. Escherichia coli β-lactamases: What really matters. Front. Microbiol. 2016, 7, 417. [Google Scholar] [CrossRef]
- Walther, B.; Tedin, K.; Lübke-Becker, A. Multidrug-resistant opportunistic pathogens challenging veterinary infection control. Vet. Microbiol. 2017, 200, 71–78. [Google Scholar] [CrossRef]
- Karkaba, A.; Grinberg, A.; Benschop, J.; Pleydell, E. Characterisation of extended-spectrum β-lactamase and AmpC β-lactamase-producing Enterobacteriaceae isolated from companion animals in New Zealand. N. Z. Vet. J. 2017, 65, 105–112. [Google Scholar] [CrossRef]
- Mandujano, A.; Cortés-Espinosa, D.V.; Vásquez-Villanueva, J.; Guel, P.; Rivera, G.; Juárez-Rendón, K.; Cruz-Pulido, W.L.; Aguilera-Arreola, G.; Guerrero, A.; Bocanegra-García, V.; et al. Extended-Spectrum β-Lactamase-Producing Escherichia coli Isolated from Food-Producing Animals in Tamaulipas, Mexico. Antibiotics 2023, 12, 1010. [Google Scholar] [CrossRef]
- Ali, T.; ur Rahman, S.; Zhang, L.; Shahid, M.; Zhang, S.; Liu, G.; Gao, J.; Han, B. ESBL-producing Escherichia coli from cows suffering mastitis in China contain clinical class 1 integrons with CTX-M linked to IS CR1. Front. Microbiol. 2016, 7, 1931. [Google Scholar] [CrossRef]
- Estany-Gestal, A.; Salgado-Barreira, A.; Vazquez-Lago, J.M. Antibiotic Use and Antimicrobial Resistance: A Global Public Health Crisis. Antibiotics 2024, 13, 900. [Google Scholar] [CrossRef] [PubMed]
- Sobur, M.A.; Sabuj, A.A.M.; Sarker, R.; Rahman, A.T.; Kabir, S.L.; Rahman, M.T. Antibiotic-resistant Escherichia coli and Salmonella spp. associated with dairy cattle and farm environment having public health significance. Vet. World 2019, 12, 984. [Google Scholar] [CrossRef]
- Khan, S.A.; Imtiaz, M.A.; Sayeed, M.A.; Shaikat, A.H.; Hassan, M.M. Antimicrobial resistance pattern in domestic animal-wildlife-environmental niche via the food chain to humans with a Bangladesh perspective; a systematic review. BMC Vet. Res. 2020, 16, 302. [Google Scholar] [CrossRef] [PubMed]
- Sarker, M.S.; Mannan, M.S.; Ali, M.Y.; Bayzid, M.; Ahad, A.; Bupasha, Z.B. Antibiotic resistance of Escherichia coli isolated from broilers sold at live bird markets in Chattogram, Bangladesh. J. Adv. Vet. Anim. Res. 2019, 6, 272–277. [Google Scholar] [CrossRef] [PubMed]
- Godambe, L.P.; Bandekar, J.; Shashidhar, R. Species specific PCR based detection of Escherichia coli from Indian foods. 3 Biotech 2017, 7, 130. [Google Scholar] [CrossRef] [PubMed]
- Bauer, A.; Kirby, W.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptivility Testing, 33rd ed.; CLSI: Pittsburgh, PA, USA, 2023; Volume 43, CLSI Supplement M100; Available online: https://clsi.org/ (accessed on 5 March 2024).
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.; Giske, C.; Harbarth, S.; Hindler, J.; Kahlmeter, G.; Olsson-Liljequist, B. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Algammal, A.M.; El-Tarabili, R.M.; Alfifi, K.J.; Al-Otaibi, A.S.; Hashem, M.E.A.; El-Maghraby, M.M.; Mahmoud, A.E. Virulence determinant and antimicrobial resistance traits of Emerging MDR Shiga toxigenic E. coli in diarrheic dogs. AMB Express 2022, 12, 34. [Google Scholar] [CrossRef]
- Koo, H.-J.; Woo, G.-J. Distribution and transferability of tetracycline resistance determinants in Escherichia coli isolated from meat and meat products. Int. J. Food Microbiol. 2011, 145, 407–413. [Google Scholar] [CrossRef]
- Lanz, R.; Kuhnert, P.; Boerlin, P. Antimicrobial resistance and resistance gene determinants in clinical Escherichia coli from different animal species in Switzerland. Vet. Microbiol. 2003, 91, 73–84. [Google Scholar] [CrossRef]
- Hasman, H.; Mevius, D.; Veldman, K.; Olesen, I.; Aarestrup, F.M. β-Lactamases among extended-spectrum β-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J. Antimicrob. Chemother. 2005, 56, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Vahedi, M.; Nasrolahei, M.; Sharif, M.; Mirabi, A. Bacteriological study of raw and unexpired pasteurized cow’s milk collected at the dairy farms and super markets in Sari city in 2011. J. Prev. Med. Hyg. 2013, 54, 120. [Google Scholar] [PubMed]
- Megersa, R.; Mathewos, M.; Fesseha, H. Isolation and identification of Escherichia coli from dairy cow raw milk in Bishoftu town, Central Ethiopia. Arch. Vet. Anim. Sci. 2019, 1, 1–7. [Google Scholar]
- Mahmud, S.; Ali, M.F.; Faruque, M.O.; Wasim, M.; Evamoni, F.Z.; Chowdhury, K.; Napis, S.; Mohiuddin, A. Prevalence and Molecular Characterization of Microbial Contaminants in Raw Cow Milk of Tangail District in Bangladesh. Curr. Nutr. Food Sci. 2022, 18, 220–230. [Google Scholar]
- Williams, E.N.; Van Doren, J.M.; Leonard, C.L.; Datta, A.R. Prevalence of Listeria monocytogenes, Salmonella spp., Shiga toxin-producing Escherichia coli, and Campylobacter spp. in raw milk in the United States between 2000 and 2019: A systematic review and meta-analysis. J. Food Prot. 2023, 86, 100014. [Google Scholar] [CrossRef]
- Agatha, T.M.; Wibawati, P.A.; Izulhaq, R.I.; Agustono, B.; Prastiya, R.A.; Wardhana, D.K.; Abdramanov, A.; Lokapirnasari, W.P.; Lamid, M. Antibiotic resistance of Escherichia coli from the milk of Ettawa crossbred dairy goats in Blitar Regency, East Java, Indonesia. Vet. World 2023, 16, 168. [Google Scholar] [CrossRef]
- Astorga, F.; Navarrete-Talloni, M.J.; Miró, M.P.; Bravo, V.; Toro, M.; Blondel, C.J.; Hervé-Claude, L.P. Antimicrobial resistance in E. coli isolated from dairy calves and bedding material. Heliyon 2019, 5, e02773. [Google Scholar] [CrossRef]
- Islam, S. Coliform and Staphylococcal Mastitis in Cows: Risk Factors and Trends in Antimicrobial Resistance. 2020. Available online: http://archive.saulibrary.edu.bd:8080/xmlui/handle/123456789/4787 (accessed on 12 January 2024).
- Collis, R.M.; Burgess, S.A.; Biggs, P.J.; Midwinter, A.C.; French, N.P.; Toombs-Ruane, L.; Cookson, A.L. Extended-spectrum beta-lactamase-producing Enterobacteriaceae in dairy farm environments: A New Zealand perspective. Foodborne Pathog. Dis. 2019, 16, 5–22. [Google Scholar] [CrossRef]
- Cheng, A.C.; Turnidge, J.; Collignon, P.; Looke, D.; Barton, M.; Gottlieb, T. Control of fluoroquinolone resistance through successful regulation, Australia. Emerg. Infect. Dis. 2012, 18, 1453. [Google Scholar] [CrossRef]
- Liu, H.; Meng, L.; Dong, L.; Zhang, Y.; Wang, J.; Zheng, N. Prevalence, antimicrobial susceptibility, and molecular characterization of Escherichia coli isolated from raw milk in dairy herds in Northern China. Front. Microbiol. 2021, 12, 730656. [Google Scholar] [CrossRef]
- Miller, S.A.; Ferreira, J.P.; LeJeune, J.T. Antimicrobial use and resistance in plant agriculture: A one health perspective. Agriculture 2022, 12, 289. [Google Scholar] [CrossRef]
- Ahmad, I.; Malak, H.A.; Abulreesh, H.H. Environmental antimicrobial resistance and its drivers: A potential threat to public health. J. Glob. Antimicrob. Resist. 2021, 27, 101–111. [Google Scholar]
- Gandra, S.; Kotwani, A. Need to improve availability of “access” group antibiotics and reduce the use of “watch” group antibiotics in India for optimum use of antibiotics to contain antimicrobial resistance. J. Pharm. Policy Pract. 2019, 12, 20. [Google Scholar] [CrossRef] [PubMed]
- Hsia, Y.; Lee, B.R.; Versporten, A.; Yang, Y.; Bielicki, J.; Jackson, C.; Newland, J.; Goossens, H.; Magrini, N.; Sharland, M. Use of the WHO Access, Watch, and Reserve classification to define patterns of hospital antibiotic use (AWaRe): An analysis of paediatric survey data from 56 countries. Lancet Glob. Health 2019, 7, e861–e871. [Google Scholar] [CrossRef]
| Antimicrobial Agents | Target Gene | Primer NAME | Primer Sequence (5′-3′) | Annealing Temp. | Amplicon Size (bp) | References |
|---|---|---|---|---|---|---|
| Tetracyclines | tetA | tetA-F | CGCCTTTCCTTTGGGTTCTCTATATC | 55 °C | 182 | [31] |
| tetA-R | CAGCCCACCGAGCACAGG | |||||
| tetB | tetB-F | GCCAGTCTTGCCAACGTTAT | 975 | |||
| tetB-R | ATAACACCGG TTGCATTGGT | |||||
| tetC | tetC-F | TTCAACCCAGTCAGCTCCTT | 560 | |||
| tetC-R | GGGAGGCAGACAAGGTATAGG | |||||
| tetD | tetD-F | GAGCGTACCGCCTGGTTC | 780 | |||
| tetD-R | TCTGATCAGCAGACAGATTGC | |||||
| Sulphonamides | sul-1 | sul-1-F | CGGCGTGGGCTACCTGAACG | 68 °C | 779 | [32] |
| sul-1-R | GCCGATCGCGTGAAGTTCCG | |||||
| sul-2 | sul-2-F | CCTGTTTCGTCCGACACAGA | 66 °C | 721 | ||
| sul-2-R | GAAGCGCAGCCGCAATTCAT | |||||
| ESBLs | blaTEM | blaTEM-F | ATAAAATTCTTGAAGACGAAA | 54 °C | 964 | [33] |
| blaTEM-R | GACAGTTACCAATGCTTAATC | |||||
| blaSHV | blaSHV-F | GCTTTCCCATGATGAGCACC | 50 °C | 854 | ||
| blaSHV-R | AGGCGGGTGACGTTGTCGC | |||||
| PampC | PampC-F | GTGAATACAGAGCCAGACGC | 50 °C | 343 | ||
| PampC-R | GTTGTTTCCGGGTGATGC | |||||
| blaOXA-1 | blaOXA-1-R | GTGTGTTTAGAATGGTGATCGCATT | 62 °C | 820 | ||
| blaOXA-1-R | GTGTGTTTAGAATGGTGATCGCATT | |||||
| blaOXA-2 | blaOXA-2-F | ACGATAGTTGTGGCAGACGAAC | 62 °C | 602 | ||
| blaOXA-2-R | ATYCTGTTTGGCGTATCRATATTC | |||||
| blaCTX-M | blaCTX-M-F | ATGTGCAGYACCAGTAARGTKATGGC | 60 °C | 593 | ||
| blaCTX-M-F | TGGGTRAARTARGTSACCAGAAYCAGCGG | |||||
| blaCMY-1 | blaCMY-1-F | GTGGTGGATGCCAGCATCC | 58 °C | 915 | ||
| blaCMY-1-R | GGTCGAGCCGGTCTTGTTGAA | |||||
| blaCMY-2 | blaCMY-2-F | GCACTTAGCCACCTATACGGCAG | 58 °C | 758 | ||
| blaCMY-2-R | GCTTTTCAAGAATGCGCCAGG | |||||
| blaACC-1 | blaACC-1-F | ATYCTGTTTGGCGTATCRATATTC | 53 °C | 818 | ||
| blaACC-1-R | AGCCTCAGCAGCCGGTTAC |
| Antimicrobial Groups | Antimicrobial Agents | Susceptible (S) N (%) | Intermediate (I) N (%) | Resistant (R) N (%) |
|---|---|---|---|---|
| Penicillins | AMP (10 µg) | 36 (26.86) | 5 (3.73) | 93 (69.40) |
| AUG (30 µg) | 36 (26.86) | 5 (3.73) | 93 (69.40) | |
| Cephalosporins | KF (30 µg) | 40 (29.85) | 1 (0.75) | 93 (69.40) |
| CL (30 µg) | 41 (30.59) | 0 (0) | 93 (69.40) | |
| FOX (30 µg) | 34 (25.37) | 44 (32.84) | 56 (41.79) | |
| CTX (30 µg) | 53 (39.55) | 49 (36.57) | 32 (23.88) | |
| CAZ (30 µg) | 53 (39.55) | 49 (36.57) | 32 (23.88) | |
| Phenicols | FFC (30 µg) | 49 (36.56) | 10 (7.46) | 75 (55.97) |
| Tetracyclines | TE (30 µg) | 40 (29.85) | 25 (18.66) | 69 (51.49) |
| DO (30 µg) | 52 (38.81) | 13 (9.70) | 69 (51.49) | |
| Aminoglycosides | CN (30 µg) | 45 (33.58) | 45 (33.58) | 44 (32.84) |
| N (30 µg) | 25 (18.65) | 40 (29.85) | 69 (51.49) | |
| Fluoroquinolones | CIP (5 µg) | 53 (39.55) | 49 (36.57) | 32 (23.88) |
| LEV (5 µg) | 80 (59.70) | 22 (16.41) | 32 (23.88) | |
| ENR (5 µg) | 34 (25.37) | 44 (32.83) | 56 (41.79) | |
| NOR (10 µg) | 85 (63.43) | 20 (14.92) | 29 (21.64) | |
| Sulfonamides | SXT (23.75 + 1.25 µg) | 40 (29.85) | 2 (1.5) | 92 (68.65) |
| Phenotypic Multidrug Resistance Patterns | No. of Isolates (%) | MAR Index |
|---|---|---|
| SXT, DO, AUG, KF, CL, FFC | 1 (0.75) | 0.35 |
| DO, AUG, CL, FFC | 1 (0.75) | 0.24 |
| ENR, NOR, SXT, TE, DO, AMP, AUG, KF, CL, FOX | 1 (0.75) | 0.59 |
| N, ENR, SXT, AMP, CL | 1 (0.75) | 0.29 |
| AUG, KF, CL, FOX, FFC | 2 (1.49) | 0.29 |
| SXT, KF, CL, FFC | 1 (0.75) | 0.24 |
| SXT, DO, AMP, KF, FFC | 1 (0.75) | 0.29 |
| ENR, NOR, SXT, AMP, CL, FOX | 1 (0.75) | 0.35 |
| TE, DO, AMP, AUG, KF, CL, FFC | 1 (0.75) | 0.41 |
| DO, AMP, CL, FOX, FFC | 1 (0.75) | 0.29 |
| ENR, NOR, DO, CL, FOX | 1 (0.75) | 0.29 |
| TE, DO, AMP, KF, CL, FFC | 1 (0.75) | 0.35 |
| TE, DO, AMP, CL, FFC | 1 (0.75) | 0.29 |
| ENR, NOR, TE, DO, AMP, CL, FOX | 1 (0.75) | 0.41 |
| N, ENR, TE, DO, AMP, CL, FFC | 1 (0.75) | 0.41 |
| CN, N, ENR, NOR, DO, AMP, KF, CL, FOX, FFC | 1 (0.75) | 0.59 |
| N, AMP, AUG, KF, CL, FOX, CTX, FFC | 1 (0.75) | 0.47 |
| SXT, DO, AMP, KF, CL, CTX, FFC | 1 (0.75) | 0.41 |
| ENR, NOR, SXT, DO, AMP, KF, CL, FOX, CTX | 1 (0.75) | 0.53 |
| N, ENR, SXT, TE, AMP, KF, CL, CTX | 1 (0.75) | 0.47 |
| AMP, AUG, KF, CL, FFC | 1 (0.75) | 0.29 |
| SXT, AMP, KF, CL, FFC | 1 (0.75) | 0.29 |
| LEV, SXT, TE, DO, AMP, KF, CL, CTX, FFC | 1 (0.75) | 0.53 |
| ENR, NOR, DO, AMP, KF, CL, FOX, FFC | 1 (0.75) | 0.47 |
| ENR, SXT, DO, AMP, KF, CL, FOX, FFC | 1 (0.75) | 0.47 |
| SXT, TE, AMP, AUG, KF, CL, FFC | 1 (0.75) | 0.41 |
| TE, AMP, KF, CL, FFC | 1 (0.75) | 0.29 |
| N, ENR, NOR, SXT, TE, DO, KF, CL, FOX | 1 (0.75) | 0.53 |
| CN, N, SXT, TE, AMP, KF, CL | 2 (1.49) | 0.41 |
| AMP, AUG, KF, CL, FOX, FFC | 1 (0.75) | 0.35 |
| SXT, AMP, AUG, KF, CL, FOX, FFC | 1 (0.75) | 0.41 |
| SXT, DO, AMP, KF, CL, FFC | 2 (1.49) | 0.35 |
| ENR, NOR, SXT, DO, AMP, CL, FOX, CTX, FFC | 1 (0.75) | 0.53 |
| ENR, SXT, DO, AMP, CL, CTX | 1 (0.75) | 0.35 |
| SXT, TE, DO, AMP, CL, CTX | 1 (0.75) | 0.35 |
| CN, N, LEV, SXT, DO, AMP, AUG, KF, CL, CTX, FFC | 1 (0.75) | 0.65 |
| CN, N, CIP, LEV, ENR, SXT, DO, AMP, AUG, KF, FOX, CTX, CAZ, FFC | 1 (0.75) | 0.82 |
| SXT, DO, AMP, AUG, KF, CL | 2 (1.49) | 0.35 |
| CN, N, CIP, LEV, ENR, NOR, SXT, DO, AMP, AUG, KF, FOX, CTX, CAZ | 1 (0.75) | 0.82 |
| SXT, TE, DO, AMP, AUG, KF, CL, CTX | 1 (0.75) | 0.47 |
| CN, N, SXT, TE, DO, AMP, AUG, CL, FFC | 1 (0.75) | 0.53 |
| CN, N, CIP, LEV, ENR, NOR, TE, DO, AUG, CL, FOX, CTX, CAZ, FFC | 1 (0.75) | 0.82 |
| CN, N, CIP, ENR, NOR, SXT, TE, DO, AUG, KF, FOX, CTX, CAZ, FFC | 1 (0.75) | 0.82 |
| CN, N, DO, AMP, AUG, KF, CL, CTX | 1 (0.75) | 0.47 |
| CN, N, CIP, ENR, SXT, DO, AMP, AUG, KF, FOX, CTX, CAZ, FFC | 1 (0.75) | 0.76 |
| LEV, SXT, DO, AMP, AUG, KF, CL, CTX | 1 (0.75) | 0.47 |
| CN, N, CIP, ENR, NOR, SXT, DO, AMP, AUG, KF, FOX, CTX, CAZ | 1 (0.75) | 0.76 |
| LEV, ENR, SXT, TE, DO, AUG, CL, FOX, CTX | 1 (0.75) | 0.53 |
| ENR, TE, DO, AUG, CL, CTX | 1 (0.75) | 0.35 |
| CN, N, CIP, SXT, LEV, ENR, NOR, TE, DO, AUG, CL, FOX, CTX, CAZ, FFC | 1 (0.75) | 0.88 |
| LEV, SXT, TE, DO, AUG, CL, CTX | 1 (0.75) | 0.41 |
| CN, N, TE, AMP, AUG, KF, FFC | 1 (0.75) | 0.41 |
| N, ENR, SXT, TE, DO, AUG, KF, CL, FOX | 2 (1.49) | 0.53 |
| N, SXT, TE, AMP, AUG, KF | 1 (0.75) | 0.35 |
| LEV, SXT, TE, DO, AMP, AUG, KF, CL, CTX | 1 (0.75) | 0.53 |
| TE, AMP, AUG, KF | 2 (1.49) | 0.24 |
| SXT, AMP, AUG, KF | 2 (1.49) | 0.24 |
| LEV, SXT, AMP, AUG, KF | 1 (0.75) | 0.29 |
| N, ENR, SXT, TE, AUG, KF, CL, FOX | 1 (0.75) | 0.47 |
| CN, N, SXT, AMP, AUG, KF | 1 (0.75) | 0.35 |
| CN, N, CIP, LEV, ENR, NOR, TE, DO, AUG, CL, FOX, CTX, CAZ | 1 (0.75) | 0.76 |
| LEV, TE, DO, AUF, CL, CTX, FFC | 1 (0.75) | 0.41 |
| N, TE, DO, AUG, CL, FFC | 1 (0.75) | 0.35 |
| CN, N, CIP, LEV, ENR, AMP, AUG, KF, FOX, CAZ, FFC | 1 (0.75) | 0.65 |
| LEV, SXT, DO, AMP, AUG, KF, CL, FFC | 3 (2.24) | 0.47 |
| CN, N, SXT, DO, AMP, AUG, KF, FFC | 1 (0.75) | 0.47 |
| CN, N, CIP, LEV, ENR, NOR, TE, DO, AUG, CL, FOX, CAZ, FFC | 1 (0.75) | 0.76 |
| CN, N, CIP, ENR, SXT, TE, DO, AUG, CL, FOX, CAZ | 1 (0.75) | 0.65 |
| N, SXT, AMP, KF, FFC | 1 (0.75) | 0.29 |
| CN, LEV, SXT, TE, DO, AUG, CL, FFC | 1 (0.75) | 0.47 |
| SXT, TE, DO, AMP, AUG, KF, CL, FFC | 2 (1.49) | 0.47 |
| CN, N, CIP, LEV, ENR, NOR, SXT, AMP, AUG, KF, FOX, CAZ | 1 (0.75) | 0.71 |
| CN, N, CIP, ENR, SXT, TE, AUG, CL, FOX, CAZ | 1 (0.75) | 0.59 |
| CN, N, LEV, DO, AMP, AUG, KF, CL | 1 (0.75) | 0.47 |
| N, SXT, TE, AUG, CL, FFC | 1 (0.75) | 0.35 |
| CN, N, CIP, ENR, SXT, TE, DO, AUG, CL, FOX, CAZ, FFC | 1 (0.75) | 0.71 |
| SXT, DO, AMP, AUG, KF, CL, FFC | 2 (1.49) | 0.41 |
| N, SXT, AMP, AUG, KF, FFC | 2 (1.49) | 0.35 |
| LEV, SXT, TE, DO, CL, FFC | 1 (0.75) | 0.35 |
| N, SXT, TE, CL, FFC | 1 (0.75) | 0.29 |
| SXT, TE, DO, CL, FFC | 2 (1.49) | 0.29 |
| CN, N, CIP, LEV, ENR, NOR, SXT, AMP, KF, FOX, CAZ, FFC | 1 (0.75) | 0.71 |
| CN, N, CIP, ENR, SXT, AMP, KF, FOX, CAZ | 1 (0.75) | 0.53 |
| CN, N, CIP, ENR, NOR, SXT, AMP, KF, FOX, CAZ | 1 (0.75) | 0.59 |
| SXT, TE, CL, FFC | 1 (0.75) | 0.24 |
| SXT, TE, DO, AUG, CL, FFC | 1 (0.75) | 0.29 |
| CN, N, CIP, LEV, ENR, SXT, AMP, AUG, KF, FOX, CTX, CAZ, FFC | 1 (0.75) | 0.76 |
| N, SXT, TE, DO, AMP, AUG, KF, FFC | 1 (0.75) | 0.47 |
| CN, N, CIP, ENR, NOR, SXT, TE, AMP, AUG, KF, FOX, CAZ, FFC | 1 (0.75) | 0.76 |
| CN, N, CIP, LEV, ENR, NOR, SXT, TE, DO, AUG, CL, FOX, CTX, CAZ | 1 (0.75) | 0.82 |
| N, AMP, AUG, KF | 2 (1.49) | 0.24 |
| CN, N, CIP, ENR, NOR, SXT, AMP, AUG, KF, FOX, CAZ, FFC | 1 (0.75) | 0.71 |
| N, SXT, TE, AMP, AUG, KF, FFC | 1 (0.75) | 0.41 |
| CN, N, CIP, ENR, SXT, TE, AMP, AUG, FOX, CTX, CAZ, FFC | 1 (0.75) | 0.71 |
| CN, N, CIP, ENR, NOR, SXT, AMP, AUG, KF, FOX, CAZ | 1 (0.75) | 0.65 |
| CN, N, CIP, LEV, ENR, SXT, TE, AUG, CL, FOX, CAZ | 1 (0.75) | 0.65 |
| TE, DO, AUG, CL, FFC | 1 (0.75) | 0.29 |
| N, TE, AUG, CL, FFC | 1 (0.75) | 0.29 |
| CN, N, CIP, ENR, NOR, AMP, AUG, KF, FOX, CAZ, FFC | 1 (0.75) | 0.65 |
| CN, N, CIP, LEV, ENR, AMP, AUG, KF, FOX, CAZ | 1 (0.75) | 0.59 |
| SXT, AMP, AUG, KF, CL | 2 (1.49) | 0.29 |
| SXT, TE, AMP, AUG, KF, CL | 1 (0.75) | 0.35 |
| N, ENR, NOR, SXT, TE, AUG, KF, CL, FOX | 1 (0.75) | 0.53 |
| ENR, SXT, TE, AUG, CL, FOX | 1 (0.75) | 0.35 |
| N, LEV, ENR, SXT, TE, AUG, CL | 1 (0.75) | 0.41 |
| SXT, TE, AUG, CL | 1 (0.75) | 0.41 |
| N, ENR, NOR, TE, AUG, KF, CL, FOX | 1 (0.75) | 0.47 |
| N, SXT, TE, AMP, KF | 1 (0.75) | 0.29 |
| TE, AMP, AUG, KF, CL, FFC | 2 (1.49) | 0.35 |
| CN, N, CIP, LEV, ENR, SXT, AMP, KF, FOX, CTX, CAZ | 1 (0.75) | 0.65 |
| CN, N, CIP, ENR, NOR, SXT, DO, AMP, KF, FOX, CAZ, FFC | 1 (0.75) | 0.71 |
| CN, N, AMP, KF, CL, CTX | 1 (0.75) | 0.35 |
| CN, N, CIP, LEV, ENR, AMP, KF, FOX, CTX, CAZ | 1 (0.75) | 0.59 |
| CN, N, CIP, LEV, ENR, TE, DO, AMP, AUG, KF, FOX, CTX, CAZ, FFC | 1 (0.75) | 0.82 |
| N, ENR, TE, AUG, KF, CL, FOX | 1 (0.75) | 0.41 |
| N, TE, AMP, AUG, KF | 1 (0.75) | 0.29 |
| Resistance Genes | No. of Resistance Genes Present | Number of Phenotypic-Resistant Isolates (n = 134) | Resistance Gene, %, 95%CI |
|---|---|---|---|
| tetA | 21 | 69 | 30.43 (20.80–42.13) |
| tetB | 4 | 69 | 5.8 (1.85–14.40) |
| tetD | 2 | 69 | 2.9 (0.2–10.57) |
| sul-1 | 23 | 92 | 25 (17.22–34.78) |
| sul-2 | 51 | 92 | 55.43 (45.26–65.17) |
| blaTEM | 69 | 93 | 74.19 (64.42–82.05) |
| blaSHV | 17 | 93 | 18.28 (11.64–27.43) |
| PampC | 35 | 93 | 37.63 (28.45–47.80) |
| blaOXA-1 | 31 | 93 | 33.33 (24.56–43.43) |
| blaOXA-2 | 38 | 93 | 40.86 (31.42–51.03) |
| blaCTX-M | 65 | 93 | 69.89 (59.90–78.31) |
| blaCMY-1 | 6 | 93 | 6.45 (2.72–13.64) |
| blaCMY-2 | 16 | 93 | 17.20 (10.77–26.24) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mim, Z.T.; Nath, C.; Sattar, A.A.; Rashid, R.; Abir, M.H.; Khan, S.A.; Kalam, M.A.; Shano, S.; Cobbold, R.; Alawneh, J.I.; et al. Epidemiology and Molecular Characterisation of Multidrug-Resistant Escherichia coli Isolated from Cow Milk. Vet. Sci. 2024, 11, 609. https://doi.org/10.3390/vetsci11120609
Mim ZT, Nath C, Sattar AA, Rashid R, Abir MH, Khan SA, Kalam MA, Shano S, Cobbold R, Alawneh JI, et al. Epidemiology and Molecular Characterisation of Multidrug-Resistant Escherichia coli Isolated from Cow Milk. Veterinary Sciences. 2024; 11(12):609. https://doi.org/10.3390/vetsci11120609
Chicago/Turabian StyleMim, Zarin Tasnim, Chandan Nath, Abdullah Al Sattar, Rijwana Rashid, Mehedy Hasan Abir, Shahneaz Ali Khan, Md Abul Kalam, Shahanaj Shano, Rowland Cobbold, John I. Alawneh, and et al. 2024. "Epidemiology and Molecular Characterisation of Multidrug-Resistant Escherichia coli Isolated from Cow Milk" Veterinary Sciences 11, no. 12: 609. https://doi.org/10.3390/vetsci11120609
APA StyleMim, Z. T., Nath, C., Sattar, A. A., Rashid, R., Abir, M. H., Khan, S. A., Kalam, M. A., Shano, S., Cobbold, R., Alawneh, J. I., & Hassan, M. M. (2024). Epidemiology and Molecular Characterisation of Multidrug-Resistant Escherichia coli Isolated from Cow Milk. Veterinary Sciences, 11(12), 609. https://doi.org/10.3390/vetsci11120609







