RAA-CRISPR/Cas12a-Mediated Rapid, Sensitive, and Onsite Detection of Newcastle Disease in Pigeons
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Virus and Clinical Samples
2.2. Reagents and Instruments
2.3. Design and Synthesis of CRISPR RNA (crRNA) and Target DNA Primers
2.4. ssDNA Reporter Probe Design and Validation
2.5. Primer Design and Recombinase-Aided Amplification (RAA)
2.6. RAA-CRISPR/Cas12a Sensitivity Analysis
2.7. RAA-CRISPR/Cas12a Specificity Analysis
2.8. RAA-CRISPR/Cas12a-LFD
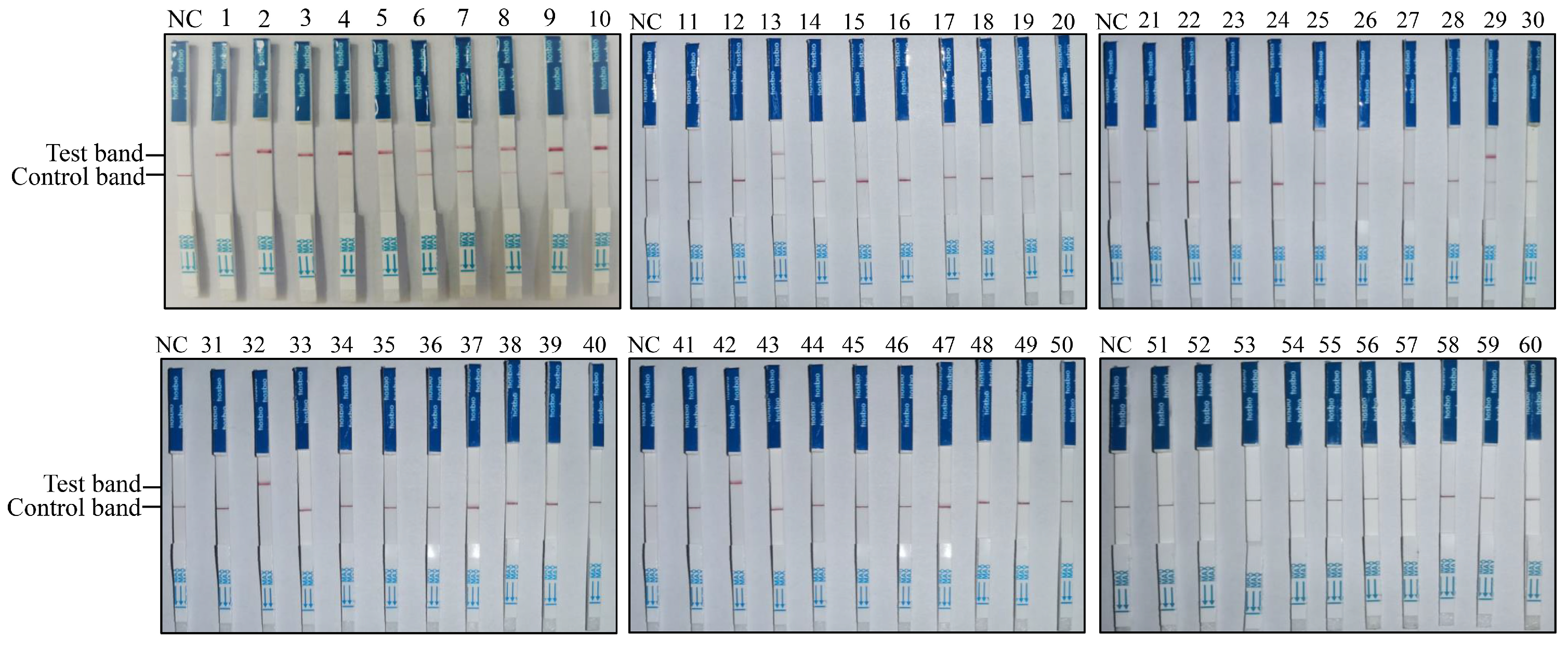
2.9. Clinical Sample Testing
2.10. Data Analysis
3. Results
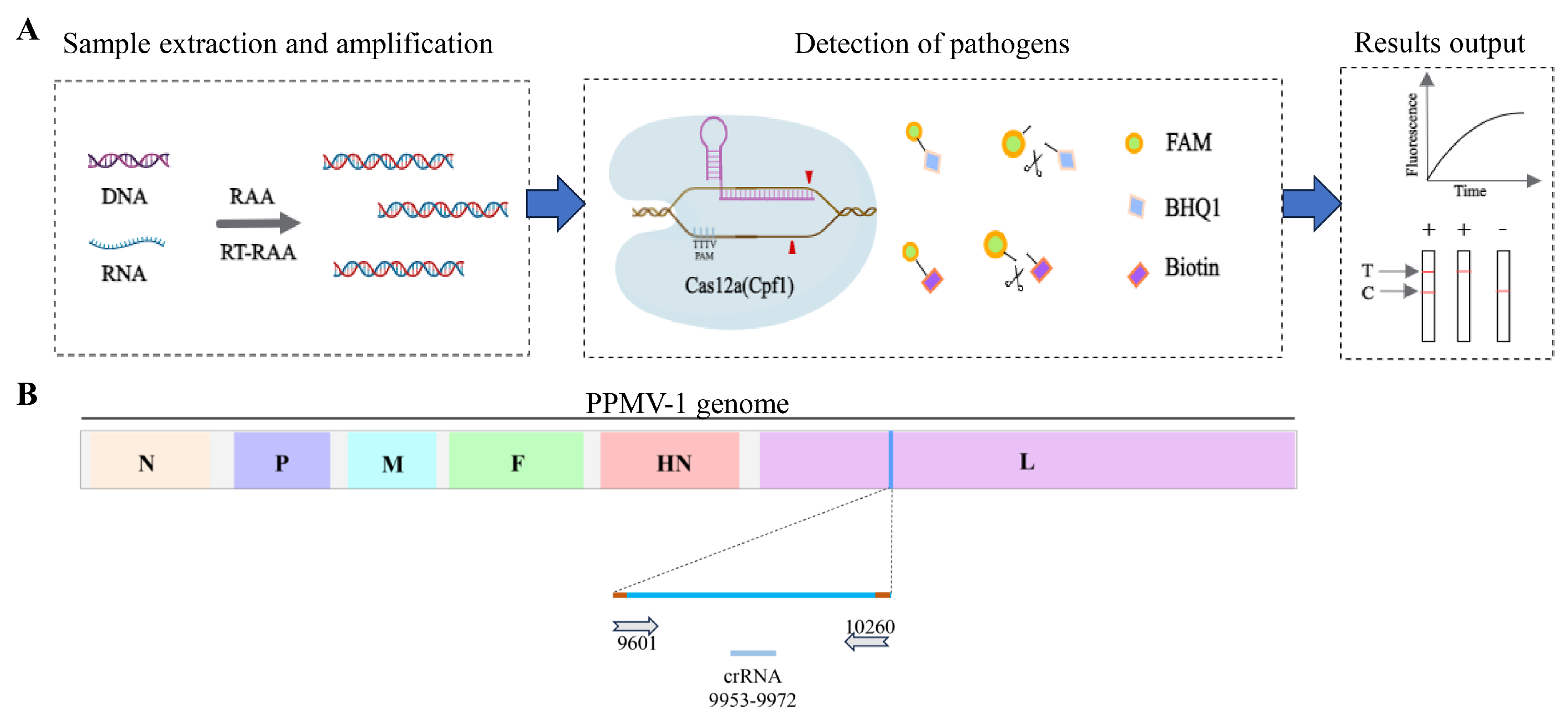
3.1. Schematic of the RAA-CRISPR/Cas12a System for the Detection of PPMV-1
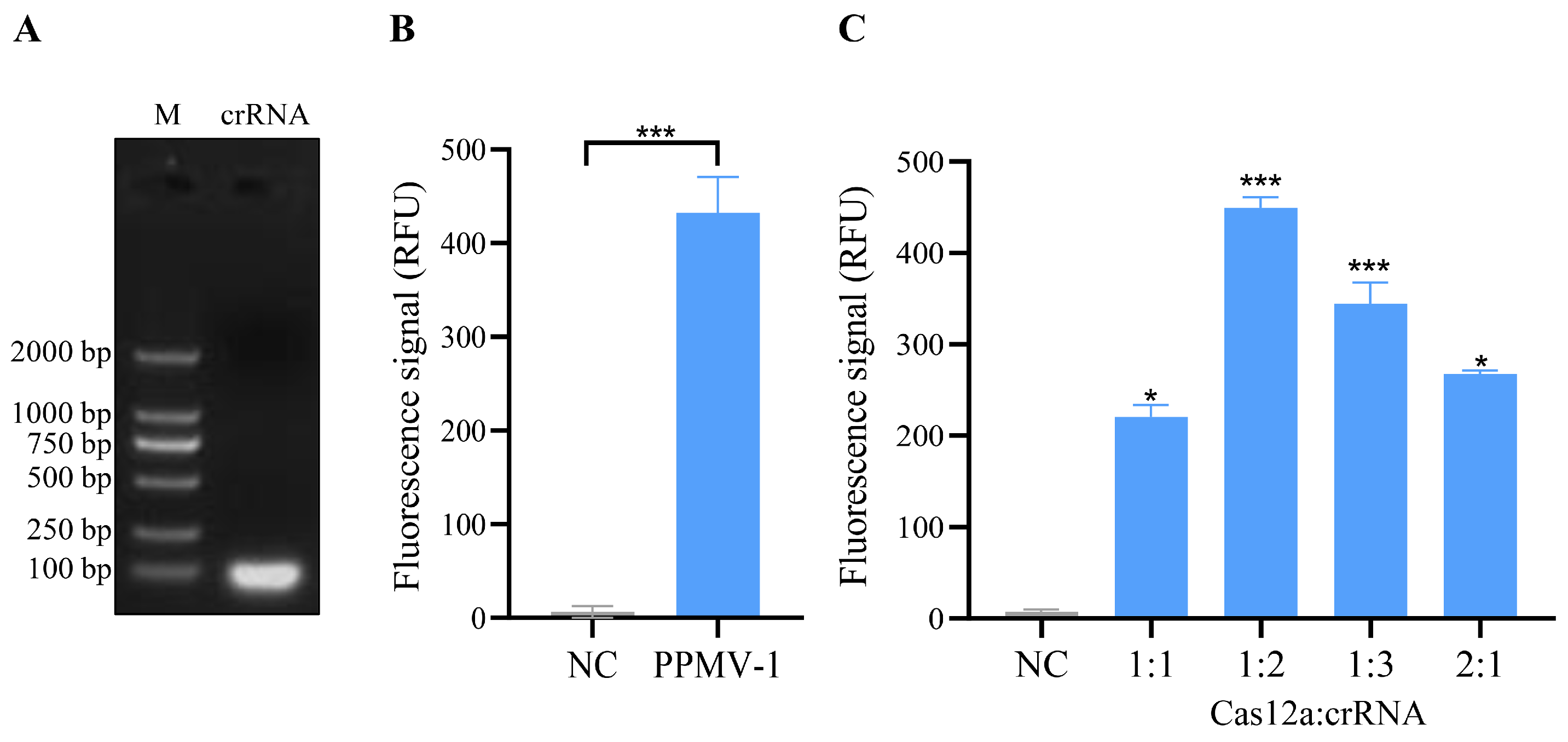
3.2. crRNA-Guided CRISPR/Cas12a Cleavage Activity and System Optimization
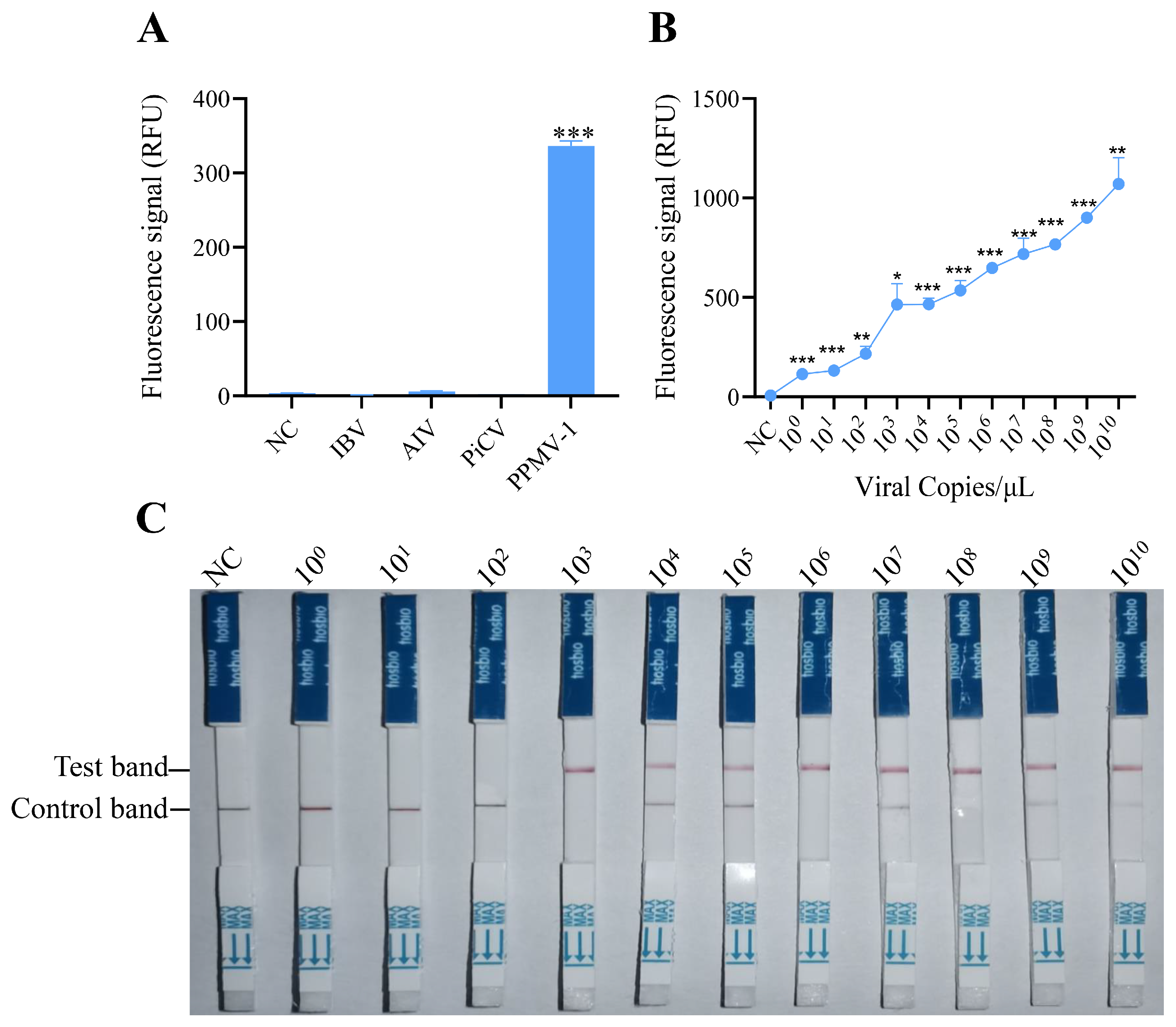
3.3. Specificity and Sensitivity of the RAA-CRISPR/Cas12a System
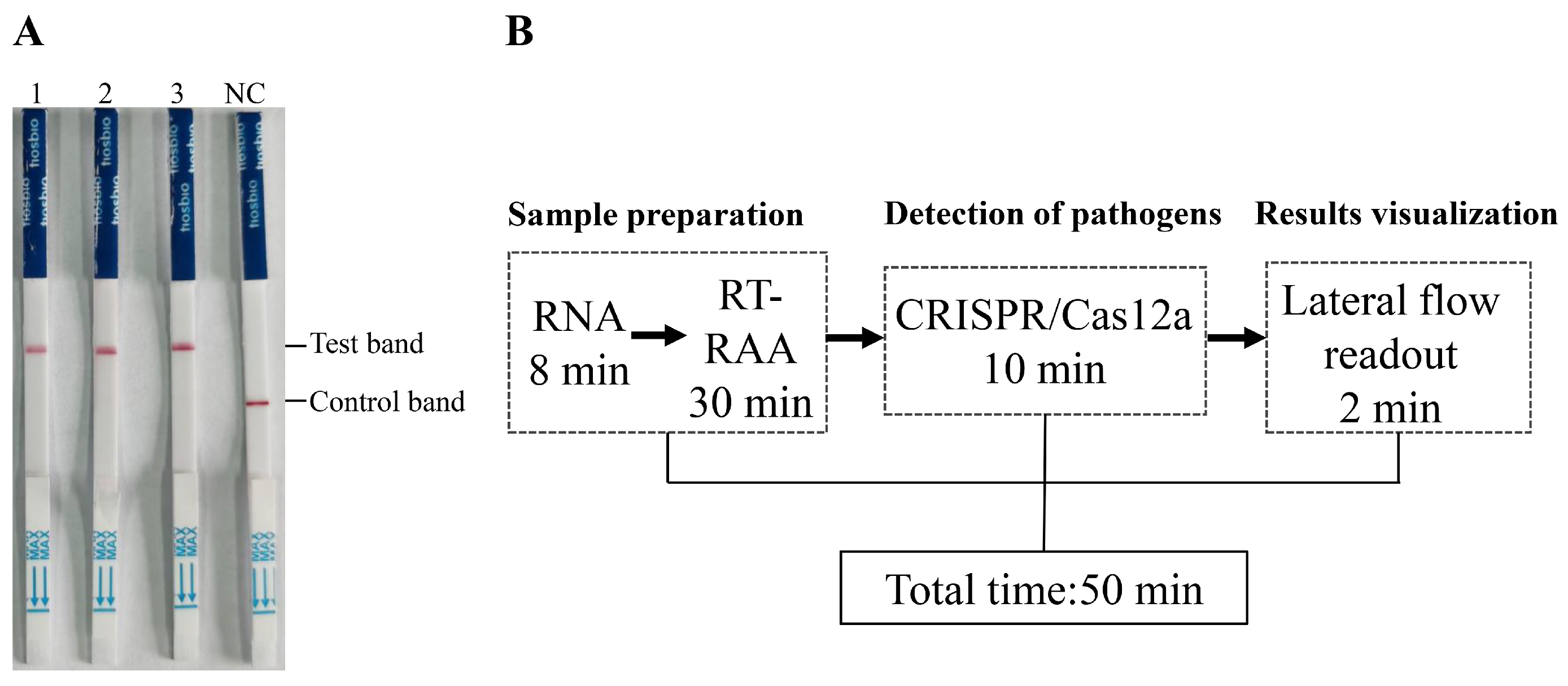
3.4. Onsite Detection Capability of RAA-CRISPR/Cas12a-LFD
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dimitrov, K.M.; Ramey, A.M.; Qiu, X.; Bahl, J.; Afonso, C.L. Temporal, geographic, and host distribution of avian paramyxovirus 1 (Newcastle disease virus). Infect. Genet. Evol. 2016, 39, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Esmaeelzadeh-Dizaji, R.; Molouki, A.; Hosseini, H.; Fallah-Mehrabadi, M.H.; Ziafati-Kafi, Z.; Takalou, A.; Eram, N.; Kumar, N.; Ashuri, A.; Sadri, N.; et al. Molecular characterization of a pigeon paramyxovirus type 1 virus isolated from Eurasian collared doves in Iran, 2017. J. Vet. Sci. 2022, 23, e29. [Google Scholar] [CrossRef] [PubMed]
- Molini, U.; Aikukutu, G.; Khaiseb, S.; Cattoli, G.; Dundon, W.G. Phylogenetic Analysis of Pigeon Paramyxoviruses Type-1 Identified in Mourning Collared-doves (Streptopelia decipiens) in Namibia, Africa. J. Wildl. Dis. 2018, 54, 601–606. [Google Scholar] [CrossRef] [PubMed]
- Zhan, T.; Lu, X.; He, D.; Gao, X.; Chen, Y.; Hu, Z.; Wang, X.; Hu, S.; Liu, X. Phylogenetic analysis and pathogenicity assessment of pigeon paramyxovirus type 1 circulating in China during 2007–2019. Transbound. Emerg. Dis. 2022, 69, 2076–2088. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Luo, Y.; Wang, J.; Shu, B.; Jiang, W.; Liu, S.; Li, Y.; Li, J.; Hou, G.; Peng, C.; et al. A molecular, epidemiological and pathogenicity analysis of pigeon paramyxovirus type 1 viruses isolated from live bird markets in China in 2014–2021. Virus Res. 2022, 318, 198846. [Google Scholar] [CrossRef]
- Zhan, T.; He, D.; Lu, X.; Liao, T.; Wang, W.; Chen, Q.; Liu, X.; Gu, M.; Wang, X.; Hu, S.; et al. Biological Characterization and Evolutionary Dynamics of Pigeon Paramyxovirus Type 1 in China. Front. Vet. Sci. 2021, 8, 721102. [Google Scholar] [CrossRef]
- Sheng, W.; Wang, K.; Gui, Y.; Qi, X.; Shen, L.; Zhang, Y.; Tang, C.; Li, X.; Tao, J.; Cao, C.; et al. Molecular characteristics and phylogenetic analysis of pigeon paramyxovirus type 1 isolates from pigeon meat farms in Shanghai (2009–2012). Sci. Rep. 2024, 14, 10741. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, W.; Li, Y.; Liu, J.; Wang, W.; Bai, J.; Yang, Z.; Liu, H.; Xiao, S. A pigeon paramyxovirus type 1 isolated from racing pigeon as an inactivated vaccine candidate provides effective protection. Poult. Sci. 2022, 101, 102097. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef]
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Zhu, M.; Meng, Q.; Wang, Y.; Wang, X. Real-time detection of Seneca Valley virus by one-tube RPA-CRISPR/Cas12a assay. Front. Cell Infect. Microbiol. 2023, 13, 1305222. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Misra, C.S.; Bindal, G.; Rangu, S.S.; Rath, D. CRISPR-Cas12a assisted specific detection of mpox virus. J. Med. Virol. 2023, 95, e28974. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zong, N.; Ye, F.; Mei, Y.; Qu, J.; Jiang, X. Dual-CRISPR/Cas12a-Assisted RT-RAA for Ultrasensitive SARS-CoV-2 Detection on Automated Centrifugal Microfluidics. Anal. Chem. 2022, 94, 9603–9609. [Google Scholar] [CrossRef]
- Liu, L.; Duan, J.J.; Wei, X.Y.; Hu, H.; Wang, Y.B.; Jia, P.P.; Pei, D.S. Generation and application of a novel high-throughput detection based on RPA-CRISPR technique to sensitively monitor pathogenic microorganisms in the environment. Sci. Total Environ. 2022, 838 Pt 2, 156048. [Google Scholar] [CrossRef]
- Qian, W.; Huang, J.; Wang, T.; Fan, C.; Kang, J.; Zhang, Q.; Li, Y.; Chen, S. Ultrasensitive and visual detection of human norovirus genotype GII.4 or GII.17 using CRISPR-Cas12a assay. Virol. J. 2022, 19, 150. [Google Scholar] [CrossRef]
- Chen, X.L.; Liu, C.C.; Li, F.X.; Zhou, J.H.; Huang, Z.H.; Zhang, H.L.; Wang, H.L.; Huang, P.; Cao, Z.G.; Chiu, S.D. Rapid and Visual Detection of Monkey B Virus Based on Recombinase Polymerase Amplification. Zoonoses 2023, 3, 39. [Google Scholar] [CrossRef]
- Li, Y.; Shang, J.; Luo, J.; Zhang, F.; Meng, G.; Feng, Y.; Jiang, W.; Yu, X.; Deng, C.; Liu, G.; et al. Rapid detection of H5 subtype avian influenza virus using CRISPR Cas13a based-lateral flow dipstick. Front. Microbiol. 2023, 14, 1283210. [Google Scholar] [CrossRef]
- Wang, H.Y.; Wu, M.C.; Chen, H.W.; Lai, Y.C.; Huang, W.H.; Chang, H.W.; Jeng, C.R.; Cheng, C.H.; Wang, P.J.; Lai, Y.H.; et al. Isolation, full sequence analysis, and in situ hybridization of pigeon paramyxovirus-1 genotype VI.2.1.1.2.2 from oriental turtle doves (Streptopelia orientalis). Poult. Sci. 2023, 102, 102974. [Google Scholar] [CrossRef]
- Tian, Y.; Xue, R.; Yang, W.; Li, Y.; Xue, J.; Zhang, G. Characterization of ten paramyxovirus type 1 viruses isolated from pigeons in China during 1996–2019. Vet. Microbiol. 2020, 244, 108661. [Google Scholar] [CrossRef] [PubMed]
- Xie, P.; Chen, L.; Zhang, Y.; Lin, Q.; Ding, C.; Liao, M.; Xu, C.; Xiang, B.; Ren, T. Evolutionary Dynamics and Age-Dependent Pathogenesis of Sub-Genotype VI.2.1.1.2.2 PPMV-1 in Pigeons. Viruses 2020, 12, 433. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Deng, Q.; Li, H.; Pan, C.; Zhai, G.; Yuan, Y.; Cheng, E.; Zhang, Y.; Mo, M.; Huang, T.; et al. Molecular characterization of two novel sub-sublineages of pigeon paramyxovirus type 1 in China. Arch. Virol. 2018, 163, 2971–2984. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Liu, X.; Xu, Y.; Han, Z.; Shao, Y.; Kong, X.; Liu, S. A comparative study of pigeons and chickens experimentally infected with PPMV-1 to determine antigenic relationships between PPMV-1 and NDV strains. Vet. Microbiol. 2014, 168, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Hossain, I.; Parvin, R.; Rahman, M.M.; Begum, J.A.; Chowdhury, E.H.; Islam, M.R.; Diel, D.G.; Nooruzzaman, M. Comparative pathogenicity of a genotype XXI.1.2 pigeon Newcastle disease virus isolate in pigeons and chickens. Microb. Pathog. 2023, 178, 106068. [Google Scholar] [CrossRef]
- Wang, X.; Ren, S.; Wang, X.; Wang, C.Y.; Fan, M.; Jia, Y.; Gao, X.; Liu, H.; Xiao, S.; Yang, Z. Genomic characterization of a wild-bird-origin pigeon paramyxovirus type 1 (PPMV-1) first isolated in the northwest region of China. Arch. Virol. 2017, 162, 749–761. [Google Scholar] [CrossRef]
- Nooruzzaman, M.; Barman, L.R.; Mumu, T.T.; Chowdhury, E.H.; Dimitrov, K.M.; Islam, M.R. A Pigeon-Derived Sub-Genotype XXI.1.2 Newcastle Disease Virus from Bangladesh Induces High Mortality in Chickens. Viruses 2021, 13, 1520. [Google Scholar] [CrossRef]
- Kommers, G.D.; King, D.J.; Seal, B.S.; Carmichael, K.P.; Brown, C.C. Pathogenesis of six pigeon-origin isolates of Newcastle disease virus for domestic chickens. Vet. Pathol. 2002, 39, 353–362. [Google Scholar] [CrossRef]
- Hurley, S.; Eden, J.S.; Bingham, J.; Rodriguez, M.; Neave, M.J.; Johnson, A.; Howard-Jones, A.R.; Kok, J.; Anazodo, A.; McMullan, B.; et al. Fatal Human Neurologic Infection Caused by Pigeon Avian Paramyxovirus-1, Australia. Emerg. Infect. Dis. 2023, 29, 2482–2487. [Google Scholar] [CrossRef]
- Liu, M.; Qu, Y.; Wang, F.; Liu, S.; Sun, H. Genotypic and pathotypic characterization of Newcastle disease virus isolated from racing pigeons in China. Poult. Sci. 2015, 94, 1476–1482. [Google Scholar] [CrossRef]
- Liang, R.; Liang, L.; Ren, X.; Jia, Y.; Han, K.; Zhao, J.; Song, C.; Cui, S. Development of a TaqMan loop-mediated isothermal amplification assay for the rapid detection of pigeon paramyxovirus type 1. Arch. Virol. 2021, 166, 1599–1605. [Google Scholar] [CrossRef] [PubMed]
- Song, H.S.; Kim, H.S.; Kim, J.Y.; Kwon, Y.K.; Kim, H.R. The Development of Novel Reverse Transcription Loop-Mediated Isothermal Amplification Assays for the Detection and Differentiation of Virulent Newcastle Disease Virus. Int. J. Mol. Sci. 2023, 24, 13847. [Google Scholar] [CrossRef] [PubMed]
- Lv, R.; Guo, J.; Zhang, Y.; Wang, X.; Li, G.; Meng, Z.; Wang, L.; Chai, S.; Li, Q.; Zhang, G. Development and Evaluation of a Blocking Lateral Flow Assay Strip for Detection of Newcastle Disease Virus Antibodies. Vet. Sci. 2023, 10, 152. [Google Scholar] [CrossRef] [PubMed]
| Primer Name | Sequence (5′→3′) |
|---|---|
| PPMV-1-crRNA-F | GAAATTAATACGACTCACTATAGGGTAATTTCTACTAAGTGTAGATTAGAATCAAATGATTTTGAT |
| PPMV-1-crRNA-R | ATCAAAATCATTTGATTCTAATCTACACTTAGTAGAAATTACCCTATAGTGAGTCGTATTAATTTC |
| PPMV-1-Target DNA-F | CCATGGGAAGAAGAATTCAGGT |
| PPMV-1-Target DNA-R | CTCGAGTCATGATTGCGGTTT |
| PPMV-1-RAA-F | AACCTCAACTAACCGCCTCTTGATAGAGTTT |
| PPMV-1-RAA-R Reporter I Reporter II | CTGCCATTACCTGGCAGTTTCTTAATCT FAM-TTATTATT-BHQ1 FAM-TTATTATT-Biotin |
| Detection Results | PCR (Positive/Total) | qPCR (Positive/Total) | RAA-CRISPR/Cas12a-LFD (Positive/Total) |
|---|---|---|---|
| Number of positive samples | 8/60 | 14/60 | 14/60 |
| Positive rate | 13% | 23% | 23% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, L.; Wang, D.; Gao, Z.; Tang, J.; Li, X.; Ren, P.; Wang, Y.; Gao, S.; Wu, X.; Guo, Y.; et al. RAA-CRISPR/Cas12a-Mediated Rapid, Sensitive, and Onsite Detection of Newcastle Disease in Pigeons. Vet. Sci. 2024, 11, 473. https://doi.org/10.3390/vetsci11100473
Liang L, Wang D, Gao Z, Tang J, Li X, Ren P, Wang Y, Gao S, Wu X, Guo Y, et al. RAA-CRISPR/Cas12a-Mediated Rapid, Sensitive, and Onsite Detection of Newcastle Disease in Pigeons. Veterinary Sciences. 2024; 11(10):473. https://doi.org/10.3390/vetsci11100473
Chicago/Turabian StyleLiang, Libin, Dou Wang, Zhen Gao, Jiao Tang, Xing Li, Pengfei Ren, Ying Wang, Shimin Gao, Xingchen Wu, Yanna Guo, and et al. 2024. "RAA-CRISPR/Cas12a-Mediated Rapid, Sensitive, and Onsite Detection of Newcastle Disease in Pigeons" Veterinary Sciences 11, no. 10: 473. https://doi.org/10.3390/vetsci11100473
APA StyleLiang, L., Wang, D., Gao, Z., Tang, J., Li, X., Ren, P., Wang, Y., Gao, S., Wu, X., Guo, Y., Yang, B., & Li, J. (2024). RAA-CRISPR/Cas12a-Mediated Rapid, Sensitive, and Onsite Detection of Newcastle Disease in Pigeons. Veterinary Sciences, 11(10), 473. https://doi.org/10.3390/vetsci11100473






