The First Report on the Complete Sequence Characterization of Bluetongue Virus Serotype 3 in the Republic of Korea
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Samples
2.2. Detection of BTV Antibodies
2.3. RNA Extraction and Real-Time RT-PCR(RT-qPCR)
2.4. Virus Isolation
2.5. Sequencing and Phylogenetic Analysis
3. Results
3.1. Detection of BTV Antibodies in Cattle and Goats
3.2. Molecular Detection and BTV Isolation
3.3. Sequence and Phylogenetic Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Caporale, M.; Di Gialleonorado, L.; Janowicz, A.; Wilkie, G.; Shaw, A.; Savini, G.; Rijn, P.A.V.; Mertens, P.; Ventura, M.D.; Palmarini, M. Virus and host factors affecting the clinical outcome of bluetongue virus infection. J. Virol. 2014, 88, 10399–10411. [Google Scholar] [CrossRef] [PubMed]
- MacLachlan, N.J.; Osburn, B.I. Impact of bluetongue virus infection on the international movement and trade of ruminants. J. Am. Vet. Med. Assoc. 2006, 228, 1346–1349. [Google Scholar] [CrossRef]
- Nomikou, K.; Hughes, J.; Wash, R.; Kellam, P.; Breard, E.; Zientara, S.; Palmarini, M.; Biek, R.; Mertens, P. Widespread reassortment shapes the evolution and epidemiology of bluetongue virus following European invasion. PLoS Pathog. 2015, 11, e1005056. [Google Scholar] [CrossRef]
- Kelso, J.K.; Milne, G.J. A spatial simulation model for the dispersal of the bluetongue vector Culicoides brevitarsis in Australia. PLoS ONE 2014, 9, e104646. [Google Scholar] [CrossRef] [PubMed]
- Purse, B.V.; Mellor, P.S.; Rogers, K.J.; Samuel, A.R.; Mertens, P.P.; Baylis, M. Climate change and the recent emergence of bluetongue in Europe. Nat. Rev. Microbiol. 2005, 3, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Jacquet, S.; Huber, K.; Pagès, N.; Talavera, S.; Burgin, L.E.; Carpenter, S.; Sanders, C.; Dicko, A.H.; Djerbal, M.; Goffredo, M.; et al. Range expansion of the Bluetongue vector, Culicoides imicola, in continental France likely due to rare wind-transport events. Sci. Rep. 2016, 6, 27247. [Google Scholar] [CrossRef] [PubMed]
- Roy, P. Orbiviruses and their replicaion. In Fields Virology, 6th ed.; Knipe, D.M., Howley, P.M., Cohen, J.I., Griffin, D.E., Lamb, R.A., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007; Volume 2, pp. 1975–1997. [Google Scholar]
- Shaw, A.E.; Ratinier, M.; Nunes, S.F.; Nomikou, K.; Caporale, M.; Golder, M.; Allan, K.; Hamers, C.; Hudelet, P.; Zientara, S.; et al. Reassortment between two serologically unrelated bluetongue virus strains is flexible and can involve any genome segment. J. Virol. 2013, 87, 543–557. [Google Scholar] [CrossRef] [PubMed]
- Maan, N.S.; Maan, S.; Belaganahalli, M.N.; Ostlund, E.N.; Johnson, D.J.; Nomikou, K.; Mertens, P.C. Identification and differentiation of the twenty six bluetongue virus serotypes by RT-PCR amplification of the serotype-specific genome segment 2. PLoS ONE 2012, 7, 32601. [Google Scholar] [CrossRef]
- WOAH Terrestrial Manual 2021 Chapter 3.1.3. Bluetongue (Infection with Bluetongue Virus). Available online: https://Woah.org/fileadmin/Home/eng/Health_standards/tahm/3.01.03_BLUETONGUE.pdf (accessed on 15 May 2021).
- Ries, C.; Sharav, T.; Tseren-Ochir, E.O.; Beer, M.; Hoffmann, B. Putative novel serotypes ‘33’ and ‘35’ in clinically healthy small ruminants in Mongolia expand the group of atypical BTV. Viruses 2020, 13, 42. [Google Scholar] [CrossRef]
- Ries, C.; Vögtlin, A.; Hüssy, D.; Jandt, T.; Gobet, H.; Hilbe, M.; Burgener, C.; Schweizer, L.; Häfliger-Speiser, S.; Beer, M.; et al. Putative novel atypical BTV serotype ‘36’ identified in small ruminants in Switzerland. Viruses 2021, 13, 721. [Google Scholar] [CrossRef]
- Golender, N.; Bumbarov, V.; Eldar, A.; Lorusso, A.; Kenigswald, G.; Varsano, J.S.; David, D.; Schainin, S.; Dagoni, I.; Gur, I.; et al. Bluetongue Serotype 3 in Israel 2013–2018: Clinical Manifestations of the Disease and Molecular Characterization of Israeli Strains. Front. Vet. Sci. 2020, 7, 112. [Google Scholar] [CrossRef] [PubMed]
- Cappai, S.; Rolesu, S.; Loi, F.; Liciardi, M.; Leone, A.; Marcacci, M.; Teodori, L.; Mangone, I.; Sghaier, S.; Portanti, O.; et al. Western Bluetongue virus serotype 3 in Sardinia, diagnosis and characterization. Transbound. Emerg. Dis. 2019, 66, 1426–1431. [Google Scholar] [CrossRef] [PubMed]
- Lorusso, A.; Sghaier, S.; Di Domenico, M.; Barbria, M.E.; Zaccaria, G.; Megdich, A.; Portanti, O.; Seliman, I.B.; Spedicato, M.; Pizzurro, F.; et al. Analysis of bluetongue serotype 3 spread in Tunisia and discovery of a novel strain related to the bluetongue virus isolated from a commercial sheep pox vaccine. Infect. Genet. Evol. 2018, 59, 63–71. [Google Scholar] [CrossRef]
- Holwerda, M.; Santman-Berends, I.-M.G.A.; Harders, F.; Engelsma, M.; Vloet, R.-P.M.; Dijkstra, E.; van Gennip, R.-G.P.; Mars, M.H.; Spierengurg, M.; Roos, L.; et al. Emergence of bluetongue virus serotype 3 in the Netherlands in September 2023. bioRxiv, 2023; preprint. [Google Scholar] [CrossRef]
- Gray, A. UK confirms cases of bluetongue serotype-3. Vet Rec. 2023. [Google Scholar] [CrossRef]
- Daniels, P.W.; Sendow, I.; Pritchard, L.I.; Sukarsih; Eaton, B.T. Regional overview of bluetongue viruses in South-East Asia: Viruses, vectors and surveillance. Vet. Ital. 2004, 40, 94–100. [Google Scholar] [PubMed]
- Saminathan, M.; Singh, K.P.; Khorajiya, J.H.; Dinesh, M.; Vineetha, S.; Maity, M.; Rahman, A.F.; Misri, J.; Malik, Y.S.; Gupta, V.K.; et al. An updated review on bluetongue virus: Epidemiology, pathobiology, and advances in diagnosis and control with special reference to India. Vet. Q. 2020, 40, 258–321. [Google Scholar] [CrossRef]
- Boyle, D.B.; Ritchie, R.A.; Broz, I.; Walker, P.J.; Melville, L.; Flanagan, D.; Davis, S.; Hunt, N.; Weir, R. Evolution of Bluetongue Virus Serotype 1 in Northern Australia over 30 Years. J. Virol. 2014, 88, 13981–13989. [Google Scholar] [CrossRef][Green Version]
- Goto, Y.; Yamaguchi, O.; Kubo, M. Epidemiological observations on bluetongue in sheep and cattle in Japan. Vet. Ital. 2004, 40, 78–82. [Google Scholar]
- Kato, T.; Shirafuji, H.; Tanaka, S.; Sato, M.; Yamakawa, M.; Tsuda, T.; Yanase, T. Bovine arboviruses in Culicoides biting midges and sentinel cattle in southern Japan from 2003 to 2013. Transbound Emerg. Dis. 2016, 63, e160–e172. [Google Scholar] [CrossRef]
- Shirafuji, H.; Yanase, T.; Kato, T.; Yamakawa, M. Genetic and phylogenetic characterization of genome segments 2 and 6 of bluetongue virus isolates in Japan from 1985 to 2008. J. Gen. Virol. 2012, 93, 1465–1473. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Miura, Y.; Inaba, Y.; Tsuda, T.; Tokuhisa, S.; Sato, K.; Akashi, H. Seroepizootiological survey on bluetongue virus infection in cattle in Japan. Natl. Inst. Anim. Health Q. 1982, 22, 154–158. [Google Scholar]
- Yang, H.; Lv, M.; Sun, M.; Lin, L.; Kou, M.; Gao, L.; Liao, D.; Xiong, H.; He, Y.; Li, H. Complete genome sequence of the first bluetongue virus serotype 7 isolate from China: Evidence for entry of African-lineage strains and reassortment between the introduced and native strains. Arch. Virol. 2016, 161, 223–227. [Google Scholar] [CrossRef]
- Sun, E.C.; Huang, L.P.; Xu, Q.Y.; Wang, H.X.; Xue, X.M.; Lu, P.; Li, W.J.; Liu, W.; Bu, Z.G.; Wu, D.L. Emergence of a novel bluetongue virus serotype, China 2014. Transbound. Emerg. Dis. 2016, 63, 585–589. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Yang, H.; Li, H.; Xiao, L.; Wang, J.; Li, N.; Zhang, N. Full-genome sequence of bluetongue virus serotype 1 (BTV-1) strain Y863, the first BTV-1 isolate of Eastern origin found in China. Genome Announc. 2013, 1, e00403-13. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Liu, N.; Xu, Q.; Sun, E.; Qin, Y.; Zhao, J.; Feng, Y.; Wu, D. Complete genomic sequence of bluetongue virus serotype 1 from China. J. Virol. 2012, 86, 1288–1289. [Google Scholar] [CrossRef][Green Version]
- Yang, H.; Zhu, J.; Li, H.; Xiao, L.; Wang, J.; Li, N.; Zhang, N.; Kirkland, P.D. Full genome sequence of bluetongue virus serotype 4 from China. J. Virol. 2012, 86, 13122–13123. [Google Scholar] [CrossRef]
- Hwang, J.M.; Kim, J.G.; Yeh, J.Y. Serological evidence of bluetongue virus infection and serotype distribution in dairy cattle in South Korea. BMC Vet. Res. 2019, 15, 255. [Google Scholar] [CrossRef]
- Seo, H.J.; Park, J.Y.; Cho, Y.S.; Cho, I.S.; Yeh, J.Y. First report of Bluetongue virus isolation in the Republic of Korea and analysis of the complete coding sequence of the segment 2 gene. Virus Genes 2015, 50, 156–159. [Google Scholar] [CrossRef]
- Jeong, D.-Y.; Ragen, P.; Juan, R.; António, A. Final Report of the Project on the Impact of Climate Change on Island and Coastal Biosphere Reserves. UNESCO Digital Library. 2015. Available online: https://unesdoc.unesco.org/ark:/48223/pf0000246972 (accessed on 15 February 2015).
- Son, W.-S. Climate Change and Tourism Sustainability in Jeju Island Landscape. Sustainability 2023, 15, 88. [Google Scholar] [CrossRef]
- Climate Change Jeju City. Available online: https://www.meteoblue.com/en/climate-change/jeju-city_south-korea_1846266 (accessed on 15 May 2021).
- Orrù, G.; Ferrando, M.L.; Meloni, M.; Liciardi, M.; Savini, G.; de Santis, P. Rapid detection and quantitation of bluetongue virus (BTV) using a molecular beacon fluorescent probe assay. J. Virol. Methods 2006, 137, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Maan, S.; Rao, S.; Maan, N.S.; Anthony, S.J.; Attoui, H.; Attoui, H.; Samuel, A.R.; Mertens, P.P.C. Rapid cDNA synthesis and sequencing techniques for the genetic study of bluetongue and other dsRNA viruses. J. Virol. Methods 2007, 143, 132–139. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for window 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Rojas, J.M.; Martín, V.; Sevilla, N. Vaccination as a Strategy to Prevent Bluetongue Virus Vertical Transmission. Pathogens 2021, 10, 1528. [Google Scholar] [CrossRef] [PubMed]
- Van Rijn, P.A. Prospects of Next-Generation Vaccines for Bluetongue Front. Vet. Sci. 2019, 6, 407. [Google Scholar] [CrossRef]
- Spedicato, M.; Compagni, E.D.; Caporale, M.; Teodor, L.; Leone, A.; Ancora, M.; Mangone, I.; Perletta, F.; Portanti, O.; Giallonardo, F.D.; et al. Reemergence of an atypical bluetongue virus strain in goats, Sardinia, Italy. Res. Vet. Sci. 2022, 10, 36–41. [Google Scholar] [CrossRef]
- Maan, S.; Maan, N.S.; Belaganahalli, M.N.; Rao, P.P.; Singh, K.P.; Hemadri, D.; Putty, K.; Kumar, A.; Batra, K.; Krishnajyothi, Y.; et al. Full-Genome Sequencing as a Basis for Molecular Epidemiology Studies of Bluetongue Virus in India. PLoS ONE 2015, 29, 0131257. [Google Scholar] [CrossRef]
- Hornyák, Á.; Malik, P.; Marton, S.; Dóró, R.; Cadar, D.; Bányai, K. Emergence of multireassortant bluetongue virus serotype 4 in Hungary. Infect. Genet. Evol. 2015, 33, 6–10. [Google Scholar] [CrossRef]
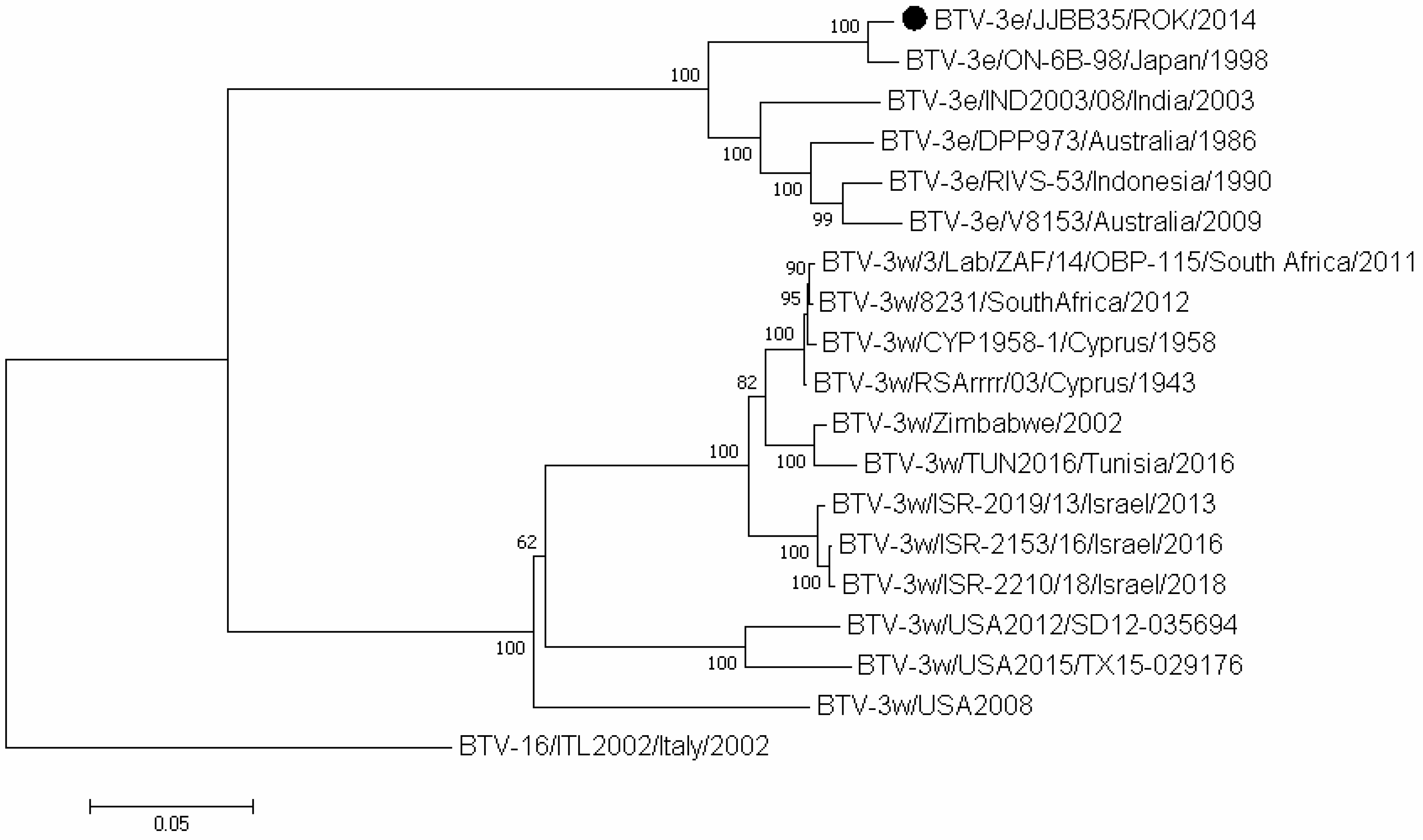
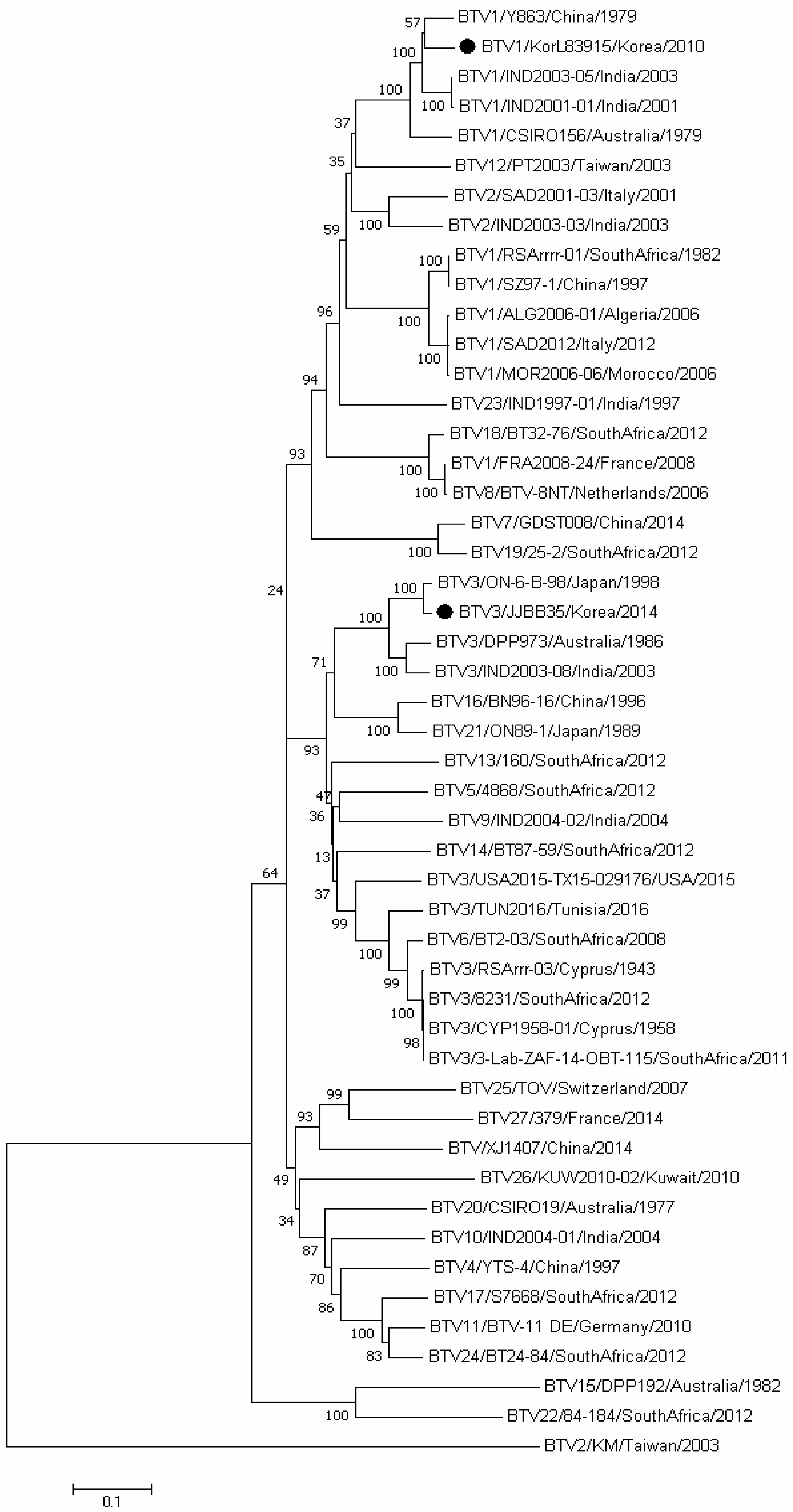
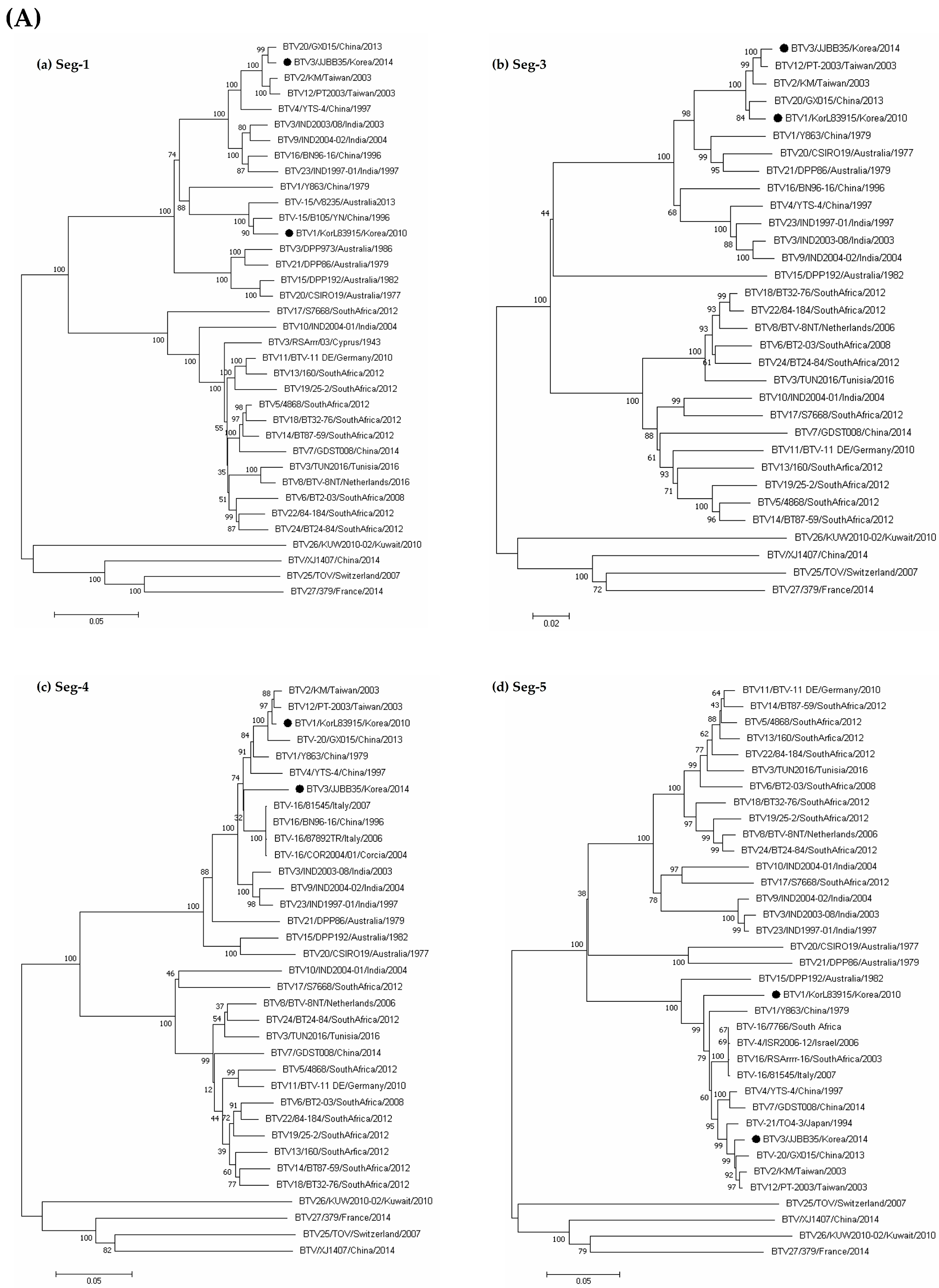
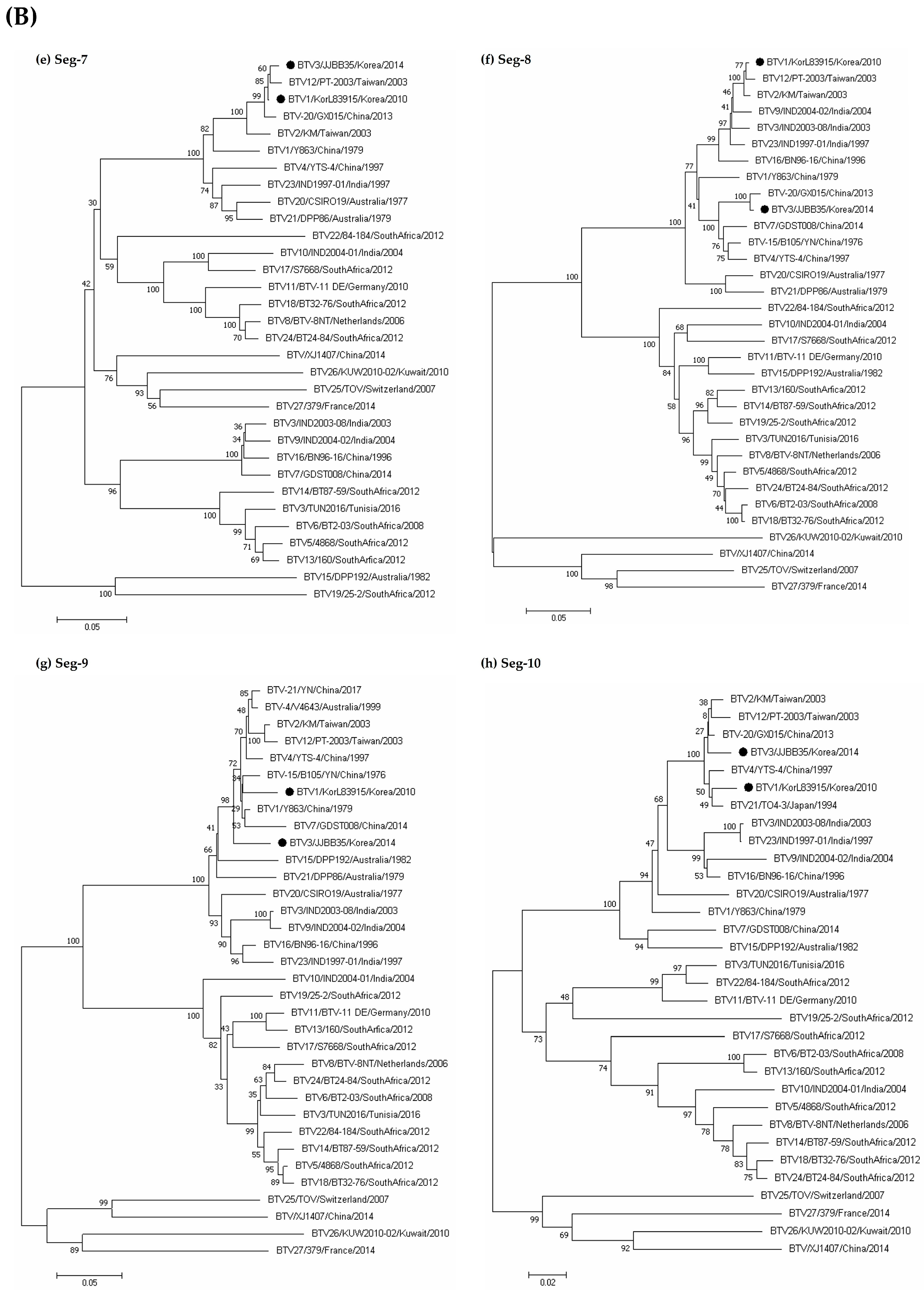
| Species | Regions | 2013 | 2014 | Total | ||
|---|---|---|---|---|---|---|
| No. of Samples | No. of Positive Samples (%) | No. of Samples | No of Positive Samples (%) | |||
| Cattle | Gyonggi-do | 378 | 3 (0.79) | 219 | 0 (0.0) | 3/597 (0.5) |
| Ganwon-do | 746 | 2 (0.27) | 140 | 0 (0.0) | 2/886 (0.23) | |
| Chungcheongbuk-do | 686 | 2 (0.29) | 60 | 32 (53.3) | 34/746 (4.56) | |
| Chungcheongnam-do | 222 | 0 (0.0) | 48 | 15 (31.3) | 15/270 (5.56) | |
| Jeollabuk-do | 170 | 0 (0.0) | 60 | 0 (0.0) | 0/230 (0.0) | |
| Jeollanam-do | 469 | 1 (0.21) | 392 | 1 (0.26) | 2/861 (0.23) | |
| Gyeongsangbuk-do | 309 | 0 (0.0) | 564 | 1 (0.18) | 1/873 (0/11) | |
| Gyeongsangnam-do | 265 | 3 (1.13) | 604 | 6 (0.99) | 9/869 (1.04) | |
| Jeju-do | 235 | 33 (14.0) | 267 | 95 (35.6) | 128/502 (25.5) | |
| Total | 3480 | 44 (1.26) | 2354 | 150 (6.37) | 194/5834 (3.33) | |
| Goat | Gyonggi-do | 144 | 0 (0.0) | - | - | 0/144 (0.0) |
| Ganwon-do | 58 | 0 (0.0) | - | - | 0/58 (0.0) | |
| Chungcheongbuk-do | 64 | 0 (0.0) | - | - | 0/64 (0.0) | |
| Chungcheongnam-do | 20 | 0 (0.0) | - | - | 0/20 (0.0) | |
| Jeollabuk-do | 25 | 0 (0.0) | - | - | 0/25 (0.0) | |
| Jeollanam-do | 100 | 1 (1.0) | 60 | 0 (0.0) | 1/160 (0.63) | |
| Gyeongsangbuk-do | 32 | 0 (0.0) | 56 | 0 (0.0) | 0/88 (0.0) | |
| Gyeongsangnam-do | 204 | 0 (0.0) | 312 | 1 (0.32) | 1/516 (0.19) | |
| Total | 647 | 1 (0.15) | 428 | 1 (0.23) | 2/1075 (0.19) | |
| Strain | Segment | Closest Sequence—nt Identity (%) |
|---|---|---|
| JJBB35 | Seg-1 | BTV-20/GX015/2013/China (99.1) |
| (BTV-3) | Seg-2 | BTV-3/ON-6/B/98/Japan (98.3) |
| Seg-3 | BTV-12/PT/2003/Taiwan (99.0) | |
| Seg-4 | BTV-16/BN96-16/1996/China (95.9) | |
| Seg-5 | BTV-12/PT/2003/Taiwan (98.9) | |
| Seg-6 | BTV-3/ON-6/B/98/Japan (98.1) | |
| Seg-7 | BTV-1/KorL83915/2010/ROK (99.1) | |
| Seg-8 | BTV-20/GX015/2013/China (99.0) | |
| Seg-9 | BTV-1/Y863/1979/China (96.1) | |
| Seg-10 | BTV-20/GX015/2013/China (98.9) | |
| KorL83915 | Seg-1 | BTV-15/B105/YN/1976/China (97.5) |
| (BTV-1) | Seg-2 | BTV-1/Kuala Lumpur/1987/Malaysia (93.9) |
| Seg-3 | BTV-20/GX015/2013/China * (97.8) | |
| Seg-4 | BTV-12/PT/2003/Taiwan (99.1) | |
| Seg-5 | BTV-16/81545/2007/Italy (94.8) | |
| Seg-6 | BTV-1/Y863/1979/China (93.8) | |
| Seg-7 | BTV-12/PT/2003/Taiwan (99.1) | |
| Seg-8 | BTV-2/KM/2003/Taiwan (99.1) | |
| Seg-9 | BTV-1/Y863/1979/China (96.5) | |
| Seg-10 | BTV-4/YTS-4/1997/China (98.7) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.-J.; Choi, J.-G.; Seong, D.-S.; Jeong, J.-U.; Kim, H.-J.; Park, S.-W.; Yun, S.-P.; Roh, I.-S. The First Report on the Complete Sequence Characterization of Bluetongue Virus Serotype 3 in the Republic of Korea. Vet. Sci. 2024, 11, 29. https://doi.org/10.3390/vetsci11010029
Kim H-J, Choi J-G, Seong D-S, Jeong J-U, Kim H-J, Park S-W, Yun S-P, Roh I-S. The First Report on the Complete Sequence Characterization of Bluetongue Virus Serotype 3 in the Republic of Korea. Veterinary Sciences. 2024; 11(1):29. https://doi.org/10.3390/vetsci11010029
Chicago/Turabian StyleKim, Hyun-Jeong, Jun-Gu Choi, Da-Seul Seong, Jong-Uk Jeong, Hye-Jung Kim, Sang-Won Park, Seung-Pil Yun, and In-Soon Roh. 2024. "The First Report on the Complete Sequence Characterization of Bluetongue Virus Serotype 3 in the Republic of Korea" Veterinary Sciences 11, no. 1: 29. https://doi.org/10.3390/vetsci11010029
APA StyleKim, H.-J., Choi, J.-G., Seong, D.-S., Jeong, J.-U., Kim, H.-J., Park, S.-W., Yun, S.-P., & Roh, I.-S. (2024). The First Report on the Complete Sequence Characterization of Bluetongue Virus Serotype 3 in the Republic of Korea. Veterinary Sciences, 11(1), 29. https://doi.org/10.3390/vetsci11010029







