Comparative Genomics of Staphylococcus rostri, an Undescribed Bacterium Isolated from Dairy Mastitis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolate Collection and Identification
2.2. Whole-Genome Sequencing Analysis
3. Results
3.1. Bacterial Species Delineation
3.2. S. rostri Isolate Distribution
3.3. Pan-Genome Analysis
3.4. Virulence Factors, Antibiotic Resistance and Mobile Genetic Elements
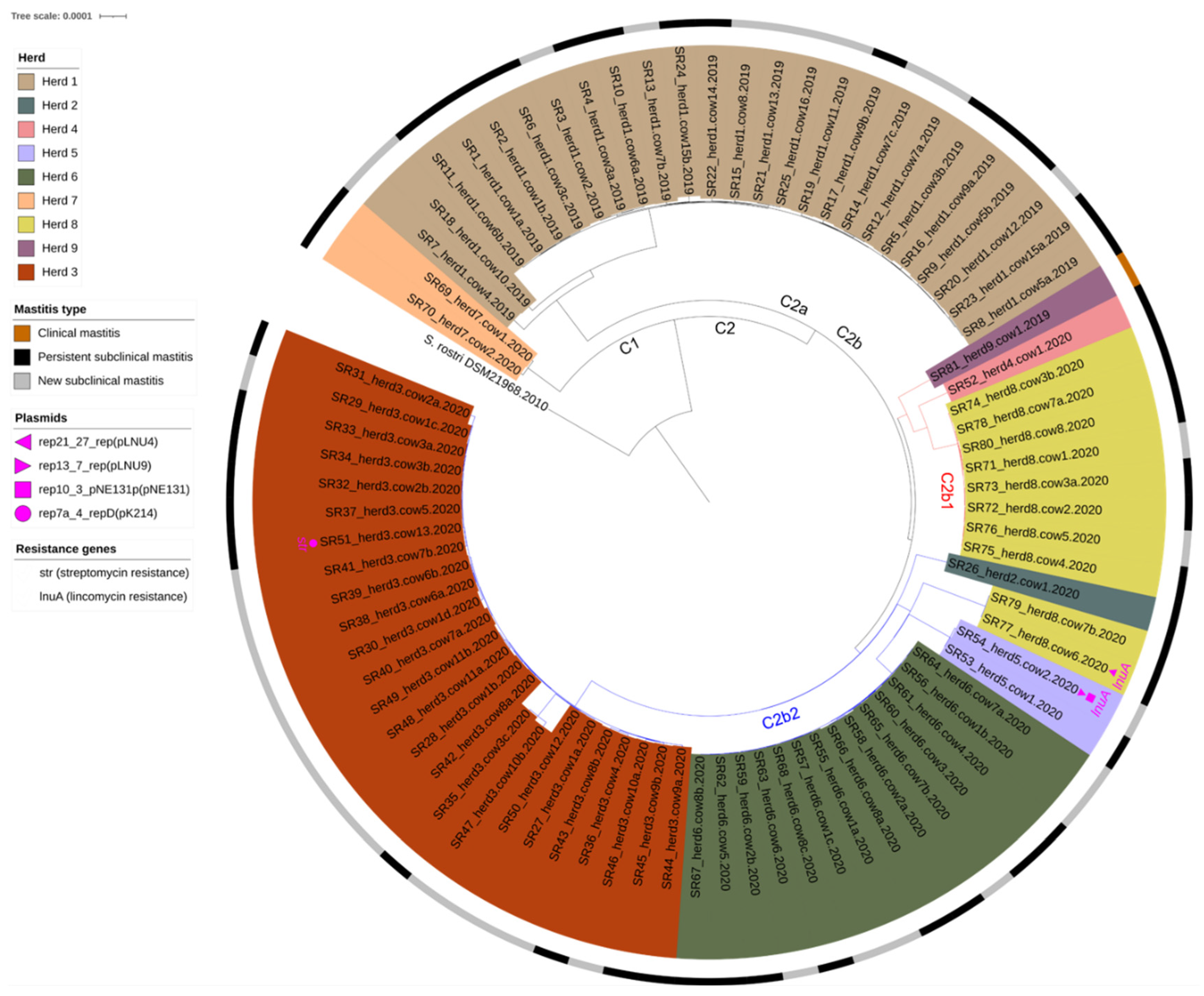
3.5. Core genome Phylogeny
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Buck, J.; Ha, V.; Naushad, S.; Nobrega, B.D.; Luby, C.; Middleton, R.J.; De Vliegher, S.; Barkema, H.W. Non-aureus Staphylococci and Bovine Udder Health: Current Understanding and Knowledge Gaps. Front. Vet. Sci. 2021, 8, 658031. [Google Scholar] [CrossRef] [PubMed]
- Tenhagen, B.A.; Köster, G.; Wallmann, J.; Heuwieser, W. Prevalence of mastitis pathogens and their resistance against antimicrobial agents in dairy cows in Brandenburg, Germany. J. Dairy Sci. 2006, 89, 2542–2551. [Google Scholar] [CrossRef]
- Pitkälä, A.; Haveri, M.; Pyörälä, S.; Myllys, V.; Honkanen-Buzalski, T. Bovine mastitis in Finland 2001—Prevalence, distribution of bacteria, and antimicrobial resistance. J. Dairy Sci. 2004, 87, 2433–2441. [Google Scholar] [CrossRef] [PubMed]
- Vanderhaeghen, W.; Piepers, S.; Leroy, F.; Van Coillie, E.; Haesebrouck, F.; De Vliegher, S. Invited review: Effect, persistence, and virulence of coagulase-negative Staphylococcus species associated with ruminant udder health. J. Dairy Sci. 2014, 97, 5275–5293. [Google Scholar] [CrossRef] [PubMed]
- Hamel, J.; Zhang, Y.; Wente, N.; Krömker, V. Non-S. aureus staphylococci (NAS) in milk samples: Infection or contamination? Vet. Microbiol. 2020, 242, 108594. [Google Scholar] [CrossRef] [PubMed]
- Astrup, L.B.; Pedersen, K.; Farre, M. Microbiological Diagnoses on Clinical Mastitis-Comparison between Diagnoses Made in Veterinary Clinics versus in Laboratory Applying MALDI-TOF MS. Antibiotics. 2022, 11, 271. [Google Scholar] [CrossRef]
- Cremonesi, P.; Monistero, V.; Moroni, P.; Barberio, A.; Almeida, R.; Latorre, A.A.; Castiglioni, B. Detection methods. In Encyclopedia of Dairy Sciences, 3rd ed.; Bansal, N., Lance, H.B., Everett, L.D.D., Harte, F., Lean, I.J., McNamara, J.P., Smithers, G.W., Tsakalidou, E., Eds.; Elsevier Ltd.: Amsterdam, The Netherlands, 2022; Volume 4, pp. 457–468. [Google Scholar]
- Nonnemann, B.; Lyhs, U.; Svennesen, L.; Kristensen, K.A.; Klaas, I.C.; Pedersen, K. Bovine mastitis bacteria resolved by MALDI-TOF mass spectrometry. J. Dairy Sci. 2019, 102, 2515–2524. [Google Scholar] [CrossRef]
- Mahmmod, Y.S.; Nonnemann, B.; Svennesen, L.; Pedersen, K.; Klaas, I.C. Typeability of MALDI-TOF assay for identification of non-aureus staphylococci associated with bovine intramammary infections and teat apex colonization. J. Dairy Sci. 2018, 101, 9430–9438. [Google Scholar] [CrossRef]
- Nováková, D.; Pantůček, R.; Hubálek, Z.; Falsen, E.; Busse, H.J.; Schumann, P.; Sedláček, I. Staphylococcus microti sp. nov., isolated from the common vole (Microtus arvalis). Int. J. Syst. Evol. Microbiol. 2010, 60, 566–573. [Google Scholar] [CrossRef]
- Riesen, A.; Perreten, V. Staphylococcus rostri sp. nov., a haemolytic bacterium isolated from the noses of healthy pigs. Int. J. Syst. Evol. Microbiol. 2010, 60, 2042–2047. [Google Scholar] [CrossRef][Green Version]
- Strube, M.L.; Hansen, J.E.; Rasmussen, S.; Pedersen, K. A detailed investigation of the porcine skin and nose microbiome using universal and Staphylococcus specific primers. Sci. Rep. 2018, 8, 12751. [Google Scholar] [CrossRef] [PubMed]
- Król, J.; Wanecka, A.; Twardoń, J.; Mrowiec, J.; Dropińska, A.; Bania, J.; Podkowik, M.; Korzeniowska-Kowal, A.; Paściak, M. Isolation of Staphylococcus microti from milk of dairy cows with mastitis. Vet. Microbiol. 2016, 182, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, S.N.; Okello, E.; Rossitto, P.V.; Lehenbauer, T.W.; Champagne, J.; Penedo, M.C.T.; Arruda, A.G.; Godden, S.; Rapnicki, P.; Gorden, P.J.; et al. Molecular epidemiology of coagulase-negative Staphylococcus species isolated at different lactation stages from dairy cattle in the United States. PeerJ 2019, 7, e6749. [Google Scholar] [CrossRef] [PubMed]
- De Visscher, A.; Piepers, S.; Haesebrouck, F.; Supré, K.; De Vliegher, S. Coagulase-negative Staphylococcus species in bulk milk: Prevalence, distribution, and associated subgroup- and species-specific risk factors. J. Dairy Sci. 2017, 100, 629–642. [Google Scholar] [CrossRef] [PubMed]
- Wuytack, A.; De Visscher, A.; Piepers, S.; Boyen, F.; Haesebrouck, F.; De Vliegher, S. Non-aureus staphylococci in fecal samples of dairy cows: First report and phenotypic and genotypic characterization. J. Dairy Sci. 2019, 102, 9345–9359. [Google Scholar] [CrossRef] [PubMed]
- Bizzini, A.; Durussel, C.; Bille, J.; Greub, G.; Prod’hom, G. Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of bacterial strains routinely isolated in a Clinical Microbiology Laboratory. J. Clin. Microbiol. 2010, 48, 1549–1554. [Google Scholar] [CrossRef]
- Bulletin of the International Dairy Federation 448/2011. Suggested Interpretation of Mastitis Terminology (Revision of Bulletin of IDF N◦ 338/1999). List of Terms and Interpretations. Available online: https://shop.fil-idf.org/products/suggestedinterpretation-of-mastitis-terminology-revision-of-bulletin-of-idf-n-3381999 (accessed on 12 September 2022).
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.; Korobeynikov, A.; Lapidus, A.; Prjibelsky, A.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling Single-Cell Genomes and Mini-Metagenomes from Chimeric MDA Products. J. Comput. Biol. 2013, 20, 714–737. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and taxonomy in diagnostics for food security: Soft-rotting enterobacterial plant pathogens. Anal. Methods 2016, 8, 12–24. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 1 May 2023).
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Akhter, S.; Aziz, R.K.; Edwards, R.A. PhiSpy: A novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res. 2012, 40, e126. [Google Scholar] [CrossRef]
- Bin, H.J.; Bolduc, B.; Zablocki, O.; Kuhn, J.H.; Roux, S.; Adriaenssens, E.M.; Brister, J.R.; Kropinski, A.M.; Krupovic, M.; Lavigne, R.; et al. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat. Biotechnol. 2019, 37, 632–639. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.D.; Zhou, S.Y.; Chen, L.H.; Yang, J. VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.F.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.R.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Zankari, E.; Allesøe, R.; Joensen, K.G.; Cavaco, L.M.; Lund, O.; Aarestrup, F.M. PointFinder: A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 2020, 72, 2764–2768. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; Garcia-Fernandez, A.; Voldby, L.M.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. PlasmidFinder and pMLST: In silico detection and typing of plasmids. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.-m.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek Vol. 2017, 110, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Åvall-Jääskeläinen, S.; Taponen, S.; Kant, R.; Paulin, L.; Blom, J.; Palva, A.; Koort, J. Comparative genome analysis of 24 bovine-associated Staphylococcus isolates with special focus on the putative virulence genes. PeerJ 2018, 2018, e4560. [Google Scholar] [CrossRef] [PubMed]
- Queraltó, C.; Álvarez, R.; Ortega, C.; Díaz-Yáñez, F.; Paredes-Sabja, D.; Gil, F. Role and Regulation of Clp Proteases: A Target against Gram-Positive Bacteria. Bacteria 2023, 2, 21–36. [Google Scholar] [CrossRef]
- Aljghami, M.E.; Barghash, M.M.; Majaesic, E.; Bhandari, V.; Houry, W.A. Cellular functions of the ClpP protease impacting bacterial virulence. Front. Mol. Biosci. 2022, 9, 1054408. [Google Scholar] [CrossRef]
- Viñes, J.; Fàbregas, N.; Pérez, D.; Cuscó, A.; Fonticoba, R.; Francino, O.; Ferrer, L.; Migura-Garcia, L. Concordance between Antimicrobial Resistance Phenotype and Genotype of Staphylococcus pseudintermedius from Healthy Dogs. Antibiotics 2022, 11, 1625. [Google Scholar] [CrossRef]
- Lüthje, P.; von Köckritz-Blickwede, M.; Schwarz, S. Identification and characterization of nine novel types of small staphylococcal plasmids carrying the lincosamide nucleotidyltransferase gene lnu(A). J. Antimicrob. Chemother. 2007, 59, 600–606. [Google Scholar] [CrossRef]
- DANMAP 2021. Use of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria from Food Animals, Food and Humans in Denmark. 4.3.2 Antimicrobial Consumption in Cattle. Available online: https://www.danmap.org/reports/2021 (accessed on 26 June 2023).
- Stegmann, R.; Perreten, V. Antibiotic resistance profile of Staphylococcus rostri, a new species isolated from healthy pigs. Vet. Microbiol. 2010, 145, 165–171. [Google Scholar] [CrossRef]
- Vanderhaeghen, W.; Vandendriessche, S.; Crombé, F.; Dispas, M.; Denis, O.; Hermans, K.; Haesebrouck, F.; Butaye, P. Species and staphylococcal cassette chromosome mec (SCCmec) diversity among methicillin-resistant non-Staphylococcus aureus staphylococci isolated from pigs. Vet. Microbiol. 2012, 158, 123–128. [Google Scholar] [CrossRef]
- Locatelli, C.; Piepers, S.; De Vliegher, S.; Barberio, A.; Supré, K.; Scaccabarozzi, L.; Pisoni, G.; Bronzo, V.; Haesebrouck, F.; Moroni, P. Effect on quarter milk somatic cell count and antimicrobial susceptibility of Staphylococcus rostri causing intramammary infection in dairy water buffaloes. J. Dairy Sci. 2013, 96, 3799–3805. [Google Scholar] [CrossRef]
- Centrale Husdyrbrugsregister—CHR. Ministeriet for Fødevarer, Landbrug og Fiskeri. Fødevarestyrelsen. Available online: https://chr.fvst.dk/chri/faces/frontpage (accessed on 21 March 2023).

| Herd | n Isolates | n Animals | n Animals One Quarter | n Animals Two Quarters | n Animals Three Quarters | n Animals All Quarters |
|---|---|---|---|---|---|---|
| herd 1 | 25 | 16 | 9 | 5 | 2 | - |
| herd 2 | 1 | 1 | 1 | - | - | - |
| herd 3 | 25 | 13 | 4 | 7 | 1 | 1 |
| herd 4 | 1 | 1 | 1 | - | - | - |
| herd 5 | 2 | 2 | 2 | - | - | - |
| herd 6 | 14 | 8 | 4 | 2 | 2 | - |
| herd 7 | 2 | 2 | 2 | - | - | - |
| herd 8 | 10 | 8 | 6 | 2 | - | - |
| herd 9 | 1 | 1 | 1 | - | - | - |
| Bacterial Species | n |
|---|---|
| Staphylococcus simulans | 14 |
| Staphylococcus epidermidis | 9 |
| Staphylococcus haemolyticus | 7 |
| Corynebacterium amycolatum | 6 |
| Lactococcus garvieae | 4 |
| Aerococcus viridans | 3 |
| Staphylococcus chromogenes | 3 |
| Streptococcus gallolyticus | 2 |
| Streptococcus uberis | 1 |
| Streptococcus canis | 1 |
| Staphylococcus muscae | 1 |
| Enterococcus faecalis | 1 |
| Escherichia coli | 1 |
| Lactococcus lactis | 1 |
| Citobacter koseri | 1 |
| Kocuria rhizophila | 1 |
| “No ID” | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kløve, D.C.; Farre, M.; Strube, M.L.; Astrup, L.B. Comparative Genomics of Staphylococcus rostri, an Undescribed Bacterium Isolated from Dairy Mastitis. Vet. Sci. 2023, 10, 530. https://doi.org/10.3390/vetsci10090530
Kløve DC, Farre M, Strube ML, Astrup LB. Comparative Genomics of Staphylococcus rostri, an Undescribed Bacterium Isolated from Dairy Mastitis. Veterinary Sciences. 2023; 10(9):530. https://doi.org/10.3390/vetsci10090530
Chicago/Turabian StyleKløve, Desiree Corvera, Michael Farre, Mikael Lenz Strube, and Lærke Boye Astrup. 2023. "Comparative Genomics of Staphylococcus rostri, an Undescribed Bacterium Isolated from Dairy Mastitis" Veterinary Sciences 10, no. 9: 530. https://doi.org/10.3390/vetsci10090530
APA StyleKløve, D. C., Farre, M., Strube, M. L., & Astrup, L. B. (2023). Comparative Genomics of Staphylococcus rostri, an Undescribed Bacterium Isolated from Dairy Mastitis. Veterinary Sciences, 10(9), 530. https://doi.org/10.3390/vetsci10090530







