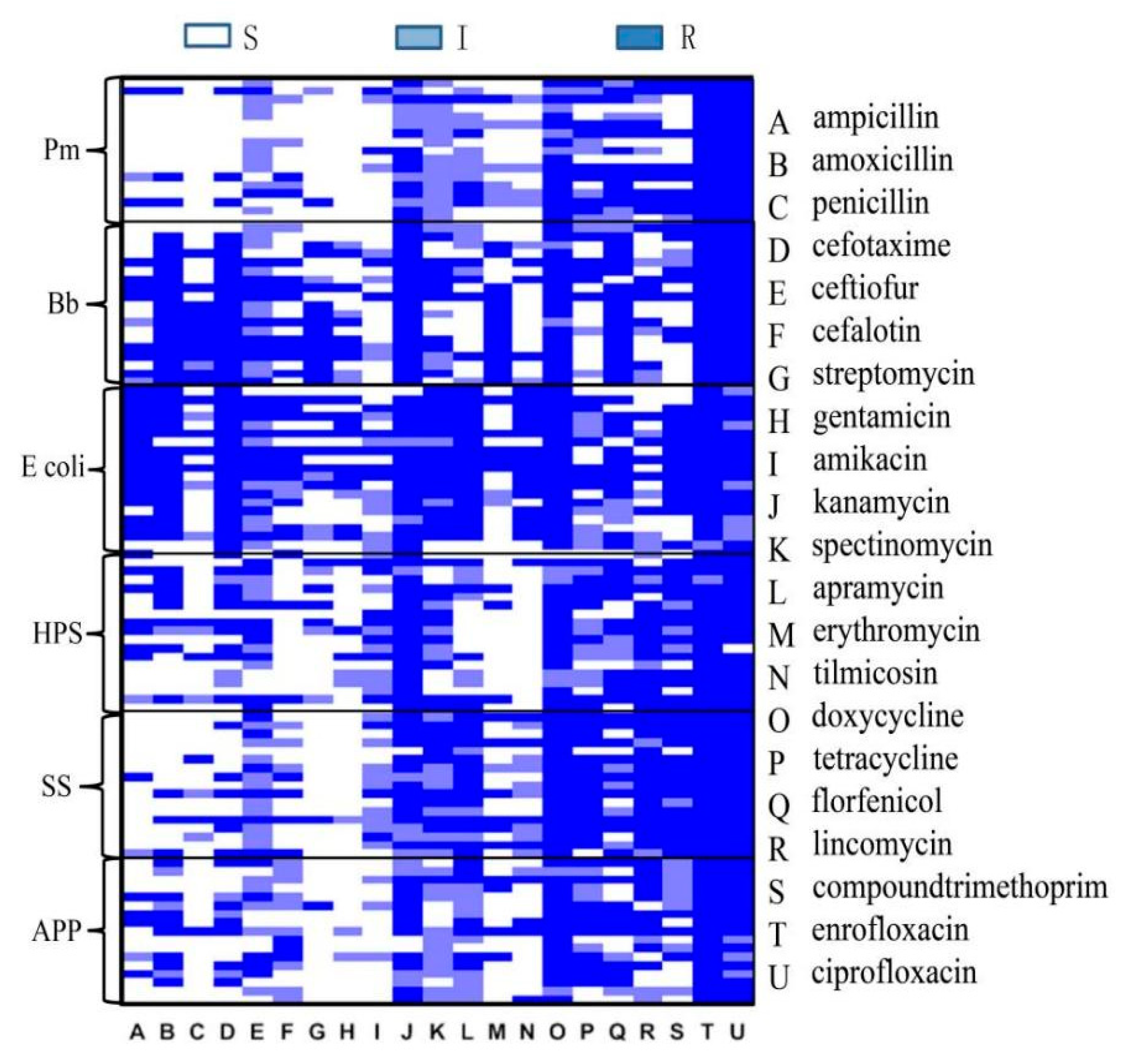
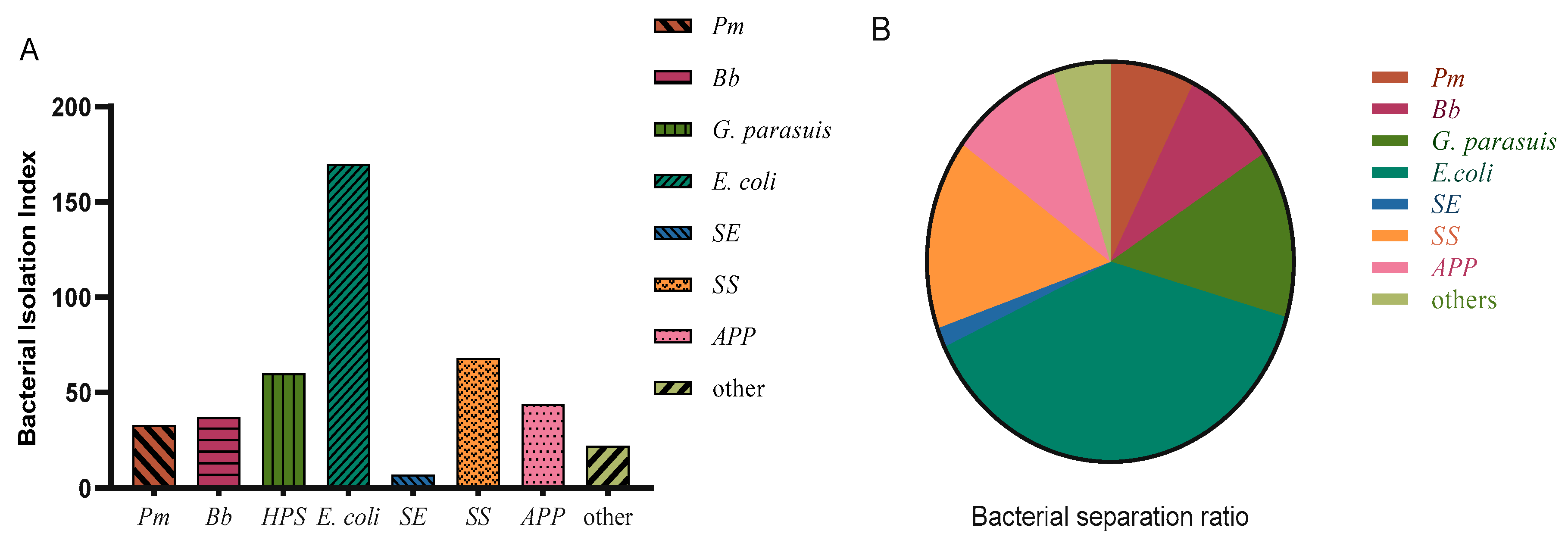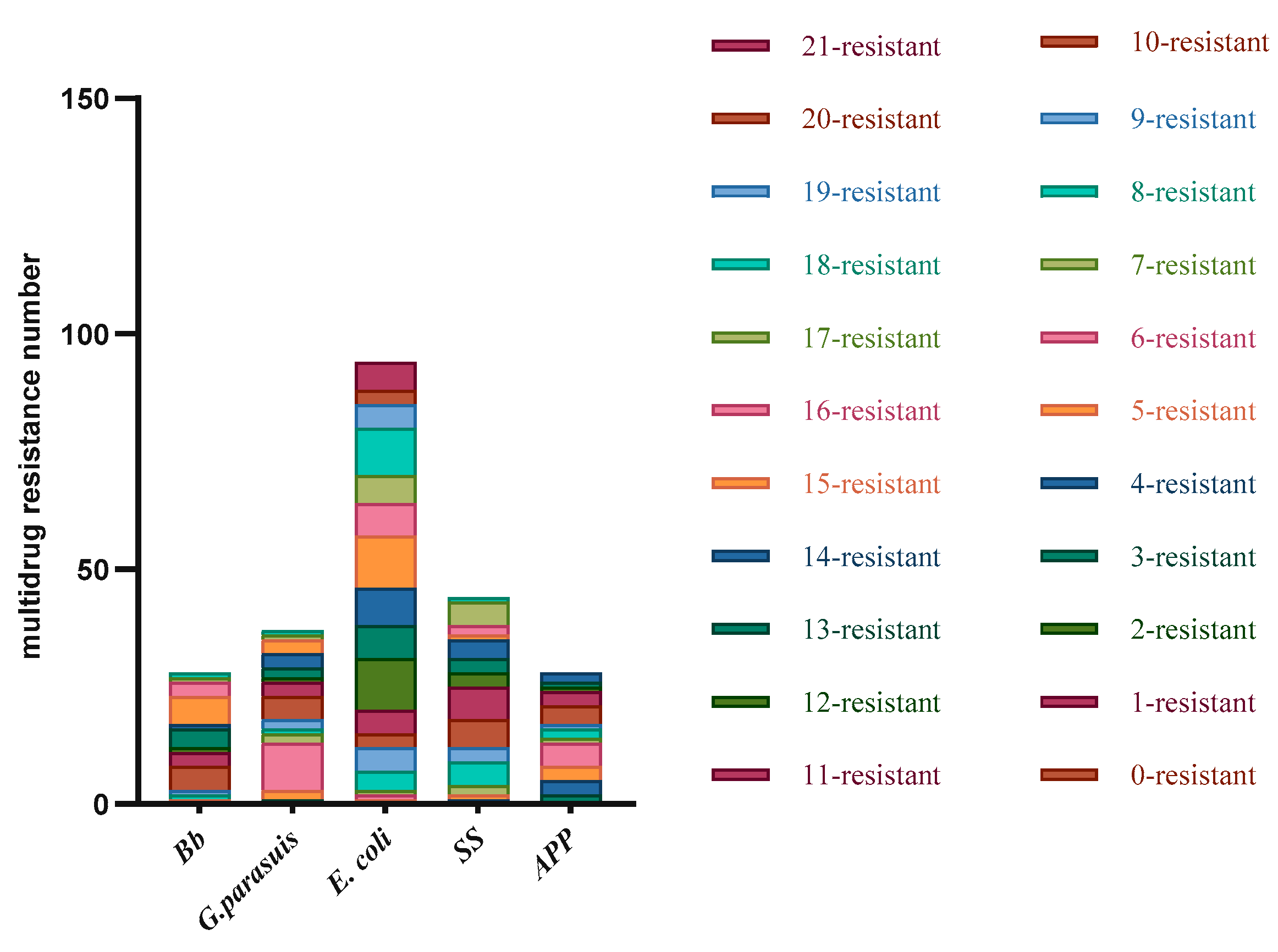3.3. Results of Disc Diffusion Testing
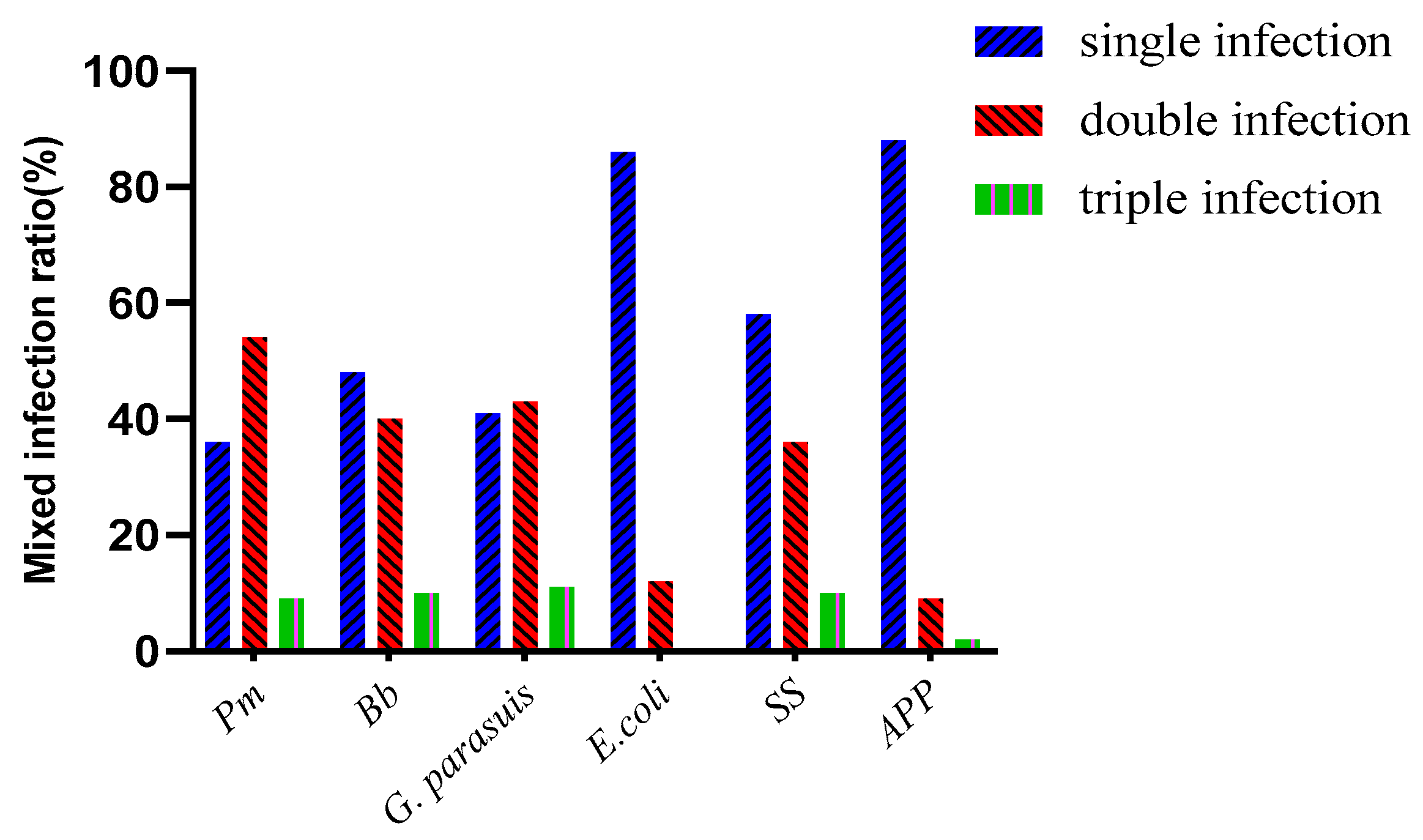
Disc diffusion testing was performed on the detected strains. The same type of bacteria detected in the same batch in the same field was tested only once, and disc diffusion testing against 21 antibiotics was performed for 228 isolated strains, including 18 strains of Pm, 28 strains of Bb, 36 strains of G. parasuis, 94 strains of E. coli, 44 strains of SS, and 28 strains of APP.
Eighteen isolated
Pm strains were tested against 21 antibacterial drugs (the numbers of resistant strains, the proportion of drug resistance to total isolates): ampicillin (3 16.67%), amoxicillin (2, 11.11%), penicillin (3, 16.67%), cephalothin (0, 0.00), ceftiofur (1, 5.56%), cefotaxime (0, 0.00), streptomycin (10, 55.56%), gentamicin (9, 50%), amikacin (5, 27.78%), kanamycin (8, 44.44%), spectinomycin (1, 5.56%), apramycin (0, 0.00), erythromycin (1, 5.56%), tilmicosin(14, 77.78%), doxycycline (0, 0.00), tetracycline (5, 27.78%), florfenicol (2, 11.11%), lincomycin (18, 100%), compound trimethoprim (11, 61.11%), enrofloxacin (2, 11.11%), and ciprofloxacin (2, 11.11%). The results reveal that
Pm was most resistant to lincomycin, with a resistance rate of 100%, and it was sensitive to cephalothin, cefotaxime, apramycin, and doxycycline, with sensitivity rates of 100%. There were five antibiotics associated with a resistance rate of 50% or above: streptomycin (55.56%), gentamicin (50%), tilmicosin (77.78%), lincomycin (100%), and compound trimethoprim (61.11%). In terms of drug types, the highest sensitivity was observed for cephalosporins, the highest resistance rate was observed for ceftiofur, at only 5.56%, and the cephalothin and cefotaxime resistance rates were 0.00%. Among the aminoglycosides, the streptomycin and gentamicin resistance rates were 55.56% and 50%, respectively, and the apramycin resistance rate was 0%. Among the tetracyclines, the tilmicosin resistance rate was 77.78%, and the doxycycline sensitivity rate was 100%. (
Table 5).
A total of 28 isolated
Bb strains were tested against 21 antibacterial drugs (the numbers of resistant strains, the proportion of drug resistance to total isolates): ampicillin (27, 96.42%), amoxicillin (13, 46.43%), penicillin (27, 96.42%), cephalothin (13, 46.43%), ceftiofur (20, 71.42%), cefotaxime (17, 60.71%), streptomycin (26, 92.86%), gentamicin (9, 32.14%), amikacin (9, 32.14%), kanamycin (9, 32.14%), spectinomycin (21, 75%), apramycin (27, 96.42%), erythromycin (13, 21.43%), tilmicosin (26, 92.86%), doxycycline (6, 28.57%), tetracycline (8, 46.43%), florfenicol (13, 42.86%), lincomycin (28, 100%), compound trimethoprim (27, 96.42%), enrofloxacin (12, 46.43%), and ciprofloxacin (5, 17.86%). The results show that
Bb was the most resistant to lincomycin, with a resistance rate of 100%. Ten antibiotics had associated resistance rates of 50% or higher: ampicillin (96.42%), penicillin (96.42%), ceftiofur (71.42%), cefotaxime (60.71%), streptomycin (92.86%), apramycin (96.42%), tilmicosin (92.86%), lincomycin (100%), and compound trimethoprim (96.42%). Among the quinolones, ciprofloxacin was associated with the highest sensitivity, with an associated resistance rate of 17.86% (
Table 6).
A total of 36 isolated
HPS strains were tested against 21 antibacterial drugs (the numbers of resistant strains, the proportion of drug resistance to total isolates): ampicillin (20, 55.56%), amoxicillin (14, 38.89%), penicillin (17, 47.22%), cephalothin (7, 19.44%), ceftiofur (8, 22.22%), cefotaxime (3, 8.33%), streptomycin (24, 66.67%), gentamicin (20, 55.56%), amikacin (22, 61.11%), kanamycin (13, 36.11%), spectinomycin (5, 13.89%), apramycin (33, 91.67%), erythromycin (16, 44.44%), tilmicosin (25, 69.44%), doxycycline (2, 5.56%), tetracycline (9, 25%), florfenicol (7, 19.44%), lincomycin (30, 83.33%), compound trimethoprim (34, 94.44%), enrofloxacin (18, 50%), and ciprofloxacin (11, 30.56%). The results reveal that
HPS was the most resistant to compound trimethoprim, with a resistance rate of 94.44%, and most sensitive to doxycycline, with a resistance rate of 5.56%. Seven antibiotics were associated with resistance rates of 50% or higher: ampicillin (55.56%), penicillin (60.71%), streptomycin (66.67%), gentamicin (55.56%), amikacin (61.11%), apramycin (91.67%), and tilmicosin (69.44%). The following cephalosporin antibiotics were associated with the highest sensitivity, with drug resistance rates of less than 25%: cephalothin (19.44%), ceftiofur (22.22%), and cefotaxime (8.33%) (
Table 7).
Ninety-four isolated
E. coli strains were tested against 21 antibacterial drugs (the numbers of resistant strains, the proportion of drug resistance to total isolates): ampicillin (87, 92.55%), amoxicillin (84, 89.36%), penicillin (92, 97.87%), cephalothin (55, 58.51%), ceftiofur (30, 31.91%), cefotaxime (15, 15.96%), streptomycin (58, 61.70%), gentamicin (62, 65.96%), amikacin (25, 26.60%), kanamycin (52, 55.32%), spectinomycin (31, 32.98%), apramycin (53, 56.38%), erythromycin (79, 84.04%), tilmicosin (94, 100%), doxycycline (86, 91.49%), tetracycline (88, 93.62%), florfenicol (69, 73.40%), lincomycin (94, 100%), compound trimethoprim (86, 91.49%), enrofloxacin (71, 75.53%), and ciprofloxacin (44, 46.80%). The results reveal that
E. coli was most resistant to tilmicosin and lincomycin, with resistance rates reaching 100%. Sensitivity to cefotaxime was the highest, and the resistance rate was 15.96%. There were 15 antibiotics associated with resistance rates of 50% or higher: ampicillin (92.55%), amoxicillin (89.36%), penicillin (97.87%), cephalothin (58.51%), streptomycin (61.70%), gentamicin (65.96%), kanamycin (55.32%), apramycin (56.38%), erythromycin (84.04%), tilmicosin (100%), doxycycline (91.49%), tetracycline (93.62%), florfenicol (73.40%), lincomycin (100%), and enrofloxacin (75.53%). Cephalosporins were associated with the highest sensitivity, with associated resistance rates of 58.51% against cephalothin, 31.91% against ceftiofur, and 15.96% against cefotaxime. In general,
E. coli exhibited the highest level of antibiotic resistance (
Table 8).
Forty-four isolated
SS strains were tested against 21 antibacterial drugs (the numbers of resistant strains, the proportion of drug resistance to total isolates): ampicillin (10, 22.72%), amoxicillin (3, 6.82%), penicillin (6, 13.64%), cephalothin (1, 2.27%), ceftiofur (3, 6.82%), cefotaxime (4, 9.09%), streptomycin (26, 59.09%), gentamicin (40, 90.91%), amikacin (43, 97.73%), kanamycin (41, 93.18%), spectinomycin (16, 36.36%), apramycin (44, 100%), erythromycin (30, 68.18%), tilmicosin (43, 97.73%), doxycycline (14,31.82%), tetracycline (37, 84.09%), florfenicol (8, 18.08%), lincomycin (43, 97.73%), compound trimethoprim (33, 75%), enrofloxacin (19, 43.18%), and ciprofloxacin (14, 31.82%). The results show that
SS was most resistant to apramycin, with a resistance rate of 100%, and had the highest sensitivity to cephalothin, with a resistance rate of 2.27%. There were nine antibiotics associated with resistance rates of 50% or higher: streptomycin (59.09%), gentamicin (90.91%), amikacin (97.73%), kanamycin (93.18%), apramycin (100%), erythromycin (68.18%), tilmicosin (97.73%), tetracycline (84.09%), and lincomycin (97.73%). SS had the highest sensitivity to cephalosporin antibiotics. The cephalothin resistance rate was 2.27%, the ceftiofur resistance rate was 6.82%, and the cefotaxime resistance rate was 9.09% (
Table 9).
Twenty-eight isolated
APP strains were tested against 21 antibacterial drugs (the numbers of resistant strains, the proportion of drug resistance to total isolates): ampicillin (11, 39.29%), amoxicillin (6, 21.42%), penicillin (13, 46.42%), cephalothin (1, 3.57%), ceftiofur (2, 7.14%), cefotaxime (0, 0.00), streptomycin (15, 53.57%), gentamicin (8, 28.57%), amikacin (15, 53.57%), kanamycin (15, 53.57%), spectinomycin (6, 21.42%), apramycin (24, 85.71%), erythromycin (3, 10.71%), tilmicosin(24, 85.71%), doxycycline (4, 14.28%), tetracycline (13, 46.42%), florfenicol (7, 25.00%), lincomycin (27, 96.43%), compound trimethoprim (16, 57.14%), enrofloxacin (11, 39.29%), and ciprofloxacin (0, 0). The results reveal that
APP was most resistant to lincomycin, with a resistance rate of 96.43%, and most sensitive to cephalothin and ciprofloxacin, with resistance rates of 0. There were six antibiotics with associated resistance rates of 50% or higher: streptomycin (53.57%), amikacin (53.57%), kanamycin (53.57%), tilmicosin (85.71%), lincomycin (96.43%), and compound trimoxazole (57.14%). Cephalosporin antibiotics were associated with the highest sensitivity, with associated resistance rates of 0% for cephalothin, 7.14% for ceftiofur, and 3.57% for cefotaxime. Two values in
Table 10 were ignored because they were zero (
Table 10).