Detection of Porcine Deltacoronavirus RNA in the Upper and Lower Respiratory Tract and Biliary Fluid and the Effect of Infection on Serum Cholesterol Levels and Blood T Cell Population Frequencies in Gnotobiotic Piglets
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Experimental Infection of Gnotobiotic Piglets with PDCoV
2.3. Sampling
2.4. RNA Extraction and RT-qPCR
2.5. Pathology and IF Staining for the Detection of PDCoV Proteins in Respiratory Tissues
2.6. Isolation of Mononuclear Cells and Flow Cytometry
2.7. Cholesterol Assay
2.8. Statistics
3. Results
3.1. Clinical Signs
3.2. RT-qPCR
3.3. Pathology and IF Staining
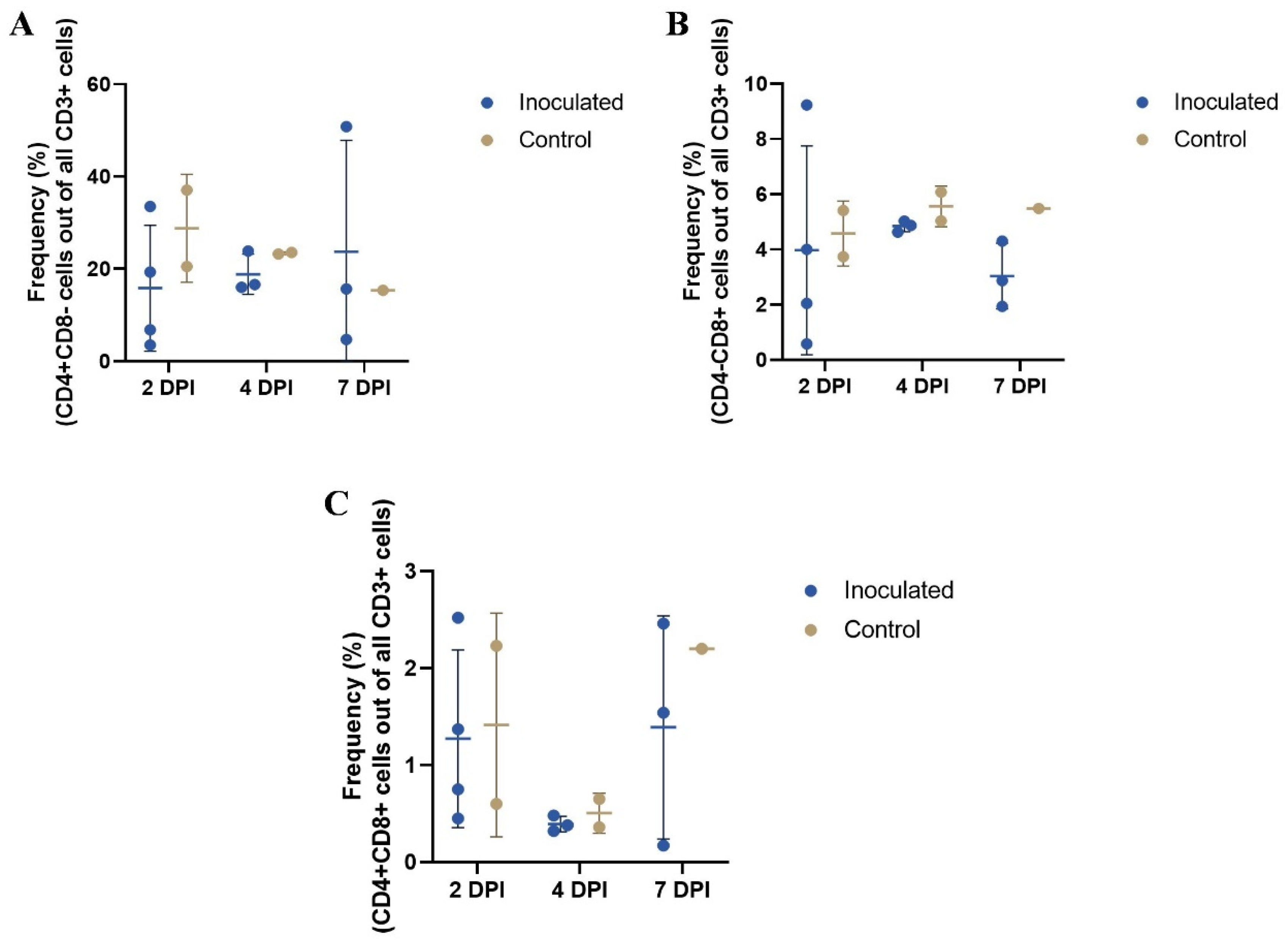
3.4. Blood T Cell Population Frequencies as Determined by Flow Cytometry
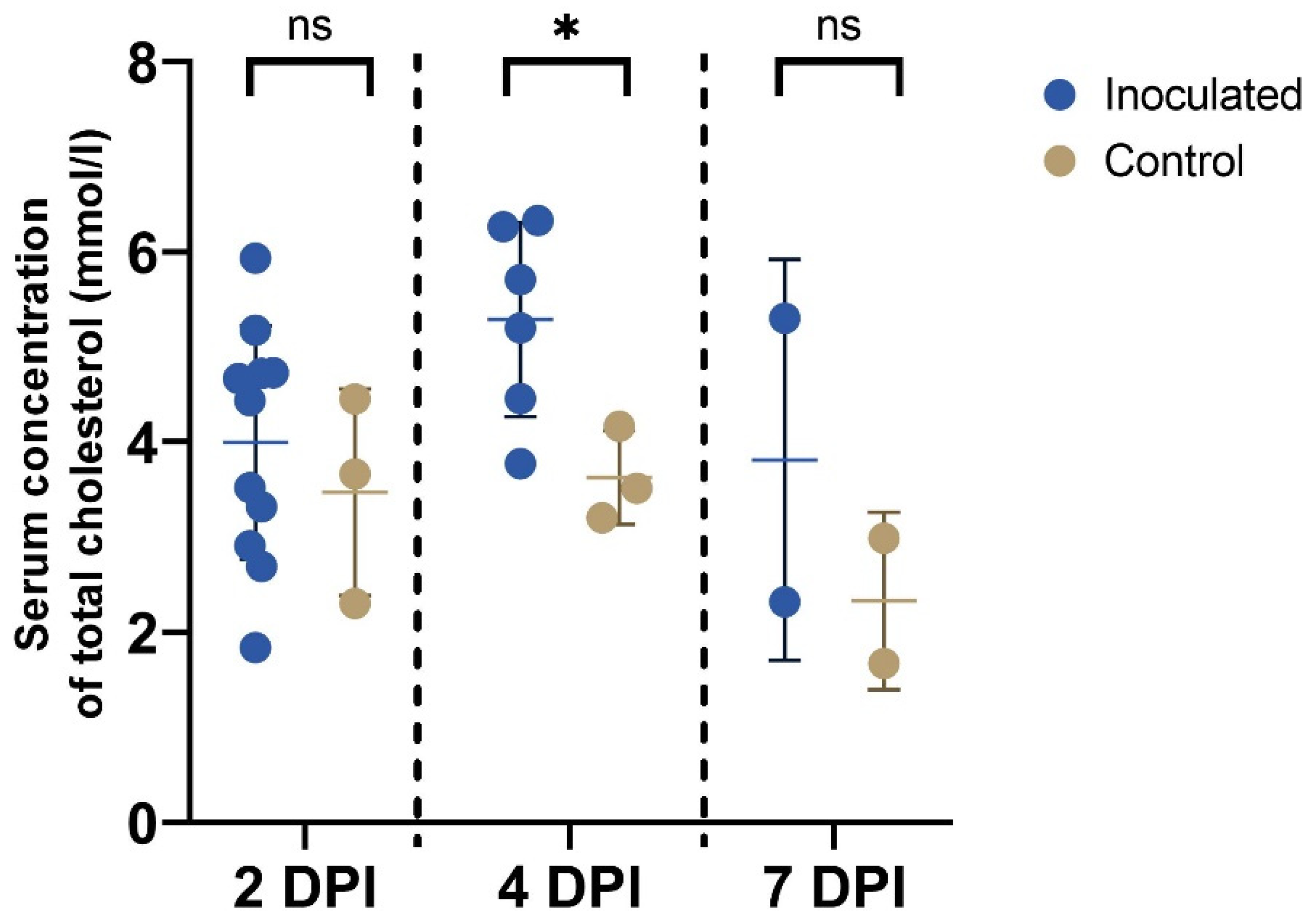
3.5. Cholesterol Assay
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jung, K.; Hu, H.; Saif, L.J. Calves Are Susceptible to Infection with the Newly Emerged Porcine Deltacoronavirus, but Not with the Swine Enteric Alphacoronavirus, Porcine Epidemic Diarrhea Virus. Arch. Virol. 2017, 162, 2357–2362. [Google Scholar] [CrossRef]
- Liang, Q.; Zhang, H.; Li, B.; Ding, Q.; Wang, Y.; Gao, W.; Guo, D.; Wei, Z.; Hu, H. Susceptibility of Chickens to Porcine Deltacoronavirus Infection. Viruses 2019, 11, 573. [Google Scholar] [CrossRef]
- Zhang, H.; Ding, Q.; Yuan, J.; Han, F.; Wei, Z.; Hu, H. Susceptibility to Mice and Potential Evolutionary Characteristics of Porcine Deltacoronavirus. J. Med. Virol. 2022, 94, 5723–5738. [Google Scholar] [CrossRef]
- Lednicky, J.A.; Tagliamonte, M.S.; White, S.K.; Elbadry, M.A.; Alam, M.M.; Stephenson, C.J.; Bonny, T.S.; Loeb, J.C.; Telisma, T.; Chavannes, S.; et al. Independent Infections of Porcine Deltacoronavirus among Haitian Children. Nature 2021, 600, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Anon. USDA to Require Reports of PED. J. Am. Vet. Med. Assoc. 2014, 244, 1234. [Google Scholar]
- Jung, K.; Hu, H.; Eyerly, B.; Lu, Z.; Chepngeno, J.; Saif, L.J. Pathogenicity of 2 Porcine Deltacoronavirus Strains in Gnotobiotic Pigs. Emerg. Infect. Dis. 2015, 21, 650–654. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Byrum, B.; Zhang, Y. Detection and Genetic Characterization of Deltacoronavirus in Pigs, Ohio, USA, 2014. Emerg. Infect. Dis. 2014, 20, 1227–1230. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lau, S.K.P.; Lam, C.S.F.; Lau, C.C.Y.; Tsang, A.K.L.; Lau, J.H.N.; Bai, R.; Teng, J.L.L.; Tsang, C.C.C.; Wang, M.; et al. Discovery of Seven Novel Mammalian and Avian Coronaviruses in the Genus Deltacoronavirus Supports Bat Coronaviruses as the Gene Source of Alphacoronavirus and Betacoronavirus and Avian Coronaviruses as the Gene Source of Gammacoronavirus and Deltacoronavirus. J. Virol. 2012, 86, 3995–4008. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Byrum, B.; Zhang, Y. Porcine Coronavirus HKU15 Detected in 9 US States, 2014. Emerg. Infect. Dis. 2014, 20, 1594–1595. [Google Scholar] [CrossRef]
- Chen, Q.; Gauger, P.; Stafne, M.; Thomas, J.; Arruda, P.; Burrough, E.; Madson, D.; Brodie, J.; Magstadt, D.; Derscheid, R.; et al. Pathogenicity and Pathogenesis of a United States Porcine Deltacoronavirus Cell Culture Isolate in 5-Day-Old Neonatal Piglets. Virology 2015, 482, 51–59. [Google Scholar] [CrossRef]
- Li, B.; Zheng, L.; Li, H.; Ding, Q.; Wang, Y.; Wei, Z. Porcine Deltacoronavirus Causes Diarrhea in Various Ages of Field-Infected Pigs in China. Biosci. Rep. 2019, 39, BSR20190676. [Google Scholar] [CrossRef]
- Zhang, M.-J.; Liu, D.-J.; Liu, X.-L.; Ge, X.-Y.; Jongkaewwattana, A.; He, Q.-G.; Luo, R. Genomic Characterization and Pathogenicity of Porcine Deltacoronavirus Strain CHN-HG-2017 from China. Arch. Virol. 2019, 164, 413–425. [Google Scholar] [CrossRef]
- Zhou, X.; Zhou, L.; Zhang, P.; Ge, X.; Guo, X.; Han, J.; Zhang, Y.; Yang, H. A Strain of Porcine Deltacoronavirus: Genomic Characterization, Pathogenicity and Its Full-length CDNA Infectious Clone. Transbound. Emerg. Dis. 2021, 68, 2130–2146. [Google Scholar] [CrossRef]
- Vitosh-Sillman, S.; Loy, J.D.; Brodersen, B.; Kelling, C.; Doster, A.; Topliff, C.; Nelson, E.; Bai, J.; Schirtzinger, E.; Poulsen, E.; et al. Experimental Infection of Conventional Nursing Pigs and Their Dams with Porcine Deltacoronavirus. J. Vet. Diagn. Investig. 2016, 28, 486–497. [Google Scholar] [CrossRef]
- Woo, P.C.; Lau, S.K.; Tsang, C.-C.; Lau, C.C.; Wong, P.-C.; Chow, F.W.; Fong, J.Y.; Yuen, K.-Y. Coronavirus HKU15 in Respiratory Tract of Pigs and First Discovery of Coronavirus Quasispecies in 5′-Untranslated Region. Emerg. Microbes Infect. 2017, 6, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhang, Y.; Liang, X.; Lou, F.; Oglesbee, M.; Krakowka, S.; Li, J. Origin, Evolution, and Virulence of Porcine Deltacoronaviruses in the United States. mBio 2015, 6, e00064-15. [Google Scholar] [CrossRef] [PubMed]
- Koszinowski, U.H.; Reddehase, M.J.; Jonjic, S. The Role of CD4 and CD8 T Cells in Viral Infections. Curr. Opin. Immunol. 1991, 3, 471–475. [Google Scholar] [CrossRef]
- Fang, P.; Zhang, J.; Zhang, H.; Xia, S.; Ren, J.; Tian, L.; Bai, D.; Fang, L.; Xiao, S. Porcine Deltacoronavirus Enters Porcine IPI-2I Intestinal Epithelial Cells via Macropinocytosis and Clathrin-Mediated Endocytosis Dependent on PH and Dynamin. J. Virol. 2021, 95, e01345-21. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.H.; Lee, C. Cholesterol Is Important for the Entry Process of Porcine Deltacoronavirus. Arch. Virol. 2018, 163, 3119–3124. [Google Scholar] [CrossRef]
- Jeon, J.H.; Lee, C. Cellular Cholesterol Is Required for Porcine Nidovirus Infection. Arch. Virol. 2017, 162, 3753–3767. [Google Scholar] [CrossRef]
- Ren, X.; Glende, J.; Yin, J.; Schwegmann-Wessels, C.; Herrler, G. Importance of Cholesterol for Infection of Cells by Transmissible Gastroenteritis Virus. Virus Res. 2008, 137, 220–224. [Google Scholar] [CrossRef]
- Wei, X.; Zeng, W.; Su, J.; Wan, H.; Yu, X.; Cao, X.; Tan, W.; Wang, H. Hypolipidemia Is Associated with the Severity of COVID-19. J. Clin. Lipidol. 2020, 14, 297–304. [Google Scholar] [CrossRef]
- Liu, H.-Y.; Gu, H.; Qu, H.; Bao, W.; Li, Y.; Cai, D. Aberrant Cholesterol Metabolic Genes Regulation in a Negative Feedback Loop Induced by an Alphacoronavirus. Front. Nutr. 2022, 9, 870680. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Wang, H.; Liao, Y.; Tan, L.; Sun, Y.; Song, C.; Liu, W.; Qiu, X.; Ding, C. Coronavirus Infection and Cholesterol Metabolism. Front. Immunol. 2022, 13, 791267. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Jung, K.; Vlasova, A.N.; Chepngeno, J.; Lu, Z.; Wang, Q.; Saif, L.J. Isolation and Characterization of Porcine Deltacoronavirus from Pigs with Diarrhea in the United States. J. Clin. Microbiol. 2015, 53, 1537–1548. [Google Scholar] [CrossRef]
- Hu, H.; Jung, K.; Vlasova, A.N.; Saif, L.J. Experimental Infection of Gnotobiotic Pigs with the Cell-Culture-Adapted Porcine Deltacoronavirus Strain OH-FD22. Arch. Virol. 2016, 161, 3421–3434. [Google Scholar] [CrossRef]
- Yuan, L.; Ward, L.A.; Rosen, B.I.; To, T.L.; Saif, L.J. Systemic and Intestinal Antibody-Secreting Cell Responses and Correlates of Protective Immunity to Human Rotavirus in a Gnotobiotic Pig Model of Disease. J. Virol. 1996, 70, 3075–3083. [Google Scholar] [CrossRef]
- Chattha, K.S.; Vlasova, A.N.; Kandasamy, S.; Rajashekara, G.; Saif, L.J. Divergent Immunomodulating Effects of Probiotics on T Cell Responses to Oral Attenuated Human Rotavirus Vaccine and Virulent Human Rotavirus Infection in a Neonatal Gnotobiotic Piglet Disease Model. J. Immunol. 2013, 191, 2446–2456. [Google Scholar] [CrossRef]
- Han, D.; Fang, Q.; Wang, X. SARS-CoV-2 Was Found in the Bile Juice from a Patient with Severe COVID-19. J. Med. Virol. 2021, 93, 102–104. [Google Scholar] [CrossRef]
- Boyer, J.L.; Soroka, C.J. Bile Formation and Secretion: An Update. J. Hepatol. 2021, 75, 190–201. [Google Scholar] [CrossRef]
- Kong, F.; Niu, X.; Liu, M.; Wang, Q. Bile Acids LCA and CDCA Inhibited Porcine Deltacoronavirus Replication in Vitro. Vet. Microbiol. 2021, 257, 109097. [Google Scholar] [CrossRef] [PubMed]
- Krishna, V.D.; Kim, Y.; Yang, M.; Vannucci, F.; Molitor, T.; Torremorell, M.; Cheeran, M.C.-J. Immune Responses to Porcine Epidemic Diarrhea Virus (PEDV) in Swine and Protection against Subsequent Infection. PLoS ONE 2020, 15, e0231723. [Google Scholar] [CrossRef] [PubMed]
- Annamalai, T.; Saif, L.J.; Lu, Z.; Jung, K. Age-Dependent Variation in Innate Immune Responses to Porcine Epidemic Diarrhea Virus Infection in Suckling versus Weaned Pigs. Vet. Immunol. Immunopathol. 2015, 168, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Zhang, Z.; Prajapati, M.; Li, Y. Lymphopenia Caused by Virus Infections and the Mechanisms Beyond. Viruses 2021, 13, 1876. [Google Scholar] [CrossRef]
- Goenka, A.; Kollmann, T.R. Development of Immunity in Early Life. J. Infect. 2015, 71, S112–S120. [Google Scholar] [CrossRef]
- Wen, K.; Li, G.; Yang, X.; Bui, T.; Bai, M.; Liu, F.; Kocher, J.; Yuan, L. CD4+ CD25− FoxP3+ Regulatory Cells Are the Predominant Responding Regulatory T Cells after Human Rotavirus Infection or Vaccination in Gnotobiotic Pigs. Immunology 2012, 137, 160–171. [Google Scholar] [CrossRef]
- Wu, J.L.; Mai, K.J.; Li, D.; Wu, R.T.; Wu, Z.X.; Tang, X.Y.; Li, Q.N.; Sun, Y.; Lan, T.; Zhang, X.B.; et al. Expression Profile Analysis of 5-Day-Old Neonatal Piglets Infected with Porcine Deltacoronavirus. BMC Vet. Res. 2019, 15, 117. [Google Scholar] [CrossRef]
- Zinellu, A.; Paliogiannis, P.; Fois, A.G.; Solidoro, P.; Carru, C.; Mangoni, A.A. Cholesterol and Triglyceride Concentrations, COVID-19 Severity, and Mortality: A Systematic Review and Meta-Analysis With Meta-Regression. Front. Public Health 2021, 9, 705916. [Google Scholar] [CrossRef]
- Campbell, N.R.C.; Wickert, W.; Magner, P.; Shumak, S.L. Dehydration during Fasting Increases Serum Lipids and Lipoproteins. Clin. Investig. Med. 1994, 17, 570–576. [Google Scholar]
- Verghese, P.B.; Arrese, E.L.; Soulages, J.L. Stimulation of Lipolysis Enhances the Rate of Cholesterol Efflux to HDL in Adipocytes. Mol. Cell. Biochem. 2007, 302, 241–248. [Google Scholar] [CrossRef]
- Li, S.; Xiao, D.; Zhao, Y.; Zhang, L.; Chen, R.; Liu, W.; Wen, Y.; Liao, Y.; Wen, Y.; Wu, R.; et al. Porcine Deltacoronavirus (PDCoV) Entry into PK-15 Cells by Caveolae-Mediated Endocytosis. Viruses 2022, 14, 496. [Google Scholar] [CrossRef]
- Bui, T.; Kocher, J.; Li, Y.; Wen, K.; Li, G.; Liu, F.; Yang, X.; LeRoith, T.; Tan, M.; Xia, M.; et al. Median Infectious Dose of Human Norovirus GII.4 in Gnotobiotic Pigs Is Decreased by Simvastatin Treatment and Increased by Age. J. Gen. Virol. 2013, 94, 2005–2016. [Google Scholar] [CrossRef]
- Li, Y.; Wu, Q.; Huang, L.; Yuan, C.; Wang, J.; Yang, Q. An Alternative Pathway of Enteric PEDV Dissemination from Nasal Cavity to Intestinal Mucosa in Swine. Nat. Commun. 2018, 9, 3811. [Google Scholar] [CrossRef]
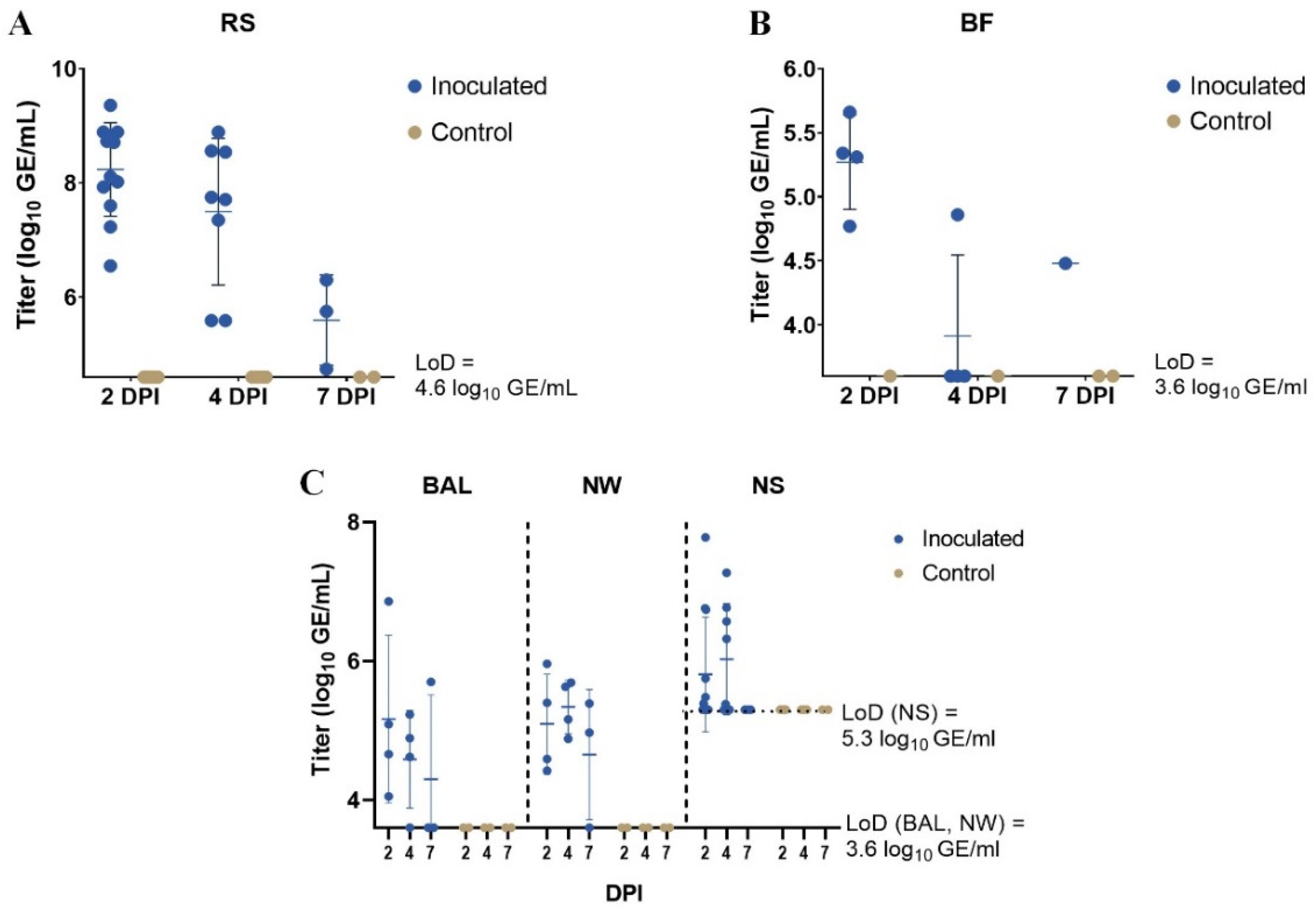
| DPI | RS | BF | BAL | NW | NS | |
|---|---|---|---|---|---|---|
| Inoculated piglets | 2 | 12/12 | 4/4 | 4/4 | 4/4 | 6/12 |
| 4 | 8/8 | 1/4 | 3/4 | 4/4 | 5/8 | |
| 7 | 3/3 | 1/1 | 1/3 | 2/3 | 0/3 | |
| Control piglets | 2 | 0/6 | 0/1 | 0/2 | 0/2 | 0/6 |
| 4 | 0/4 | 0/1 | 0/2 | 0/2 | 0/4 | |
| 7 | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 |
| CD4+CD8− | CD4−CD8+ | CD4+CD8+ | ||||
|---|---|---|---|---|---|---|
| I | C | I | C | I | C | |
| 2 DPI | 15.8% (13.6%) | 28.8% (11.7%) | 4.0% 3.8%) | 4.6% (1.2%) | 1.3% (0.9%) | 1.4% (1.2%) |
| 4 DPI | 18.9% (4.4%) | 23.4% (0.3%) | 4.8% (0.2%) | 5.6% (0.7%) | 0.4% (0.1%) | 0.5% (0.2%) |
| 7 DPI | 23.7% (24.1%) | 15.4% * | 3.0% (1.2%) | 5.5% * | 1.4% (1.2%) | 2.2% * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bedsted, A.E.; Jung, K.; Saif, L.J. Detection of Porcine Deltacoronavirus RNA in the Upper and Lower Respiratory Tract and Biliary Fluid and the Effect of Infection on Serum Cholesterol Levels and Blood T Cell Population Frequencies in Gnotobiotic Piglets. Vet. Sci. 2023, 10, 117. https://doi.org/10.3390/vetsci10020117
Bedsted AE, Jung K, Saif LJ. Detection of Porcine Deltacoronavirus RNA in the Upper and Lower Respiratory Tract and Biliary Fluid and the Effect of Infection on Serum Cholesterol Levels and Blood T Cell Population Frequencies in Gnotobiotic Piglets. Veterinary Sciences. 2023; 10(2):117. https://doi.org/10.3390/vetsci10020117
Chicago/Turabian StyleBedsted, Amalie Ehlers, Kwonil Jung, and Linda J. Saif. 2023. "Detection of Porcine Deltacoronavirus RNA in the Upper and Lower Respiratory Tract and Biliary Fluid and the Effect of Infection on Serum Cholesterol Levels and Blood T Cell Population Frequencies in Gnotobiotic Piglets" Veterinary Sciences 10, no. 2: 117. https://doi.org/10.3390/vetsci10020117
APA StyleBedsted, A. E., Jung, K., & Saif, L. J. (2023). Detection of Porcine Deltacoronavirus RNA in the Upper and Lower Respiratory Tract and Biliary Fluid and the Effect of Infection on Serum Cholesterol Levels and Blood T Cell Population Frequencies in Gnotobiotic Piglets. Veterinary Sciences, 10(2), 117. https://doi.org/10.3390/vetsci10020117







