Development of a TaqMan-Probe-Based Multiplex Real-Time PCR for the Simultaneous Detection of African Swine Fever Virus, Porcine Circovirus 2, and Pseudorabies Virus in East China from 2020 to 2022
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Special Primers and Probes Design
2.2. Positive DNA/cDNA and Field Specimens
2.3. Construction of the Recombinant Plasmid
2.4. Optimization of Reaction Conditions for the Multiplex Real-Time qPCR
2.5. Establishment of Standard Curves of the Multiplex Real-Time qPCR
2.6. Specificity, Sensitivity, and Repeatability of the Multiplex Real-Time qPCR
2.7. Field Specimen Detection
3. Results
3.1. Optimization of the Multiplex Real-Time qPCR Reaction Conditions
3.2. Standard Curves
3.3. Specificity of the Multiplex Real-Time qPCR
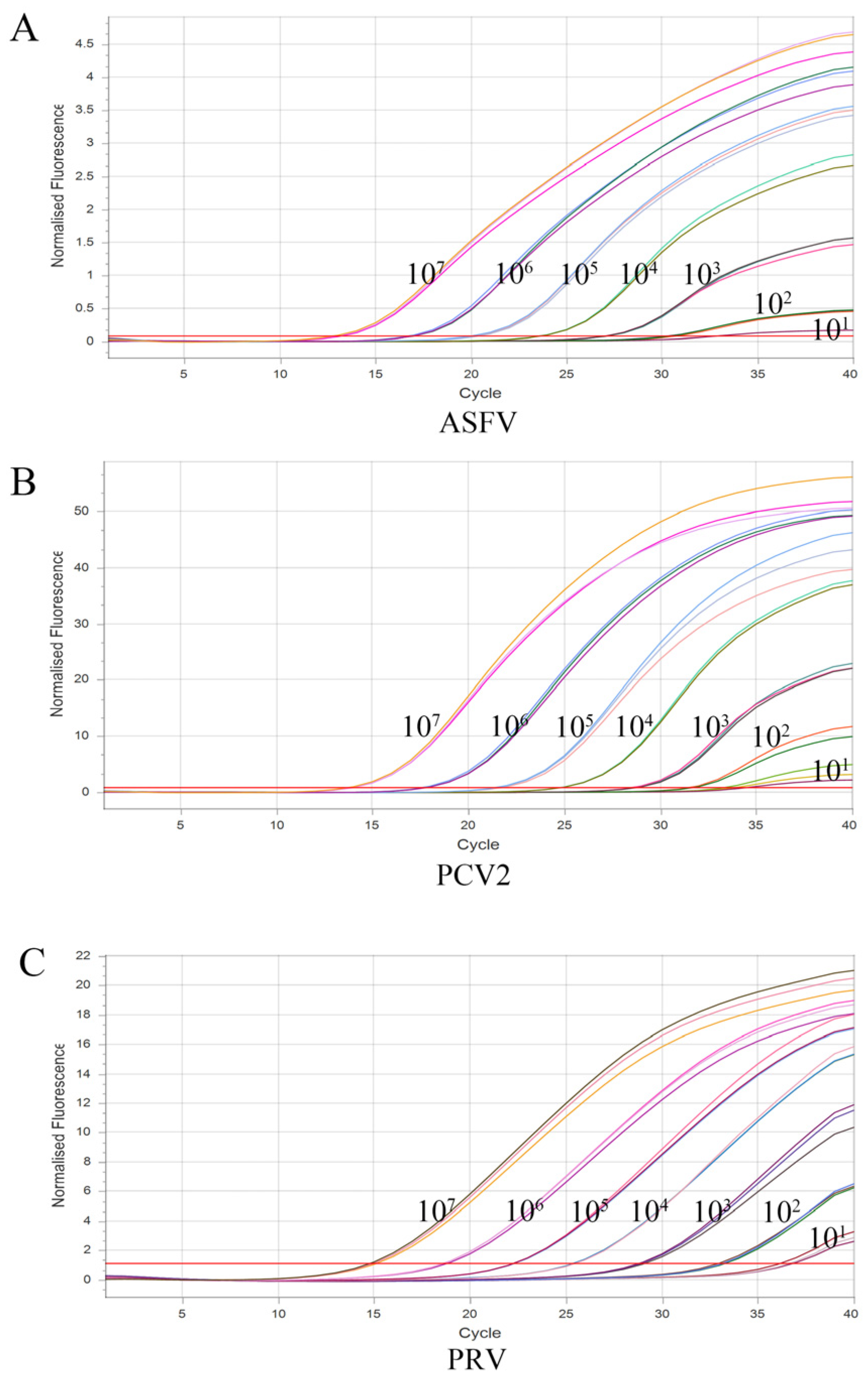
3.4. Sensitivity of the Multiplex Real-Time qPCR
3.5. Repeatability of the Multiplex Real-Time qPCR
3.6. The Detection of Field Specimens
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- VanderWaal, K.; Deen, J. Global trends in infectious diseases of swine. Proc. Natl. Acad. Sci. USA 2018, 115, 11495–11500. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, S.; Zhang, L.; Niu, G.; Zhang, X.; Yang, L.; Ji, W.; Ren, L. Coinfection of Porcine Circovirus 2 and Pseudorabies Virus Enhances Immunosuppression and Inflammation through NF-kappa B, JAK/STAT, MAPK, and NLRP3 Pathways. Int. J. Mol. Sci. 2022, 23, 4469. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chen, J.; Wu, X.; Ma, D.; Zhang, X.; Li, R.; Han, C.; Liu, H.; Yin, X.; Du, Q.; et al. PCV2 targets cGAS to inhibit type I interferon induction to promote other DNA virus infection. PLoS Pathog. 2021, 17, e1009940. [Google Scholar] [CrossRef] [PubMed]
- Mighell, E.; Ward, M.P. African Swine Fever spread across Asia, 2018–2019. Transbound. Emerg. Dis. 2021, 68, 2722–2732. [Google Scholar] [CrossRef]
- Ge, X.; Wang, F.; Guo, X.; Yang, H. Porcine circovirus type 2 and its associated diseases in China. Virus Res. 2012, 164, 100–106. [Google Scholar] [CrossRef]
- Liu, Q.; Kuang, Y.; Li, Y.; Guo, H.; Zhou, C.; Guo, S.; Tan, C.; Wu, B.; Chen, H.; Wang, X. The Epidemiology and Variation in Pseudorabies Virus: A Continuing Challenge to Pigs and Humans. Viruses 2022, 14, 1463. [Google Scholar] [CrossRef]
- Dixon, L.K.; Sun, H.; Roberts, H. African swine fever. Antivir. Res. 2019, 165, 34–41. [Google Scholar] [CrossRef]
- Huang, Y.; Li, J.; Zhang, J.; Jin, Z. Dynamical analysis of the spread of African swine fever with the live pig price in China. Math. Biosci. Eng. MBE 2021, 18, 8123–8148. [Google Scholar] [CrossRef]
- Dixon, L.K.; Chapman, D.A.; Netherton, C.L.; Upton, C. African swine fever virus replication and genomics. Virus Res. 2013, 173, 3–14. [Google Scholar] [CrossRef]
- Wang, Z.; Ai, Q.; Huang, S.; Ou, Y.; Gao, Y.; Tong, T.; Fan, H. Immune Escape Mechanism and Vaccine Research Progress of African Swine Fever Virus. Vaccines 2022, 10, 344. [Google Scholar] [CrossRef]
- Liu, S.; Luo, Y.; Wang, Y.; Li, S.; Zhao, Z.; Bi, Y.; Sun, J.; Peng, R.; Song, H.; Zhu, D.; et al. Cryo-EM Structure of the African Swine Fever Virus. Cell Host Microbe 2019, 26, 836–843. [Google Scholar] [CrossRef]
- Rakibuzzaman, A.G.M.; Ramamoorthy, S. Comparative immunopathogenesis and biology of recently discovered porcine circoviruses. Transbound. Emerg. Dis. 2021, 68, 2957–2968. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, L.; Li, X.; Niu, G.; Ren, L. Recent Progress on Epidemiology and Pathobiology of Porcine Circovirus 3. Viruses 2021, 13, 1944. [Google Scholar] [CrossRef]
- Ouyang, T.; Zhang, X.; Liu, X.; Ren, L. Co-Infection of Swine with Porcine Circovirus Type 2 and Other Swine Viruses. Viruses 2019, 11, 185. [Google Scholar] [CrossRef]
- Wen, L.; He, K. Genomic Rearrangement and Recombination of Porcine Circovirus Type 2 and Porcine Circovirus-Like Virus P1 in China. Front. Vet. Sci. 2021, 8, 736366. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, X.; Xie, C.; Ding, S.; Yang, H.; Guo, S.; Li, J.; Qin, L.; Ban, F.; Wang, D.; et al. A Novel Human Acute Encephalitis Caused by Pseudorabies Virus Variant Strain. Clin. Infect. Dis. 2021, 73, e3690–e3700. [Google Scholar] [CrossRef]
- He, W.; Auclert, L.Z.; Zhai, X.; Wong, G.; Zhang, C.; Zhu, H.; Xing, G.; Wang, S.; He, W.; Li, K.; et al. Interspecies Transmission, Genetic Diversity, and Evolutionary Dynamics of Pseudorabies Virus. J. Infect. Dis. 2019, 219, 1705–1715. [Google Scholar] [CrossRef]
- Tong, W.; Liu, F.; Zheng, H.; Liang, C.; Zhou, Y.J.; Jiang, Y.F.; Shan, T.L.; Gao, F.; Li, G.X.; Tong, G.Z. Emergence of a Pseudorabies virus variant with increased virulence to piglets. Vet. Microbiol. 2015, 181, 236–240. [Google Scholar] [CrossRef]
- An, T.Q.; Peng, J.M.; Tian, Z.J.; Zhao, H.Y.; Li, N.; Liu, Y.M.; Chen, J.Z.; Leng, C.L.; Sun, Y.; Chang, D.; et al. Pseudorabies virus variant in Bartha-K61-vaccinated pigs, China, 2012. Emerg. Infect. Dis. 2013, 19, 1749–1755. [Google Scholar] [CrossRef]
- Sun, Y.; Luo, Y.; Wang, C.H.; Yuan, J.; Li, N.; Song, K.; Qiu, H.J. Control of swine pseudorabies in China: Opportunities and limitations. Vet. Microbiol. 2016, 183, 119–124. [Google Scholar] [CrossRef]
- Zhao, K.; Shi, W.; Han, F.; Xu, Y.; Zhu, L.; Zou, Y.; Wu, X.; Zhu, H.; Tan, F.; Tao, S.; et al. Specific, simple and rapid detection of porcine circovirus type 2 using the loop-mediated isothermal amplification method. Virol. J. 2011, 8, 126. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Qin, X.; Sun, Y.; Cong, G.; Li, Y.; Zhang, Z. Development of Isothermal Recombinase Polymerase Amplification Assay for Rapid Detection of Porcine Circovirus Type 2. Bio. Res. Int. 2017, 2017, 8403642. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Yang, Y.; Ru, Y.; Qin, X.; Li, M.; Zhang, Z.; Zhang, R.; Li, Y.; Zhang, Z.; Li, Y. The Development of a Real-Time Recombinase-Aid Amplification Assay for Rapid Detection of African Swine Fever Virus. Front. Microbiol. 2022, 13, 846770. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Li, L.; Zhao, Y.; Liu, Y.; Liu, C.; Wang, Q.; Dong, Y.; Wang, S.; Chi, T.; Song, F.; et al. Clinical Validation of Two Recombinase-Based Isothermal Amplification Assays (RPA/RAA) for the Rapid Detection of African Swine Fever Virus. Front. Microbiol. 2020, 11, 1696. [Google Scholar] [CrossRef] [PubMed]
- Ilya, T.; Monoldorova, S.; Kang, S.; Yun, S.; Byeon, H.; Mariia, N.; Jeon, B. Development of a Real-Time Recombinase Polymerase Amplification Assay for the Rapid Detection of African Swine Fever Virus Genotype I and II. Pathogens 2022, 11, 439. [Google Scholar] [CrossRef]
- Wang, D.; Yu, J.; Wang, Y.; Zhang, M.; Li, P.; Liu, M.; Liu, Y. Development of a real-time loop-mediated isothermal amplification (LAMP) assay and visual LAMP assay for detection of African swine fever virus (ASFV). J. Virol. Methods 2020, 276, 113775. [Google Scholar] [CrossRef]
- En, F.; Wei, X.; Jian, L.; Qin, C. Loop-mediated isothermal amplification establishment for detection of pseudorabies virus. J. Virol. Methods 2008, 151, 35–39. [Google Scholar] [CrossRef]
- Lei, L.; Liao, F.; Tan, L.; Duan, D.; Zhan, Y.; Wang, N.; Wang, Y.; Peng, X.; Wang, K.; Huang, X.; et al. LAMP Coupled CRISPR-Cas12a Module for Rapid, Sensitive and Visual Detection of Porcine Circovirus 2. Animals 2022, 12, 2413. [Google Scholar] [CrossRef]
- Hu, L.; Lin, X.Y.; Yang, Z.X.; Yao, X.P.; Li, G.L.; Peng, S.Z.; Wang, Y. A multiplex PCR for simultaneous detection of classical swine fever virus, African swine fever virus, highly pathogenic porcine reproductive and respiratory syndrome virus, porcine reproductive and respiratory syndrome virus and pseudorabies in swines. Pol. J. Vet. Sci. 2015, 18, 715–723. [Google Scholar] [CrossRef]
- Zhao, J.; Shi, B.; Huang, X.; Peng, M.; Zhang, X.; He, D.; Pang, R.; Zhou, B.; Chen, P. A multiplex RT-PCR assay for rapid and differential diagnosis of four porcine diarrhea associated viruses in field specimens from pig farms in East China from 2010 to 2012. J. Virol. Methods 2013, 194, 107–112. [Google Scholar] [CrossRef]
- Chamberlain, J.S.; Gibbs, R.A.; Ranier, J.E.; Nguyen, P.N.; Caskey, C.T. Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res. 1988, 16, 11141–11156. [Google Scholar] [CrossRef]
- Haines, F.J.; Hofmann, M.A.; King, D.P.; Drew, T.W.; Crooke, H.R. Development and validation of a multiplex, real-time RT PCR assay for the simultaneous detection of classical and African swine fever viruses. PLoS ONE 2013, 8, e71019. [Google Scholar] [CrossRef]
- Zou, J.; Liu, H.; Chen, J.; Zhang, J.; Li, X.; Long, Y.; Jiang, Y.; Li, W.; Zhou, B. Development of a TaqMan-Probe-Based Multiplex Real-Time PCR for the Simultaneous Detection of Porcine Circovirus 2, 3, and 4 in East China from 2020 to 2022. Vet. Sci. 2023, 10, 29. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, L.; Noll, L.; Stoy, C.; Porter, E.; Fu, J.; Feng, Y.; Peddireddi, L.; Liu, X.; Dodd, K.A.; et al. Development of a real-time PCR assay for detection of African swine fever virus with an endogenous internal control. Transbound. Emerg. Dis. 2020, 67, 2446–2454. [Google Scholar] [CrossRef]
- Huang, Y.L.; Pang, V.F.; Pan, C.H.; Chen, T.H.; Jong, M.H.; Huang, T.S.; Jeng, C.R. Development of a reverse transcription multiplex real-time PCR for the detection and genotyping of classical swine fever virus. J. Virol. Methods 2009, 160, 111–118. [Google Scholar] [CrossRef]
- Sun, Y.; Xu, Z.; Gao, H.; Xu, S.; Liu, J.; Xing, J.; Kuang, Q.; Chen, Y.; Wang, H.; Zhang, G. Detection of a Novel African Swine Fever Virus with Three Large-Fragment Deletions in Genome, China. Microbiol. Spectr. 2022, 10, e215522. [Google Scholar] [CrossRef]
- Sun, E.; Huang, L.; Zhang, X.; Zhang, J.; Shen, D.; Zhang, Z.; Wang, Z.; Huo, H.; Wang, W.; Huangfu, H.; et al. Genotype I African swine fever viruses emerged in domestic pigs in China and caused chronic infection. Emerg. Microbes Infect. 2021, 10, 2183–2193. [Google Scholar] [CrossRef]
- Salogni, C.; Lazzaro, M.; Giacomini, E.; Giovannini, S.; Zanoni, M.; Giuliani, M.; Ruggeri, J.; Pozzi, P.; Pasquali, P.; Boniotti, M.B.; et al. Infectious agents identified in aborted swine fetuses in a high-density breeding area: A three-year study. J. Vet. Diagn. Investig. 2016, 28, 550–554. [Google Scholar] [CrossRef]
- Kadam, U.S.; Lossie, A.C.; Schulz, B.; Irudayaraj, J. Gene Expression Analysis Using Conventional and Imaging Methods. In DNA and RNA Nanobiotechnologies. In Medicine: Diagnosis and Treatment of Diseases; Erdmann, V.A., Barciszewski, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 141–162. [Google Scholar]
- Zhao, K.; Shi, K.; Zhou, Q.; Xiong, C.; Mo, S.; Zhou, H.; Long, F.; Wei, H.; Hu, L.; Mo, M. The Development of a Multiplex Real-Time Quantitative PCR Assay for the Differential Detection of the Wild-Type Strain and the MGF505-2R, EP402R and I177L Gene-Deleted Strain of the African Swine Fever Virus. Animals 2022, 12, 1754. [Google Scholar] [CrossRef]
- Cheng, T.Y.; Henao-Diaz, A.; Poonsuk, K.; Buckley, A.; van Geelen, A.; Lager, K.; Harmon, K.; Gauger, P.; Wang, C.; Ambagala, A.; et al. Pseudorabies (Aujeszky’s disease) virus DNA detection in swine nasal swab and oral fluid specimens using a gB-based real-time quantitative PCR. Prev. Vet. Med. 2021, 189, 105308. [Google Scholar] [CrossRef]
- Zhao, S.; Lin, H.; Chen, S.; Yang, M.; Yan, Q.; Wen, C.; Hao, Z.; Yan, Y.; Sun, Y.; Hu, J.; et al. Sensitive detection of Porcine circovirus-2 by droplet digital polymerase chain reaction. J. Vet. Diagn. Investig. 2015, 27, 784–788. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, N.; Luo, Y.; Liu, Y.; Miao, F.; Chen, T.; Zhang, S.; Cao, P.; Li, X.; Tian, K.; et al. Emergence of African Swine Fever in China, 2018. Transbound. Emerg. Dis. 2018, 65, 1482–1484. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Lu, Q.; Wang, F.; Xing, G.; Feng, H.; Jin, Q.; Guo, Z.; Teng, M.; Hao, H.; Li, D.; et al. Phylogenetic analysis of porcine circovirus type 2 (PCV2) between 2015 and 2018 in Henan Province, China. BMC Vet. Res. 2020, 16, 6. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Hou, C.Y.; Zhang, Y.H.; Li, H.X.; Chen, X.M.; Pan, J.J.; Chen, H.Y. Simultaneous detection and genetic characterization of porcine circovirus 2 and 4 in Henan province of China. Gene 2022, 808, 145991. [Google Scholar] [CrossRef]
- Nan, W.; Wu, J.; Hu, H.; Peng, G.; Tan, S.; Deng, Z. Prevalence and genetic diversity of porcine circovirus type 2 in northern Guangdong Province during 2016–2021. Front. Vet. Sci. 2022, 9, 932612. [Google Scholar] [CrossRef]
- Liu, Y.; Gong, Q.; Nie, L.; Wang, Q.; Ge, G.; Li, D.; Ma, B.; Sheng, C.; Su, N.; Zong, Y.; et al. Prevalence of porcine circovirus 2 throughout China in 2015–2019: A systematic review and meta-analysis. Microb. Pathog. 2020, 149, 104490. [Google Scholar] [CrossRef]


| Primers and Probes | Sequences (5′-3′) | Length (Base Pair) | Use |
|---|---|---|---|
| ASFV-p72F | CGAAGGGAATGGATACTGA | 563 | Amplification of p72 |
| ASFV-p72R | TGTACCCGGCACAAAGA | ||
| ASFV-p72qF | ACGTTCGCTGCGTATCATTT | 112 | qPCR for detection of p72 |
| ASFV-p72qR | GAGGTATCGGTGGAGGGAAC | ||
| ASFV-probe | FAM- TGCACAAGCCGCACCAAAGCA-TAMRA | ||
| PCV2-CapF | TTACACGGATATTGTATTCCTGGTCG | 295 | Amplification of Cap |
| PCV2-CapR | GTGGGCTCCAGTGCTGTTATTCTA | ||
| PCV2-CapQF | AGTCTCAGCCACAGCTGATT | 128 | qPCR for detection of Cap |
| PCV2-CapQR | TCCTCCCGCCATACCAT | ||
| PCV2-probe | Cy5-AGCCCTTCTCCTACCACTCCCGCT-BHQ2 | ||
| PRV-gEF | GCGGCTGTTTGTGCTG | 413 | Amplification of gE |
| PRV-gER | CATAGTTGGGTCCATTCGT | ||
| PRV-gEqF | GCTCCTTCGTGATGACGTG | 131 | qPCR for detection of gE |
| PRV-gEqR | GTACACCGGAGAGAGCATGT | ||
| PRV-Probe | Texas Red-CTGCGTGCTGTGCTCCCGGC-BHQ2 | ||
| β-ActinqF | CCCTGGAGAAGAGCTACGAG | 175 | qPCR for detection of β-Actin [34] |
| β-ActinqR | AGGTCCTTCCTGATGTCCAC | ||
| β-Actin-probe | HEX-CGGCAACGAGCGCTTCCGGT-BHQ1 |
| Province | Number | Characteristic | Specimen Type |
|---|---|---|---|
| Jiangsu | 144 | Sows | Lymph nodes, sera, and/or blood |
| Aborted piglets | Lymph nodes and/or placentas | ||
| Anhui | 107 | Sows | Lymph nodes, sera, and/or blood |
| Aborted piglets | Lymph nodes and/or placentas | ||
| Zhejiang | 72 | Sows | Lymph nodes, sera, and/or blood |
| Shandong | 60 | Sows | Lymph nodes, sera, and/or blood |
| Swine Pathogens | Ct Value a | |||
|---|---|---|---|---|
| FAM | HEX | Cy5 | Texas Red | |
| ASFV | 25.22 0.19 | - | - | - |
| PCV2 | - | - | 27.25 0.21 | - |
| PRV | - | - | - | 24.73 0.24 |
| PCV1 | - | - | - | - |
| PCV3 | - | - | - | - |
| PCV4 | - | - | - | - |
| CSFV | - | - | - | - |
| JEV | - | - | - | - |
| PPV | - | - | - | - |
| PRRSV | - | - | - | - |
| Chlamydia suis | - | - | - | - |
| Toxoplasma gondii | - | - | - | - |
| Plasmid | Concentration (Copies/μL) | Intra-Assay | Inter-Assay | ||||
|---|---|---|---|---|---|---|---|
| Mean | S.D. | CV (%) | Mean | S.D. | CV (%) | ||
| pEasy-ASFV | 107 | 15.36 | 0.132 | 0.86 | 15.21 | 0.058 | 0.39 |
| 106 | 18.19 | 0.078 | 0.43 | 18.12 | 0.012 | 0.06 | |
| 105 | 21.44 | 0.083 | 0.39 | 21.57 | 0.073 | 0.34 | |
| 104 | 25.47 | 0.276 | 1.08 | 25.05 | 0.493 | 1.96 | |
| pEasy-PCV2 | 107 | 15.53 | 0.104 | 0.67 | 15.47 | 0.048 | 0.31 |
| 106 | 18.8 | 0.139 | 0.74 | 18.88 | 0.063 | 0.33 | |
| 105 | 20.85 | 0.237 | 1.14 | 21.47 | 0.380 | 1.73 | |
| 104 | 24.76 | 0.297 | 1.20 | 25.04 | 0.203 | 0.81 | |
| pEasy-PRV | 107 | 15.53 | 0.104 | 0.67 | 15.49 | 0.027 | 0.18 |
| 106 | 18.80 | 0.139 | 0.74 | 18.77 | 0.134 | 0.71 | |
| 105 | 20.85 | 0.237 | 1.14 | 20.81 | 0.127 | 0.61 | |
| 104 | 24.76 | 0.297 | 1.20 | 24.70 | 0.134 | 0.54 | |
| Virus | Positive Specimens a | Infection Rate (%) |
|---|---|---|
| ASFV | 86 | 22.45 |
| PCV2 | 109 | 28.46 |
| PRV | 11 | 2.87 |
| ASFV + PCV2 | 20 | 5.22 |
| ASFV + PRV | 1 | 0.26 |
| PRV + PCV2 | 7 | 1.83 |
| ASFV + PCV2 + PRV | 1 | 0.26 |
| β-Actin | 383 | 100 |
| Total | 383 | / |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Zou, J.; Liu, R.; Chen, J.; Li, X.; Zheng, H.; Li, L.; Zhou, B. Development of a TaqMan-Probe-Based Multiplex Real-Time PCR for the Simultaneous Detection of African Swine Fever Virus, Porcine Circovirus 2, and Pseudorabies Virus in East China from 2020 to 2022. Vet. Sci. 2023, 10, 106. https://doi.org/10.3390/vetsci10020106
Liu H, Zou J, Liu R, Chen J, Li X, Zheng H, Li L, Zhou B. Development of a TaqMan-Probe-Based Multiplex Real-Time PCR for the Simultaneous Detection of African Swine Fever Virus, Porcine Circovirus 2, and Pseudorabies Virus in East China from 2020 to 2022. Veterinary Sciences. 2023; 10(2):106. https://doi.org/10.3390/vetsci10020106
Chicago/Turabian StyleLiu, Huaicheng, Jianwen Zou, Rongchao Liu, Jing Chen, Xiaohan Li, Haixue Zheng, Long Li, and Bin Zhou. 2023. "Development of a TaqMan-Probe-Based Multiplex Real-Time PCR for the Simultaneous Detection of African Swine Fever Virus, Porcine Circovirus 2, and Pseudorabies Virus in East China from 2020 to 2022" Veterinary Sciences 10, no. 2: 106. https://doi.org/10.3390/vetsci10020106
APA StyleLiu, H., Zou, J., Liu, R., Chen, J., Li, X., Zheng, H., Li, L., & Zhou, B. (2023). Development of a TaqMan-Probe-Based Multiplex Real-Time PCR for the Simultaneous Detection of African Swine Fever Virus, Porcine Circovirus 2, and Pseudorabies Virus in East China from 2020 to 2022. Veterinary Sciences, 10(2), 106. https://doi.org/10.3390/vetsci10020106







