Genomic Analysis of Lymphoma Risk in Bullmastiff Dogs
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Inclusion of Dogs
2.2. DNA Extraction and Genotyping
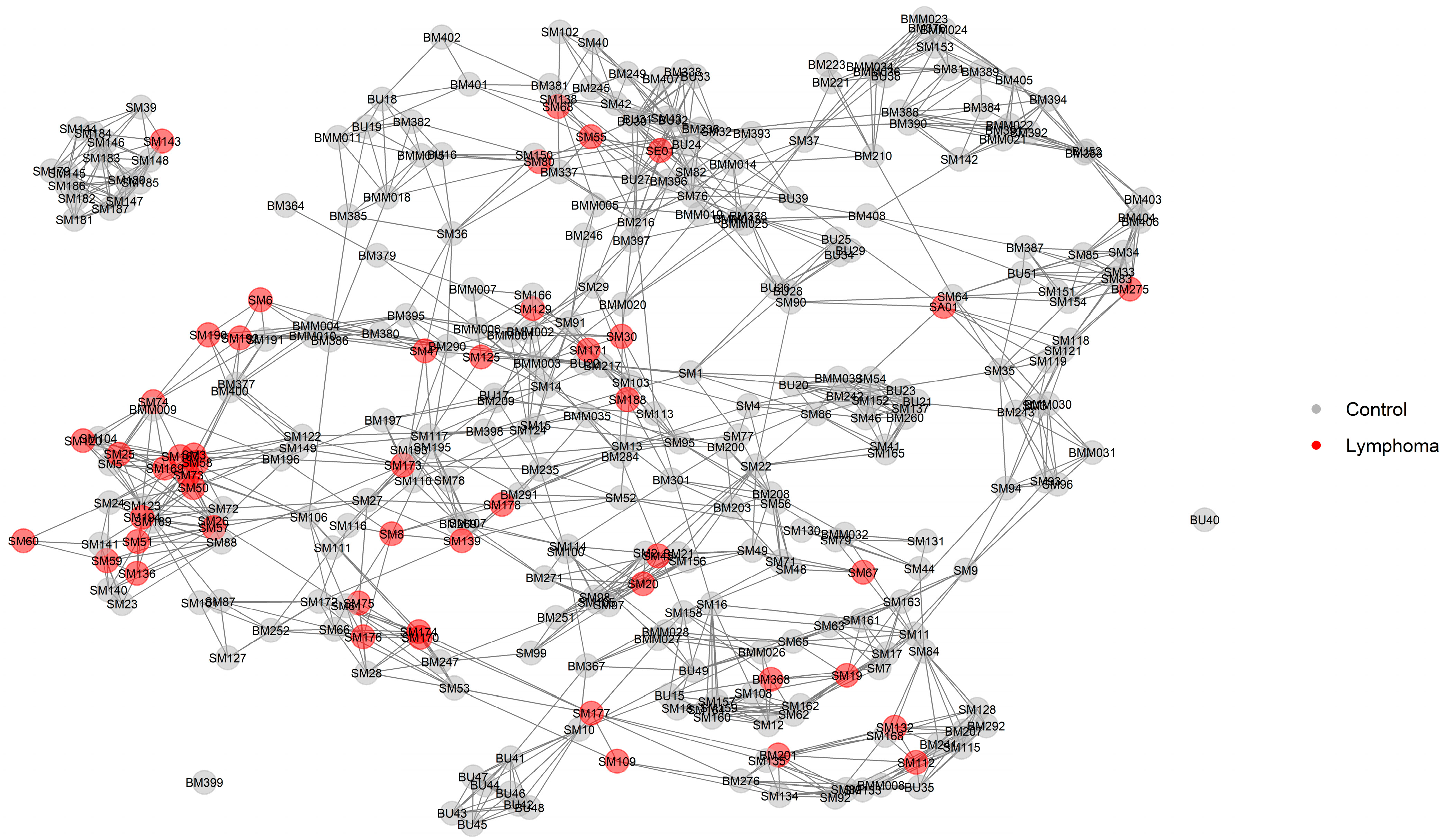
2.3. Relationship Networks (NetView)
2.4. Genome-Wide Association Analysis
2.5. Haplotype Analysis
2.6. Composite Selection Signals (CSS) Analysis
2.7. DNA Sequencing
3. Results
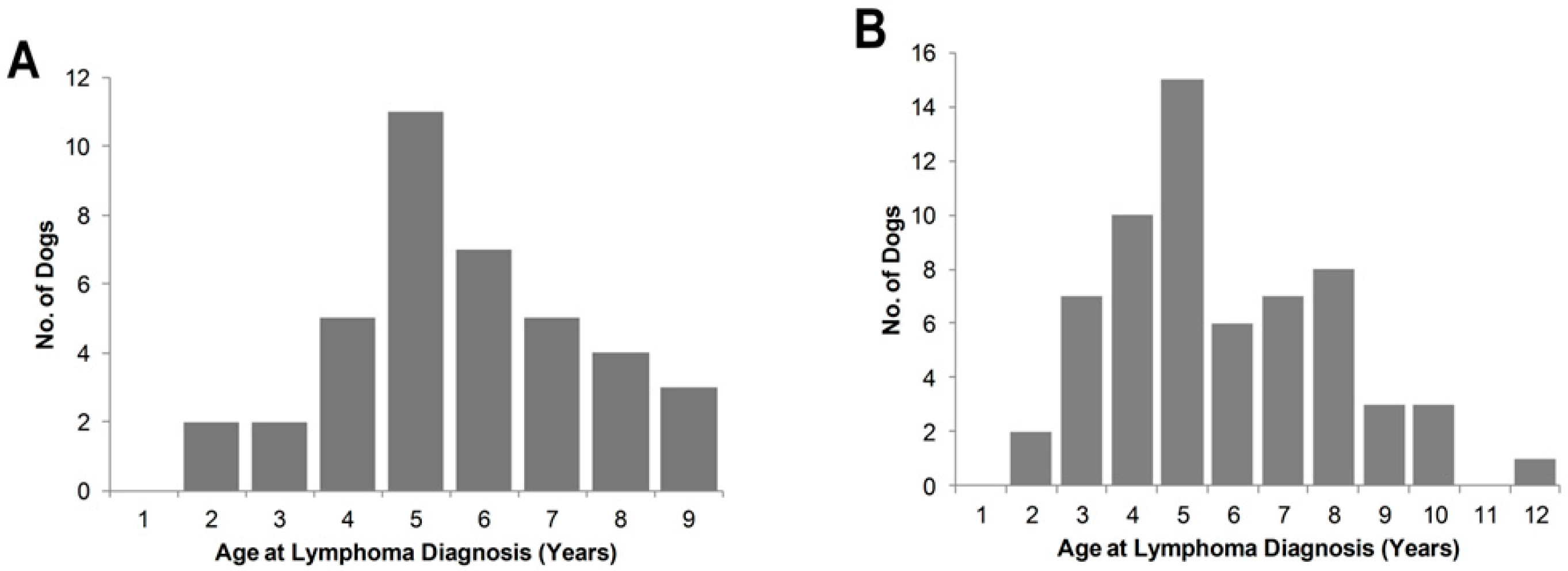
3.1. Lymphoma Cases
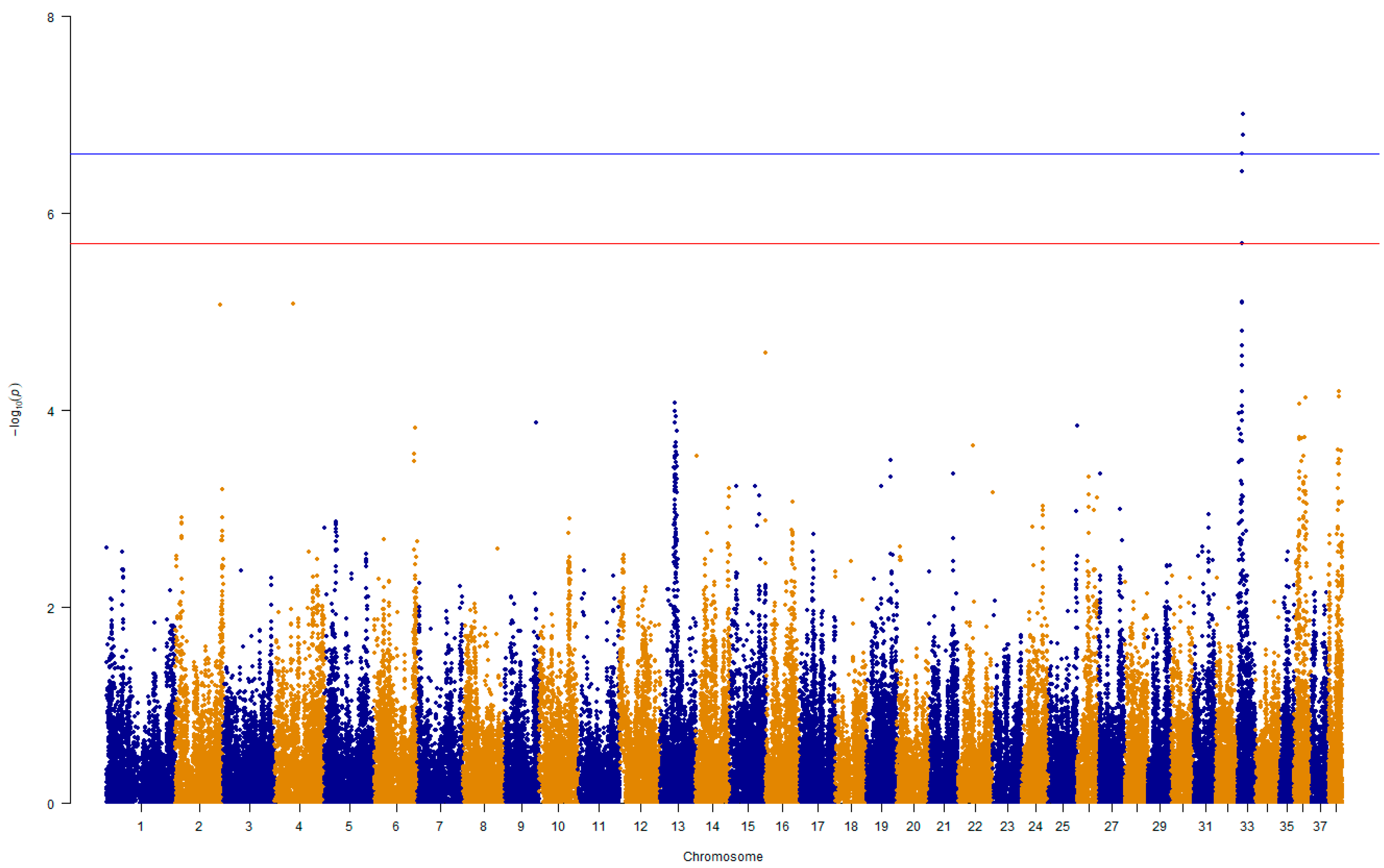
3.2. Genome-Wide Association Analysis
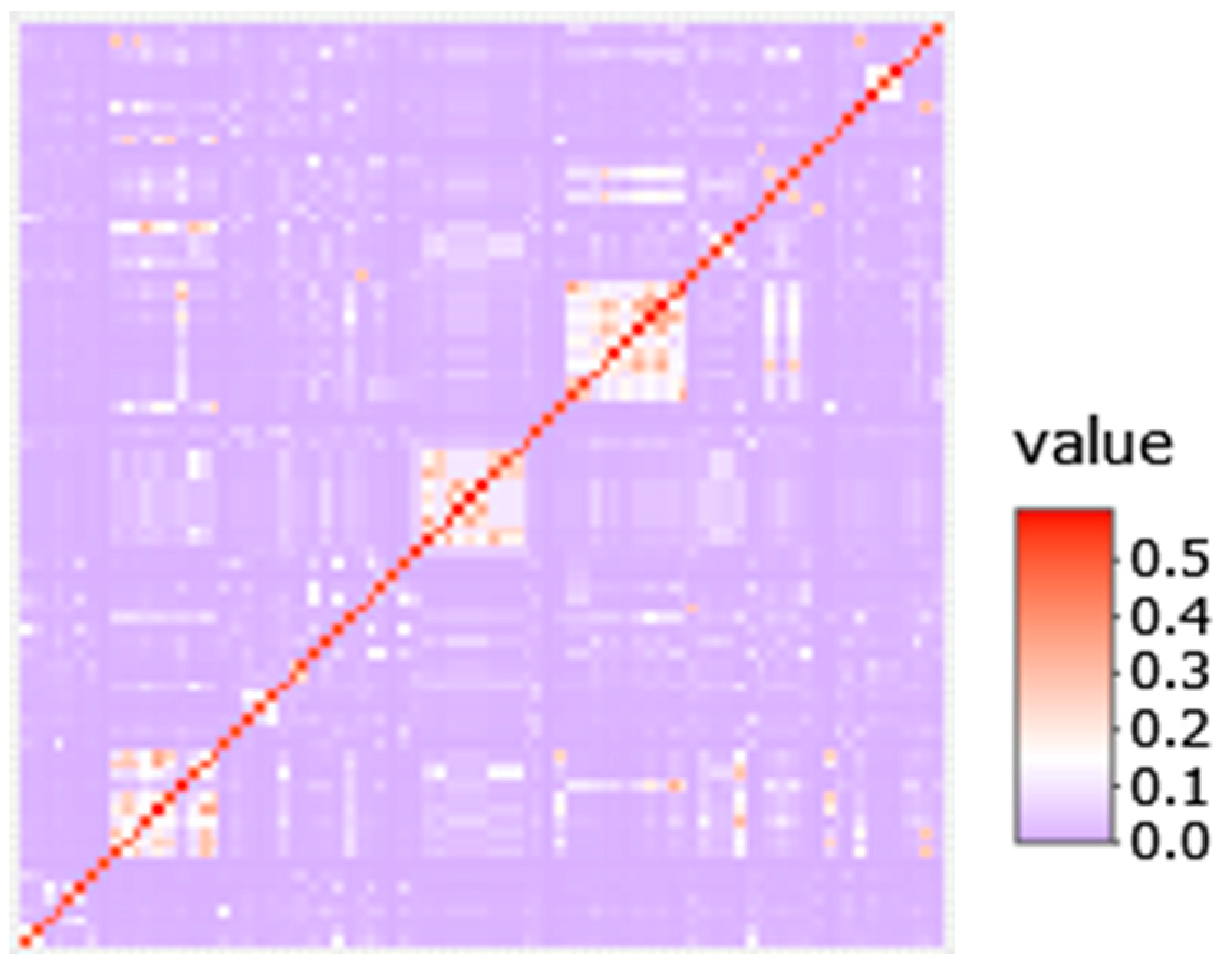
3.3. Genomic Signatures in Lymphoma-Affected Bullmastiff Dogs
3.4. DNA Sequence Variants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gruntzig, K.; Graf, R.; Hassig, M.; Welle, M.; Meier, D.; Lott, G.; Erni, D.; Schenker, N.S.; Guscetti, F.; Boo, G.; et al. The Swiss Canine Cancer Registry: A Retrospective Study on the Occurrence of Tumours in Dogs in Switzerland from 1955 to 2008. J. Comp. Pathol. 2015, 152, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Merlo, D.F.; Tanara, G.; Bocchini, V.; Rossi, L.; Pellegrino, C.; Ceppi, M.; Cardellino, U.; Capurro, C.; Ratto, A.; Sambucco, P.L.; et al. Cancer incidence in pet dogs: Findings of the Animal Tumor Registry of Genoa, Italy. J. Vet. Intern. Med. 2008, 22, 976–984. [Google Scholar] [CrossRef] [PubMed]
- Villamil, J.A.; Henry, C.J.; Hahn, A.W.; Bryan, J.N.; Tyler, J.W.; Caldwell, C.W. Hormonal and sex impact on the epidemiology of canine lymphoma. J. Cancer Epidemiol. 2009, 2009, 591753. [Google Scholar] [CrossRef] [PubMed]
- Marconato, L. The staging and treatment of multicentric high-grade lymphoma in dogs: A review of recent developments and future prospects. Vet. J. 2011, 188, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Bennett, P.; Williamson, P.; Taylor, R. Review of Canine Lymphoma Treated with Chemotherapy-Outcomes and Prognostic Factors. Vet. Sci. 2023, 10, 342. [Google Scholar] [CrossRef] [PubMed]
- Lurie, D.M.; Lucroy, M.D.; Griffey, S.M.; Simonson, E.; Madewell, B.R. T-cell-derived malignant lymphoma in the boxer breed. Vet. Comp. Oncol. 2004, 2, 171–175. [Google Scholar] [CrossRef]
- Modiano, J.F.; Breen, M.; Burnett, R.C.; Parker, H.G.; Inusah, S.; Thomas, R.; Avery, P.R.; Lindblad-Toh, K.; Ostrander, E.A.; Cutter, G.C.; et al. Distinct B-cell and T-cell lymphoproliferative disease prevalence among dog breeds indicates heritable risk. Cancer Res. 2005, 65, 5654–5661. [Google Scholar] [CrossRef]
- Comazzi, S.; Marelli, S.; Cozzi, M.; Rizzi, R.; Finotello, R.; Henriques, J.; Pastor, J.; Ponce, F.; Rohrer-Bley, C.; Rutgen, B.C.; et al. Breed-associated risks for developing canine lymphoma differ among countries: An European canine lymphoma network study. BMC Vet. Res. 2018, 14, 232. [Google Scholar] [CrossRef]
- Bennett, P.F.; Taylor, R.; Williamson, P. Demographic risk factors for lymphoma in Australian dogs: 6201 cases. J. Vet. Intern. Med. 2018, 32, 2054–2060. [Google Scholar] [CrossRef]
- Onions, D.E. A prospective survey of familial canine lymphosarcoma. J. Natl. Cancer Inst. 1984, 72, 909–912. [Google Scholar]
- Edwards, D.S.; Henley, W.E.; Harding, E.F.; Dobson, J.M.; Wood, J.L.N. Breed incidence of lymphoma in a UK population of insured dogs. Vet. Comp. Oncol. 2003, 1, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Pastor, M.; Chalvet-Monfray, K.; Marchal, T.; Keck, G.; Magnol, J.P.; Fournel-Fleury, C.; Ponce, F. Genetic and environmental risk indicators in canine non-Hodgkin’s lymphomas: Breed associations and geographic distribution of 608 cases diagnosed throughout France over 1 year. J. Vet. Intern. Med. 2009, 23, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Dobson, J.M. Breed-predispositions to cancer in pedigree dogs. ISRN Vet. Sci. 2013, 2013, 941275. [Google Scholar] [CrossRef] [PubMed]
- Walkey, B. The Bullmastiff Fancier’s Manual; Coast Arts Publishing: Sechelt, BC, Canada, 1992. [Google Scholar]
- Adams, V.J.; Evans, K.M.; Sampson, J.; Wood, J.L.N. Methods and mortality results of a health survey of purebred dogs in the UK. J. Small Anim. Pract. 2010, 51, 512–524. [Google Scholar] [CrossRef] [PubMed]
- Gavazza, A.; Presciuttini, S.; Barale, R.; Lubas, G.; Gugliucci, B. Association between canine malignant lymphoma, living in industrial areas, and use of chemicals by dog owners. J. Vet. Intern. Med. 2001, 15, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.; Seiser, E.L.; Motsinger-Reif, A.; Borst, L.; Valli, V.E.; Kelley, K.; Suter, S.E.; Argyle, D.; Burgess, K.; Bell, J.; et al. Refining tumor-associated aneuploidy through ‘genomic recoding’ of recurrent DNA copy number aberrations in 150 canine non-Hodgkin lymphomas. Leuk. Lymphoma 2011, 52, 1321–1335. [Google Scholar] [CrossRef] [PubMed]
- Pittaway, C.; Schofield, I.; Dobson, J.; O’Neill, D.G.; Brodbelt, D.C. Incidence and risk factors for the diagnosis of lymphoma in dogs in UK primary-care practice. J. Small Anim. Pr. 2019, 60, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Elvers, I.; Turner-Maier, J.; Swofford, R.; Koltookian, M.; Johnson, J.; Stewart, C.; Zhang, C.Z.; Schumacher, S.E.; Beroukhim, R.; Rosenberg, M.; et al. Exome sequencing of lymphomas from three dog breeds reveals somatic mutation patterns reflecting genetic background. Genome Res. 2015, 25, 1634–1645. [Google Scholar] [CrossRef]
- Evans, J.M.; Parker, H.G.; Rutteman, G.R.; Plassais, J.; Grinwis, G.C.M.; Harris, A.C.; Lana, S.E.; Ostrander, E.A. Multi-omics approach identifies germline regulatory variants associated with hematopoietic malignancies in retriever dog breeds. PLoS Genet. 2021, 17, e1009543. [Google Scholar] [CrossRef]
- Hayward, J.J.; Castelhano, M.G.; Oliveira, K.C.; Corey, E.; Balkman, C.; Baxter, T.L.; Casal, M.L.; Center, S.A.; Fang, M.; Garrison, S.J.; et al. Complex disease and phenotype mapping in the domestic dog. Nat. Commun. 2016, 7, 10460. [Google Scholar] [CrossRef]
- Hédan, B.; Cadieu, É.; Rimbault, M.; Vaysse, A.; Dufaure de Citres, C.; Devauchelle, P.; Botherel, N.; Abadie, J.; Quignon, P.; Derrien, T.; et al. Identification of common predisposing loci to hematopoietic cancers in four dog breeds. PLoS Genet. 2021, 17, e1009395. [Google Scholar] [CrossRef] [PubMed]
- Labadie, J.D.; Elvers, I.; Feigelson, H.S.; Magzamen, S.; Yoshimoto, J.; Dossey, J.; Burnett, R.; Avery, A.C. Genome-wide association analysis of canine T zone lymphoma identifies link to hypothyroidism and a shared association with mast-cell tumors. BMC Genom. 2020, 21, 464. [Google Scholar] [CrossRef] [PubMed]
- Soh, P.X.Y.; Khatkar, M.S.; Williamson, P. Lymphoma in Border Collies: Genome-Wide Association and Pedigree Analysis. Vet. Sci. 2023, 10, 581. [Google Scholar] [CrossRef]
- Tonomura, N.; Elvers, I.; Thomas, R.; Megquier, K.; Turner-Maier, J.; Howald, C.; Sarver, A.L.; Swofford, R.; Frantz, A.M.; Ito, D.; et al. Genome-wide Association Study Identifies Shared Risk Loci Common to Two Malignancies in Golden Retrievers. PLoS Genet. 2015, 11, e1004922. [Google Scholar] [CrossRef]
- Mortlock, S.A.; Khatkar, M.S.; Williamson, P. Comparative Analysis of Genome Diversity in Bullmastiff Dogs. PLoS ONE 2016, 11, e0147941. [Google Scholar] [CrossRef]
- Sinnwell, J.P.; Therneau, T.M.; Schaid, D.J. The kinship2 R package for pedigree data. Hum. Hered. 2014, 78, 91–93. [Google Scholar] [CrossRef] [PubMed]
- Rainer, J.; Taliun, D.; D’Elia, Y.; Pattaro, C.; Domingues, F.S.; Weichenberger, C.X. FamAgg: An R package to evaluate familial aggregation of traits in large pedigrees. Bioinformatics 2016, 32, 1583–1585. [Google Scholar] [CrossRef]
- Coster, A.J.R.P.V. pedigree: Pedigree Functions. 2013, Volume 1. Available online: https://www.imsbio.co.jp/RGM/R_package_list?package=pedigree&data_source=all&init=true (accessed on 19 November 2023).
- Browning, B.L.; Zhou, Y.; Browning, S.R. A One-Penny Imputed Genome from Next-Generation Reference Panels. Am. J. Hum. Genet. 2018, 103, 338–348. [Google Scholar] [CrossRef]
- Soh, P.X.Y.; Hsu, W.T.; Khatkar, M.S.; Williamson, P. Evaluation of genetic diversity and management of disease in Border Collie dogs. Sci. Rep. 2021, 11, 6243. [Google Scholar] [CrossRef]
- Steinig, E.J.; Neuditschko, M.; Khatkar, M.S.; Raadsma, H.W.; Zenger, K.R. netview p: A network visualization tool to unravel complex population structure using genome-wide SNPs. Mol. Ecol. Resour. 2016, 16, 216–227. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef]
- Turner, D. qqman: An R package for visualizing GWAS results using Q-Q and manhattan plots. J. Open Source Softw. 2018, 3, 731. [Google Scholar] [CrossRef]
- He, F.; Ding, S.; Wang, H.; Qin, F. IntAssoPlot: An R Package for Integrated Visualization of Genome-Wide Association Study Results with Gene Structure and Linkage Disequilibrium Matrix. Front. Genet. 2020, 11, 260. [Google Scholar] [CrossRef]
- Sabik, O.L.; Calabrese, G.M.; Taleghani, E.; Ackert-Bicknell, C.L.; Farber, C.R. Identification of a Core Module for Bone Mineral Density through the Integration of a Co-expression Network and GWAS Data. Cell Rep. 2020, 32, 108145. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef]
- Hsu, W.T.; Williamson, P.; Khatkar, M.S. Analysis of dog breed diversity using a composite selection index. Sci. Rep. 2023, 13, 1674. [Google Scholar] [CrossRef]
- Mortlock, S.A.; Williamson, P.; Khatkar, M.S. Copy number variation and variant discovery in Bullmastiff dogs. Anim. Genet. 2019, 50, 177–181. [Google Scholar] [CrossRef]
- McLaren, W.; Pritchard, B.; Rios, D.; Chen, Y.; Flicek, P.; Cunningham, F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics 2010, 26, 2069–2070. [Google Scholar] [CrossRef]
- Modiano, J.F.; Breen, M.; Valli, V.E.O.; Wojcieszyn, J.W.; Cutter, G.R. Predictive value of p16 or Rb inactivation in a model of naturally occurring canine non-Hodgkin’s lymphoma. Leukemia 2007, 21, 184–187. [Google Scholar] [CrossRef] [PubMed]
- Hezroni, H.; Koppstein, D.; Schwartz, M.G.; Avrutin, A.; Bartel, D.P.; Ulitsky, I. Principles of Long Noncoding RNA Evolution Derived from Direct Comparison of Transcriptomes in 17 Species. Cell Rep. 2015, 11, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Beck-Engeser, G.B.; Lum, A.M.; Huppi, K.; Caplen, N.J.; Wang, B.B.; Wabl, M. Pvt1-encoded microRNAs in oncogenesis. Retrovirology 2008, 5, 14. [Google Scholar] [CrossRef] [PubMed]
- Gabay, M.; Li, Y.; Felsher, D.W. MYC Activation Is a Hallmark of Cancer Initiation and Maintenance. Cold Spring Harb. Perspect. Med. 2014, 4, a014241. [Google Scholar] [CrossRef]
- Tolomeo, D.; Agostini, A.; Visci, G.; Traversa, D.; Storlazzi, C.T. PVT1: A long non-coding RNA recurrently involved in neoplasia-associated fusion transcripts. Gene 2021, 779, 145497. [Google Scholar] [CrossRef] [PubMed]
- Cerhan, J.R.; Berndt, S.I.; Vijai, J.; Ghesquieres, H.; McKay, J.; Wang, S.S.; Wang, Z.M.; Yeager, M.; Conde, L.; de Bakker, P.I.W.; et al. Genome-wide association study identifies multiple susceptibility loci for diffuse large B cell lymphoma. Nat. Genet. 2014, 46, 1233–1238. [Google Scholar] [CrossRef] [PubMed]
- Tseng, Y.Y.; Moriarity, B.S.; Gong, W.; Akiyama, R.; Tiwari, A.; Kawakami, H.; Ronning, P.; Reuland, B.; Guenther, K.; Beadnell, T.C.; et al. PVT1 dependence in cancer with MYC copy-number increase. Nature 2014, 512, 82–86. [Google Scholar] [CrossRef]
- Hung, K.L.; Yost, K.E.; Xie, L.; Shi, Q.; Helmsauer, K.; Luebeck, J.; Schöpflin, R.; Lange, J.T.; Chamorro González, R.; Weiser, N.E.; et al. ecDNA hubs drive cooperative intermolecular oncogene expression. Nature 2021, 600, 731–736. [Google Scholar] [CrossRef]
- Mortlock, S.-A.; Wei, J.; Williamson, P. T-Cell Activation and Early Gene Response in Dogs. PLoS ONE 2015, 10, e0121169. [Google Scholar] [CrossRef]
- Daly, J.A.; Mortlock, S.A.; Taylor, R.M.; Williamson, P. Cluster Analysis of Tumor Suppressor Genes in Canine Leukocytes Identifies Activation State. Bioinform. Biol. Insights 2015, 9, 59–67. [Google Scholar] [CrossRef]
- Thomas, R.; Smith, K.C.; Ostrander, E.A.; Galibert, F.; Breen, M. Chromosome aberrations in canine multicentric lymphomas detected with comparative genomic hybridisation and a panel of single locus probes. Br. J. Cancer 2003, 89, 1530–1537. [Google Scholar] [CrossRef] [PubMed]
- Arico, A.; Ferraresso, S.; Bresolin, S.; Marconato, L.; Comazzi, S.; Kronnie, G.T.; Aresu, L. Array-Based Comparative Genomic Hybridization Analysis Reveals Chromosomal Copy Number Aberrations Associated with Clinical Outcome in Canine Diffuse Large B-Cell Lymphoma. PLoS ONE 2014, 9, e111817. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.X.; Fan, L.; Wang, L.; Wu, J.Z.; Miao, K.R.; Liang, J.H.; Gong, Q.X.; Wang, Z.; Young, K.H.; Xu, W.; et al. MYC or BCL2 copy number aberration is a strong predictor of outcome in patients with diffuse large B-cell lymphoma. Oncotarget 2015, 6, 18374–18388. [Google Scholar] [CrossRef]
- Zhou, K.G.; Xu, D.M.; Cao, Y.; Wang, J.; Yang, Y.F.; Huang, M. C-MYC Aberrations as Prognostic Factors in Diffuse Large B-cell Lymphoma: A Meta-Analysis of Epidemiological Studies. PLoS ONE 2014, 9, e95020. [Google Scholar] [CrossRef] [PubMed]
- Valentino, C.; Kendrick, S.; Johnson, N.; Gascoyne, R.; Chan, W.C.; Weisenburger, D.; Braziel, R.; Cook, J.R.; Tubbs, R.; Campo, E.; et al. Colorimetric In Situ Hybridization Identifies MYC Gene Signal Clusters Correlating with Increased Copy Number, mRNA, and Protein in Diffuse Large B-Cell Lymphoma. Am. J. Clin. Pathol. 2013, 139, 242–254. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.O.; Jeon, Y.K.; Paik, J.H.; Kim, W.Y.; Kim, Y.A.; Kim, J.E.; Kim, C.W. MYC translocation and an increased copy number predict poor prognosis in adult diffuse large B-cell lymphoma (DLBCL), especially in germinal centre-like B cell (GCB) type. Histopathology 2008, 53, 205–217. [Google Scholar] [CrossRef]
- He, Y.; Jing, Y.; Wei, F.; Tang, Y.; Yang, L.; Luo, J.; Yang, P.; Ni, Q.; Pang, J.; Liao, Q.; et al. Long non-coding RNA PVT1 predicts poor prognosis and induces radioresistance by regulating DNA repair and cell apoptosis in nasopharyngeal carcinoma. Cell Death Dis. 2018, 9, 235. [Google Scholar] [CrossRef] [PubMed]
- Bialik, P.; Wozniak, K. SUMO proteases as potential targets for cancer therapy. Postep. Hig. I Med. Dosw. (Online) 2017, 71, 997–1004. [Google Scholar] [CrossRef]
- Gonzalez-Prieto, R.; Cuijpers, S.A.; Kumar, R.; Hendriks, I.A.; Vertegaal, A.C. c-Myc is targeted to the proteasome for degradation in a SUMOylation-dependent manner, regulated by PIAS1, SENP7 and RNF4. Cell Cycle 2015, 14, 1859–1872. [Google Scholar] [CrossRef]
- Nogai, H.; Wenzel, S.S.; Hailfinger, S.; Grau, M.; Kaergel, E.; Seitz, V.; Wollert-Wulf, B.; Pfeifer, M.; Wolf, A.; Frick, M.; et al. IkappaB-zeta controls the constitutive NF-kappaB target gene network and survival of ABC DLBCL. Blood 2013, 122, 2242–2250. [Google Scholar] [CrossRef] [PubMed]
- Mudaliar, M.A.; Haggart, R.D.; Miele, G.; Sellar, G.; Tan, K.A.; Goodlad, J.R.; Milne, E.; Vail, D.M.; Kurzman, I.; Crowther, D.; et al. Comparative gene expression profiling identifies common molecular signatures of NF-κB activation in canine and human diffuse large B cell lymphoma (DLBCL). PLoS ONE 2013, 8, e72591. [Google Scholar] [CrossRef] [PubMed]
- Willems, M.; Dubois, N.; Musumeci, L.; Bours, V.; Robe, P.A. IκBζ: An emerging player in cancer. Oncotarget 2016, 7, 66310–66322. [Google Scholar] [CrossRef] [PubMed]
- Arthur, S.E.; Jiang, A.; Grande, B.M.; Alcaide, M.; Cojocaru, R.; Rushton, C.K.; Mottok, A.; Hilton, L.K.; Lat, P.K.; Zhao, E.Y.; et al. Genome-wide discovery of somatic regulatory variants in diffuse large B-cell lymphoma. Nat. Commun. 2018, 9, 4001. [Google Scholar] [CrossRef]
- Avery, A.C. The Genetic and Molecular Basis for Canine Models of Human Leukemia and Lymphoma. Front. Oncol. 2020, 10, 23. [Google Scholar] [CrossRef]
- Pasqualucci, L.; Klein, U. NF-κB Mutations in Germinal Center B-Cell Lymphomas: Relation to NF-κB Function in Normal B Cells. Biomedicines 2022, 10, 2450. [Google Scholar] [CrossRef] [PubMed]
- Hsu, W.-T.; Williamson, P.; Khatkar, M.S. Identification of Genomic Signatures in Bullmastiff Dogs Using Composite Selection Signals Analysis of 23 Purebred Clades. Animals 2023, 13, 1149. [Google Scholar] [CrossRef]
- Hsu, W.T.; Williamson, P.; Khatkar, M.S. Genomic signatures in dog breeds at high risk of lymphoma. 2023; submitted. [Google Scholar]
- Truve, K.; Dickinson, P.; Xiong, A.; York, D.; Jayashankar, K.; Pielberg, G.; Koltookian, M.; Muren, E.; Fuxelius, H.H.; Weishaupt, H.; et al. Utilizing the Dog Genome in the Search for Novel Candidate Genes Involved in Glioma Development-Genome Wide Association Mapping followed by Targeted Massive Parallel Sequencing Identifies a Strongly Associated Locus. PLoS Genet. 2016, 12, e1006000. [Google Scholar] [CrossRef]
- Leeb, T.; Bannasch, D.; Schoenebeck, J.J. Identification of Genetic Risk Factors for Monogenic and Complex Canine Diseases. Annu. Rev. Anim. Biosci. 2023, 11, 183–205. [Google Scholar] [CrossRef]
- Meadows, J.R.S.; Kidd, J.M.; Wang, G.D.; Parker, H.G.; Schall, P.Z.; Bianchi, M.; Christmas, M.J.; Bougiouri, K.; Buckley, R.M.; Hitte, C.; et al. Genome sequencing of 2000 canids by the Dog10K consortium advances the understanding of demography, genome function and architecture. Genome Biol. 2023, 24, 187. [Google Scholar] [CrossRef]
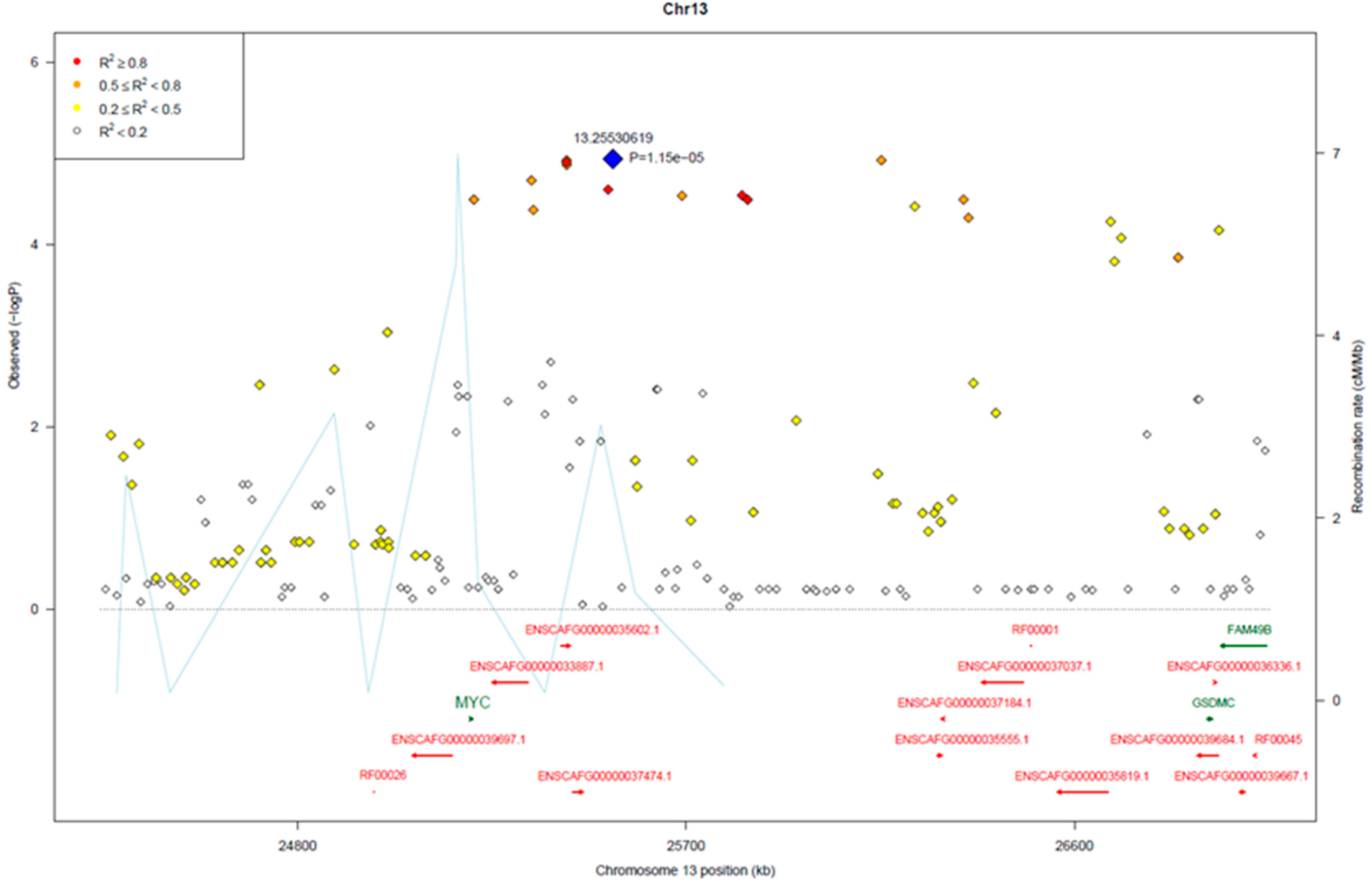
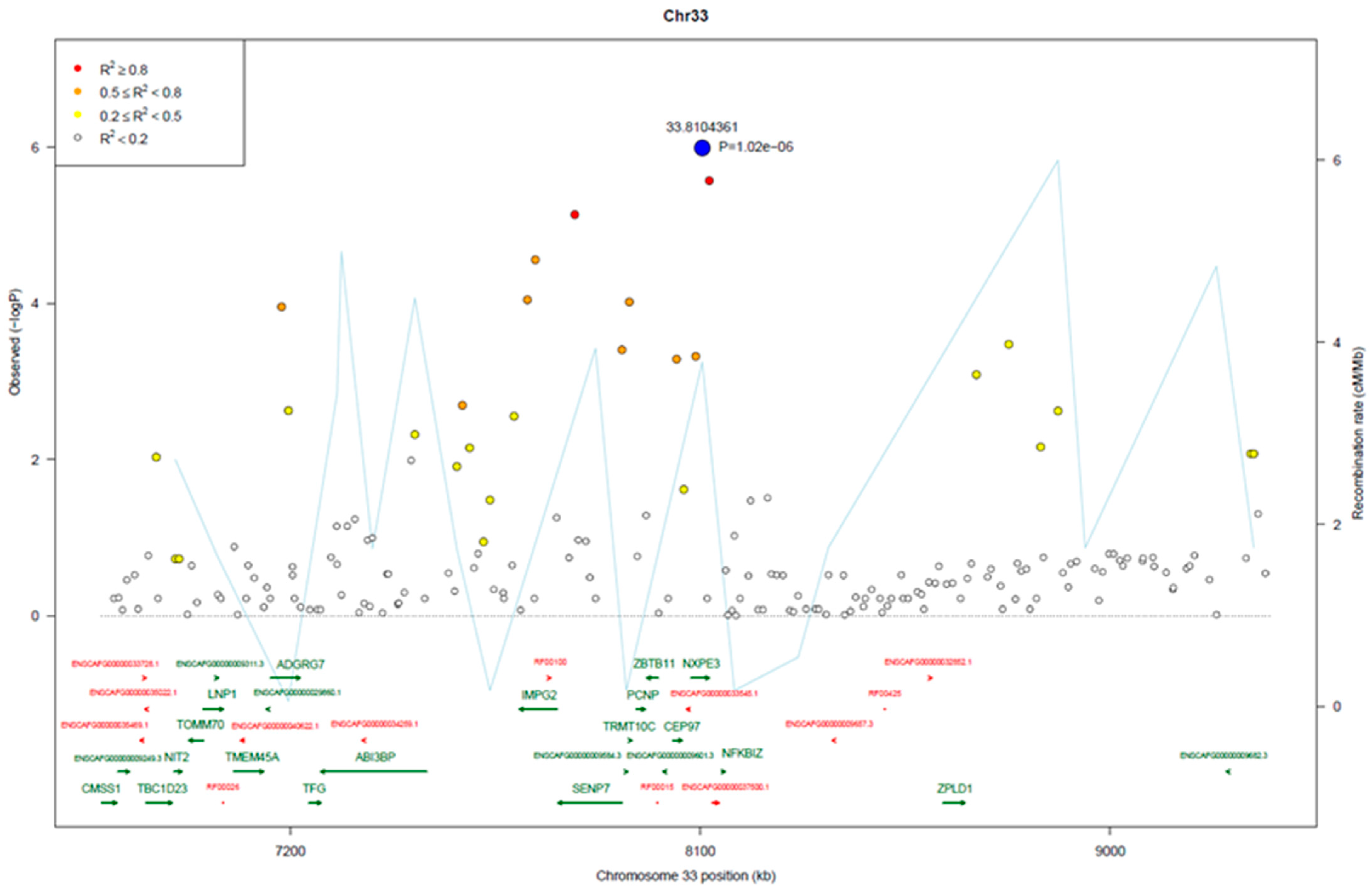
| Location | Type | Ref | Allele | Gene | Exon Number | Consequence | A.A. Change |
|---|---|---|---|---|---|---|---|
| 25205559^ 25205560 | Ins | - | T | MYC | 3/3 | 3 prime UTR | - |
| 26907214 | SNP | A | G | GSDMC | 1/12 | missense | H/R |
| 28304328 | SNP | G | A | LDH | 1/1 | missense | D/N |
| 28304341 | SNP | T | C | LDH | 1/1 | missense | V/A |
| 29249168 | SNP | C | T | TMEM71 | 1/11 | 5 prime UTR | - |
| 29681948 | SNP | G | A | WISP1 | 4/4 | 3 prime UTR | - |
| 29682320 | SNP | A | T | WISP1 | 4/4 | 3 prime UTR | - |
| 29689113 | SNV | A | T | NDRG1 | 16/16 | 3 prime UTR | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mortlock, S.A.; Asada, M.C.; Soh, P.X.Y.; Hsu, W.-T.; Lee, C.; Bennett, P.F.; Taylor, R.M.; Khatkar, M.S.; Williamson, P. Genomic Analysis of Lymphoma Risk in Bullmastiff Dogs. Vet. Sci. 2023, 10, 703. https://doi.org/10.3390/vetsci10120703
Mortlock SA, Asada MC, Soh PXY, Hsu W-T, Lee C, Bennett PF, Taylor RM, Khatkar MS, Williamson P. Genomic Analysis of Lymphoma Risk in Bullmastiff Dogs. Veterinary Sciences. 2023; 10(12):703. https://doi.org/10.3390/vetsci10120703
Chicago/Turabian StyleMortlock, Sally A., Monica C. Asada, Pamela Xing Yi Soh, Wei-Tse Hsu, Carol Lee, Peter F. Bennett, Rosanne M. Taylor, Mehar S. Khatkar, and Peter Williamson. 2023. "Genomic Analysis of Lymphoma Risk in Bullmastiff Dogs" Veterinary Sciences 10, no. 12: 703. https://doi.org/10.3390/vetsci10120703
APA StyleMortlock, S. A., Asada, M. C., Soh, P. X. Y., Hsu, W.-T., Lee, C., Bennett, P. F., Taylor, R. M., Khatkar, M. S., & Williamson, P. (2023). Genomic Analysis of Lymphoma Risk in Bullmastiff Dogs. Veterinary Sciences, 10(12), 703. https://doi.org/10.3390/vetsci10120703







