Identification of Prototheca from the Cerebrospinal Fluid of a Cat with Neurological Signs
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Samples
2.2. Nucleic Acid Extraction
2.3. Screening for Prototheca spp.
2.4. Quantitative Real Time PCR (qPCR), Specifically for P. bovis
2.5. Screening for Other Pathogens
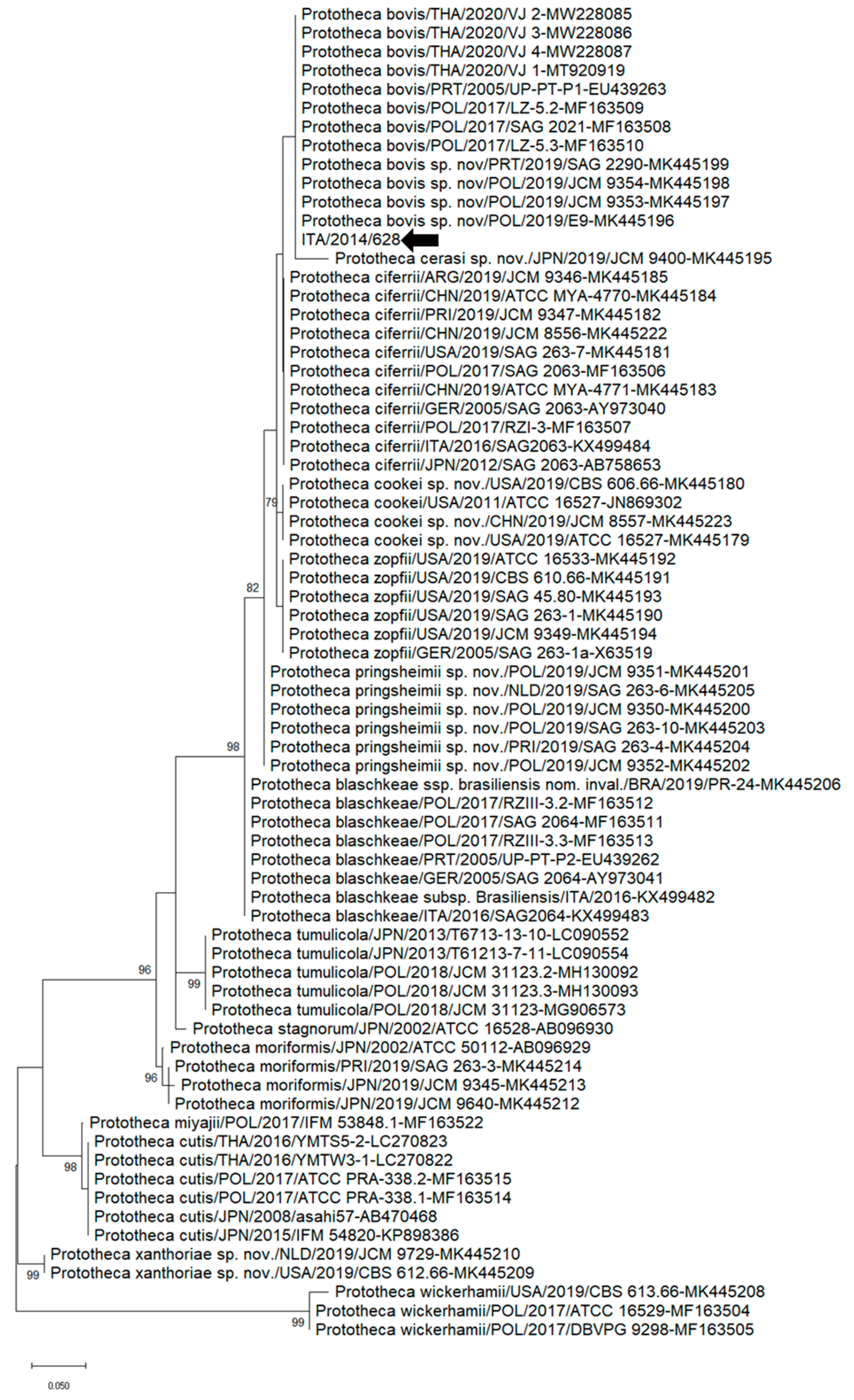
2.6. Sequence and Phylogenetic Analyses
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shave, C.D.; Millyard, L.; May, R.C. Now for Something Completely Different: Prototheca, Pathogenic Algae. PLoS Pathog. 2021, 17, e1009362. [Google Scholar] [CrossRef] [PubMed]
- Jagielski, T.; Gawor, J.; Bakuła, Z.; Decewicz, P.; Maciszewski, K.; Karnkowska, A. Cytb as a New Genetic Marker for Differentiation of Prototheca Species. J. Clin. Microbiol. 2018, 56, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Kano, R. Emergence of Fungal-Like Organisms: Prototheca. Mycopathologia 2019, 185, 747–754. [Google Scholar] [CrossRef] [PubMed]
- Irrgang, A.; Murugaiyan, J.; Weise, C.; Azab, W.; Roesler, U. Well-Known Surface and Extracellular Antigens of Pathogenic Microorganisms among the Immunodominant Proteins of the Infectious Microalgae Prototheca Zopfii. Front. Cell. Infect. Microbiol. 2015, 5, 67. [Google Scholar] [CrossRef] [PubMed]
- Lass-Flörl, C.; Mayr, A. Human Protothecosis. Clin. Microbiol. Rev. 2007, 20, 230–242. [Google Scholar] [CrossRef] [PubMed]
- Jagielski, T.; Dyląg, M.; Roesler, U.; Murugaiyan, J. Isolation of Infectious Microalga Prototheca wickerhamii from a Carp (Cyprinus carpio)—A First Confirmed Case Report of Protothecosis in a Fish. J. Fish. Dis. 2017, 40, 1417–1421. [Google Scholar] [CrossRef]
- Jagielski, T.; Roeske, K.; Bakuła, Z.; Piech, T.; Wlazło, Ł.; Bochniarz, M.; Woch, P.; Krukowski, H. A Survey on the Incidence of Prototheca Mastitis in Dairy Herds in Lublin Province, Poland. J. Dairy Sci. 2019, 102, 619–628. [Google Scholar] [CrossRef]
- Masuda, M.; Jagielski, T.; Danesi, P.; Falcaro, C.; Bertola, M.; Krockenberger, M.; Malik, R.; Kano, R. Protothecosis in Dogs and Cats—New Research Directions. Mycopathologia 2021, 186, 143–152. [Google Scholar] [CrossRef]
- Jagielski, T.; Lagneau, P.-E. Protothecosis. A Pseudofungal Infection. J. Mycol. Med. 2007, 17, 261–270. [Google Scholar] [CrossRef]
- Pressler, B.M. Infectious Diseases of the Dog and Cat. In Protothecosis and Chlorellosis; Greene, C.E., Ed.; Saunders: St. Louis, MO, USA, 2012; Volume 1, pp. 696–701. [Google Scholar]
- Stenner, V.J.; MacKay, B.; King, T.; Barrs, V.R.D.; Irwin, P.; Abraham, L.; Swift, N.; Langer, N.; Bernays, M.; Hampson, E.; et al. Protothecosis in 17 Australian Dogs and a Review of the Canine Literature. Med. Mycol. 2007, 45, 249–266. [Google Scholar] [CrossRef]
- Hirose, N.; Hua, Z.; Kato, Y.; Zhang, Q.; Li, R.; Nishimura, K.; Masuda, M. Molecular Characterization of Prototheca Strains Isolated in China Revealed the First Cases of Protothecosis Associated with Prototheca Zopfii Genotype 1. Med. Mycol. 2018, 56, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Masuda, M.; Hirose, N.; Ishikawa, T.; Ikawa, Y.; Nishimura, K. Prototheca miyajii sp. Nov., Isolated from a Patient with Systemic Protothecosis. Int. J. Syst. Evol. Microbiol. 2016, 66, 1510–1520. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.D.; Sahni, A.K.; Sen, S.; Gupta, R.M.; Basu, A. Outbreak of Prototheca wickerhamii Algaemia and Sepsis in a Tertiary Care Chemotherapy Oncology Unit. Med. J. Armed Forces India 2018, 74, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Wolf, A.M. Fungal Diseases of the Nasal Cavity of the Dog and Cat. Vet. Clin. N. Am. Small Anim. Pract. 1992, 22, 1119–1132. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, W.; Chandler, F.W.; Holzinger, E.A.; Plue, R.E.; Dickinson, R.O. Protothecosis in a Cat: First Recorded Case. Med. Mycol. 1976, 14, 281–286. [Google Scholar] [CrossRef]
- Finnie, J.W.; Coloe, P.J. Cutaneous Protothecosis in a Cat. Aust. Vet. J. 1981, 57, 307–308. [Google Scholar] [CrossRef]
- Coloe, P.J.; Allison, J.F. Protothecosis in a Cat. J. Am. Vet. Med. Assoc. 1982, 180, 78–79. [Google Scholar]
- Dillberger, J.E.; Homer, B.; Daubert, D.; Altman, N.H. Protothecosis in Two Cats. J. Am. Vet. Med. Assoc. 1988, 192, 1557–1559. [Google Scholar]
- Endo, S.; Sekiguchi, M.; Kishimoto, Y.; Kano, R.; Aoki, S.; Sichinohe, T.; Hasegawa, A. The First Case of Feline Prototheca wickerhamii Infection in Japan. J. Vet. Med. Sci. 2010, 72, 1351–1353. [Google Scholar] [CrossRef]
- Huth, N.; Wenkel, R.F.; Roschanski, N.; Rösler, U.; Plagge, L.; Schöniger, S. Prototheca Zopfii Genotype 2-Induced Nasal Dermatitis in a Cat. J. Comp. Pathol. 2015, 152, 287–290. [Google Scholar] [CrossRef]
- Roesler, U.; Möller, A.; Hensel, A.; Baumann, D.; Truyen, U. Diversity within the Current Algal Species Prototheca Zopfii: A Proposal for Two Prototheca Zopfii Genotypes and Description of a Novel Species, Prototheca blaschkeae sp. Nov. Int. J. Syst. Evol. Microbiol. 2006, 56, 1419–1425. [Google Scholar] [CrossRef] [PubMed]
- Sobukawa, H.; Yamaguchi, S.; Kano, R.; Ito, T.; Suzuki, K.; Onozaki, M.; Hasegawa, A.; Kamata, H. Short Communication: Molecular Typing of Prototheca Zopfii from Bovine Mastitis in Japan. J. Dairy Sci. 2012, 95, 4442–4446. [Google Scholar] [CrossRef] [PubMed]
- Bacova, R.; Kralik, P.; Kucharovicova, I.; Seydlova, R.; Moravkova, M. A novel TaqMan qPCR assay for rapid detection and quantification of pro-inflammatory microalgae Prototheca spp. in milk samples. Med. Mycol. 2021, 59, 784–792. [Google Scholar] [CrossRef] [PubMed]
- Gut, M.; Leutenegger, C.M.; Huder, J.B.; Pedersen, N.C.; Lutz, H. One-Tube Fluorogenic Reverse Transcription-Polymerase Chain Reaction for the Quantitation of Feline Coronaviruses. J. Virol. Methods 1999, 77, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Stiles, J.; Bienzle, D.; Render, J.A.; Buyukmihci, N.C.; Johnson, E.C. Use of Nested Polymerase Chain Reaction (PCR) for Detection of Retroviruses from Formalin-Fixed, Paraffin-Embedded Uveal Melanomas in Cats. Vet. Ophthalmol. 1999, 2, 113–116. [Google Scholar] [CrossRef]
- Endo, Y.; Cho, K.-W.; Nishigaki, K.; Momoi, Y.; Nishimura, Y.; Mizuno, T.; Goto, Y.; Watari, T.; Tsujimoto, H.; Hasegawa, A. Molecular Characteristics of Malignant Lymphomas in Cats Naturally Infected with Feline Immunodeficiency Virus. Vet. Immunol. Immunopathol. 1997, 57, 153–167. [Google Scholar] [CrossRef]
- Decaro, N.; Desario, C.; Lucente, M.S.; Amorisco, F.; Campolo, M.; Elia, G.; Cavalli, A.; Martella, V.; Buonavoglia, C. Specific Identification of Feline Panleukopenia Virus and Its Rapid Differentiation from Canine Parvoviruses Using Minor Groove Binder Probes. J. Virol. Methods 2008, 147, 67–71. [Google Scholar] [CrossRef]
- Claerebout, E.; Losson, B.; Cochez, C.; Casaert, S.; Dalemans, A.-C.; De Cat, A.; Madder, M.; Saegerman, C.; Heyman, P.; Lempereur, L. Ticks and Associated Pathogens Collected from Dogs and Cats in Belgium. Parasit. Vectors 2013, 6, 183. [Google Scholar] [CrossRef]
- Reitt, K.; Hilbe, M.; Voegtlin, A.; Corboz, L.; Haessig, M.; Pospischil, A. Aetiology of Bovine Abortion in Switzerland from 1986 to 1995? A Retrospective Study with Emphasis on Detection of Neospora Caninum and Toxoplasma Gondii by PCR. J. Vet. Med. Ser. A 2007, 54, 15–22. [Google Scholar] [CrossRef]
- Hashimoto, A.; Koga, H.; Kohno, S.; Miyazaki, Y.; Kaku, M.; Hara, K. Rapid Detection and Identification of Mycobacteria by Combined Method of Polymerase Chain Reaction and Hybridization Protection Assay. J. Infect. 1996, 33, 71–77. [Google Scholar] [CrossRef]
- Stecher, B.; Chaffron, S.; Käppeli, R.; Hapfelmeier, S.; Freedrich, S.; Weber, T.C.; Kirundi, J.; Suar, M.; McCoy, K.D.; von Mering, C.; et al. Like Will to Like: Abundances of Closely Related Species Can Predict Susceptibility to Intestinal Colonization by Pathogenic and Commensal Bacteria. PLoS Pathog. 2010, 6, e1000711. [Google Scholar] [CrossRef] [PubMed]
- Helps, C.; Reeves, N.; Egan, K.; Howard, P.; Harbour, D. Detection of Chlamydophila felis and feline herpesvirus by multi-plex real-time PCR analysis. J. Clin. Microbiol. 2003, 41, 2734–2736. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Meyer, R.E. Euthanasia and Humane Killing. In Veterinary Anesthesia and Analgesia, 5th ed.; Grimm, K.A., Lamont, L.A., Tranquilli, W.J., Greene, S.A., Robertson, S.A., Eds.; Wiley Blackwell: Chichester, UK, 2015; pp. 130–143. [Google Scholar]
- Pore, R.S.; Barnett, E.A.; Barnes, W.C.; Walker, J.D. Prototheca Ecology. Mycopathologia 1983, 81, 49–62. [Google Scholar] [CrossRef]
- Satoh, K.; Ooe, K.; Nagayama, H.; Makimura, K. Prototheca cutis sp. Nov., a Newly Discovered Pathogen of Protothecosis Isolated from Inflamed Human Skin. Int. J. Syst. Evol. Microbiol. 2010, 60, 1236–1240. [Google Scholar] [CrossRef]
- Severgnini, M.; Lazzari, B.; Capra, E.; Chessa, S.; Luini, M.; Bordoni, R.; Castiglioni, B.; Ricchi, M.; Cremonesi, P. Genome Sequencing of Prototheca Zopfii Genotypes 1 and 2 Provides Evidence of a Severe Reduction in Organellar Genomes. Sci. Rep. 2018, 8, 14637. [Google Scholar] [CrossRef]
- Thomas, J.; Preston, N. Generalised Protothecosis in a Collie Dog. Aust. Vet. J. 1990, 67, 25–27. [Google Scholar] [CrossRef]
- Schultze, A.E.; Ring, R.D.; Morgan, R.V.; Patton, C.S. Clinical, Cytologic and Histopathologic Manifestations of Protothecosis in Two Dogs. Vet. Ophthalmol. 1998, 1, 239–243. [Google Scholar] [CrossRef]
- Lane, L.V.; Meinkoth, J.H.; Brunker, J.; Smith, S.K.; Snider, T.A.; Thomas, J.; Bradway, D.; Love, B.C. Disseminated Protothecosis Diagnosed by Evaluation of CSF in a Dog. Vet. Clin. Pathol. 2012, 41, 147–152. [Google Scholar] [CrossRef]
- Vince, A.R.; Pinard, C.; Ogilvie, A.T.; Tan, E.O.; Abrams-Ogg, A.C.G. Protothecosis in a Dog. Can. Vet. J. 2014, 55, 950–954. [Google Scholar] [PubMed]
- Carfora, V.; Noris, G.; Caprioli, A.; Iurescia, M.; Stravino, F.; Franco, A. Evidence of a Prototheca Zopfii Genotype 2 Disseminated Infection in a Dog with Cutaneous Lesions. Mycopathologia 2017, 182, 603–608. [Google Scholar] [CrossRef] [PubMed]
- Sonne, L.; de Oliveira, E.C.; Argenta, F.F.; Monteggia, R.S.; Ferreiro, L.; Rösler, U.; Kershaw, O.; Driemeier, D. Prototheca Zopfii Genotype 2 Disseminated Infection in a Dog with Neurological Signs. Ciência Rural. 2017, 47, e20160877. [Google Scholar] [CrossRef]
- Ribeiro, M.G.; Rodrigues de Farias, M.; Roesler, U.; Roth, K.; Rodigheri, S.M.; Ostrowsky, M.A.; Salerno, T.; Siqueira, A.K.; Fernandes, M.C. Phenotypic and Genotypic Characterization of Prototheca Zopfii in a Dog with Enteric Signs. Res. Vet. Sci. 2009, 87, 479–481. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.; MacEwan, I.; Fluen, T.; Hardcastle, M. Disseminated Protothecosis with Central Nervous System Involvement in a Dog in New Zealand. N. Z. Vet. J. 2022, 70, 238–243. [Google Scholar] [CrossRef] [PubMed]
- Kessell, A.E.; McNair, D.; Munday, J.S.; Savory, R.; Halliday, C.; Malik, R. Successful Treatment of Multifocal Pedal Prototheca wickerhamii Infection in a Feline Immunodeficiency Virus-Positive Cat with Multiple Bowenoid in Situ Carcinomas Containing Papillomaviral DNA Sequences. J. Feline Med. Surg. Open Rep. 2017, 3, 205511691668859. [Google Scholar] [CrossRef]
- Mayorga, J.; Barba-Gómez, J.F.; Verduzco-Martínez, A.P.; Munoz-Estrada, V.F.; Welsh, O. Protothecosis. Clin. Dermatol. 2012, 3, 432–436. [Google Scholar] [CrossRef] [PubMed]
- Todd, J.R.; Matsumoto, T.; Ueno, R.; Murugaiyan, J.; Britten, A.; King, J.W.; Odaka, Y.; Oberle, A.; Weise, C.; Roesler, U.; et al. Medical Phycology 2017. Med. Mycol. 2018, 56, S188–S204. [Google Scholar] [CrossRef]
- Jinatham, V.; Cantoni, D.M.; Brown, I.R.; Vichaslip, T.; Suwannahitatorn, P.; Popluechai, S.; Tsaousis, A.D.; Gentekaki, E. Prototheca bovis, a Unicellular Achlorophyllous Trebouxiophyte Green Alga in the Healthy Human Intestine. J. Med. Microbiol. 2021, 70, 001415. [Google Scholar] [CrossRef]
- Hillesheim, P.B.; Bahrami, S. Cutaneous Protothecosis. Arch. Pathol. Lab. Med. 2011, 135, 941–944. [Google Scholar] [CrossRef]
- Silveira, C.S.; Cesar, D.; Keating, M.K.; DeLeon-Carnes, M.; Armién, A.G.; Luhers, M.; Riet-Correa, F.; Giannitti, F. A Case of Prototheca zopfii Genotype 1 Infection in a Dog (Canis lupus Familiaris). Mycopathologia 2018, 183, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, H.; Kano, R.; Hirai, A.; Murakami, M.; Yanai, T.; Namihira, Y.; Chiba, J.; Hasegawa, A. An Isolate of Prototheca wickerhamii from Systemic Canine Protothecosis. Vet. Microbiol. 2006, 118, 305–311. [Google Scholar] [CrossRef] [PubMed]
| Pathogen | Assay | Primers and Probes | Oligonucleotide Sequence | Reference(s) |
|---|---|---|---|---|
| Prototheca spp. | PCR | Proto 18S-4F Proto 18S-4R-1 | 5′-GACATGGCGAGGATTGACAGA-3′ 5′-ATCACAGACCTGTTATC-3′ | [22] [23] |
| Prototheca bovis | qPCR | PZg2F SPZg2 PZg2R | 5′-GACGATGATCCTAGTTATGGTGTAC-3′ | [24] |
| 5′Fam-TGGTAGAAGACAAATAATGTACCAAAACCA-BHQ13′ | ||||
| 5′-TATAAAAGCAAGTCCAGTTACAGCAC-3′ | ||||
| feline infectious peritonitis | qPCR | FCoV1128f FCoV1200p FCoV1229r | 5′-GATTTGATTTGGCAATGCTAGATTT-3′ 5′Fam-TCCATTGTTGGCTCGTCATAGCGGA-Tamra3′ 5′-AACAATCACTAGATCCAGACGTTAGCT-3′ | [25] |
| feline leukemia virus | PCR | 118for 119rev | 5′-TTACTCAAGTATGTTCCCATG-3′ 5′-CTGGGGAGCCTG GAGACTGCT-3′ | [26] |
| feline immunodeficiency virus | PCR | 158for 159rev | 5′-GAGTAGATACWTGGTTRCAAG-3′ 5′-CATCCTAATTCTTGCATAGC-3′ 5′-CAAAATGTGGATGGTGGAAY-3′ 5′-ACCATTCCWATAGCAGTRGC-3′ | [27] |
| nPCR | 160for 161rev | |||
| feline panleukopenia virus | qPCR | FPV/CPV-For FPV-Pb CPV-Pb FPV/CPV-Rev | 5′-ACAAGATAAAAGACGTGGTGTAACTCAA-3′ 5′Vic-ATGGGAAATACAGACTATAT-MGB3′ 5′Fam-ATGGGAAATACAAACTATAT-MGB3′ 5′-CAACCTCAGCTGGTCTCATAATAGT-3′ | [28] |
| rickettsia | PCR | RSFG 877 RSFG1258 | 5′-GGGGGCCTGCTCACGGCGG-3′ 5′-ATTGCAAAAAGTACAGTGAACA-3′ | [29] |
| neospora | qPCR | Neo For Neo Probe Neo Rev | 5′-GCATCGGAGGACACTGCT-3′ 5′Fam-CTGACTCTGAACACCGGAGGCACG-Tamra3′ 5′-ATGTCGTAAATCGGAGTTGCTTC-3′ | [30] |
| toxoplasma | qPCR | Tox For Tox Probe Tox Rev | 5′-GTCCTATCGCAACGGAGTTCTT-3′ 5′Fam-CCAGACGTGGATTTCCGTTGGTTCC-Tamra3′ 5′-TTCGTCCGTCGTAATATCAGGC-3′ | |
| mycobacterium | PCR | Myc For Myc Rev | 5′-CATGCAAGTCGAACGGAAAG-3′ 5′-CGGTGCTTCTTCTCCACCTA-3′ 5′-TACTCGAGTGGCGAACGGGT-3′ 5′-CGGACCTTCGTCGATGGTGA-3′ | [31] |
| nPCR | Myc NFor Myc NRev | |||
| bacterial 16S rDNA | PCR | B-V5 A-V6 | 5′-ATTAGATACCCYGGTAGTCC-3′ 5′-ACGAGCTGACGACARCCATG-3′ | [32] |
| Internal control | PCR | feline 28S rDNA Fw | 5’-AGCAGGAGG TGTTGGAAGAG-3′ | [33] |
| feline 28S rDNA Rv | 5′-AGGGAGAGCCTAAATCAAAGG-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lanave, G.; Pellegrini, F.; Palermo, G.; Zini, E.; Mercuriali, E.; Zagarella, P.; Bányai, K.; Camero, M.; Martella, V. Identification of Prototheca from the Cerebrospinal Fluid of a Cat with Neurological Signs. Vet. Sci. 2023, 10, 681. https://doi.org/10.3390/vetsci10120681
Lanave G, Pellegrini F, Palermo G, Zini E, Mercuriali E, Zagarella P, Bányai K, Camero M, Martella V. Identification of Prototheca from the Cerebrospinal Fluid of a Cat with Neurological Signs. Veterinary Sciences. 2023; 10(12):681. https://doi.org/10.3390/vetsci10120681
Chicago/Turabian StyleLanave, Gianvito, Francesco Pellegrini, Giuseppe Palermo, Eric Zini, Edy Mercuriali, Paolo Zagarella, Krisztián Bányai, Michele Camero, and Vito Martella. 2023. "Identification of Prototheca from the Cerebrospinal Fluid of a Cat with Neurological Signs" Veterinary Sciences 10, no. 12: 681. https://doi.org/10.3390/vetsci10120681
APA StyleLanave, G., Pellegrini, F., Palermo, G., Zini, E., Mercuriali, E., Zagarella, P., Bányai, K., Camero, M., & Martella, V. (2023). Identification of Prototheca from the Cerebrospinal Fluid of a Cat with Neurological Signs. Veterinary Sciences, 10(12), 681. https://doi.org/10.3390/vetsci10120681







