Detection of Necrosis in Digitised Whole-Slide Images for Better Grading of Canine Soft-Tissue Sarcomas Using Machine-Learning
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset and Slide Annotation
2.2. Pre-Processing
2.3. DenseNet161
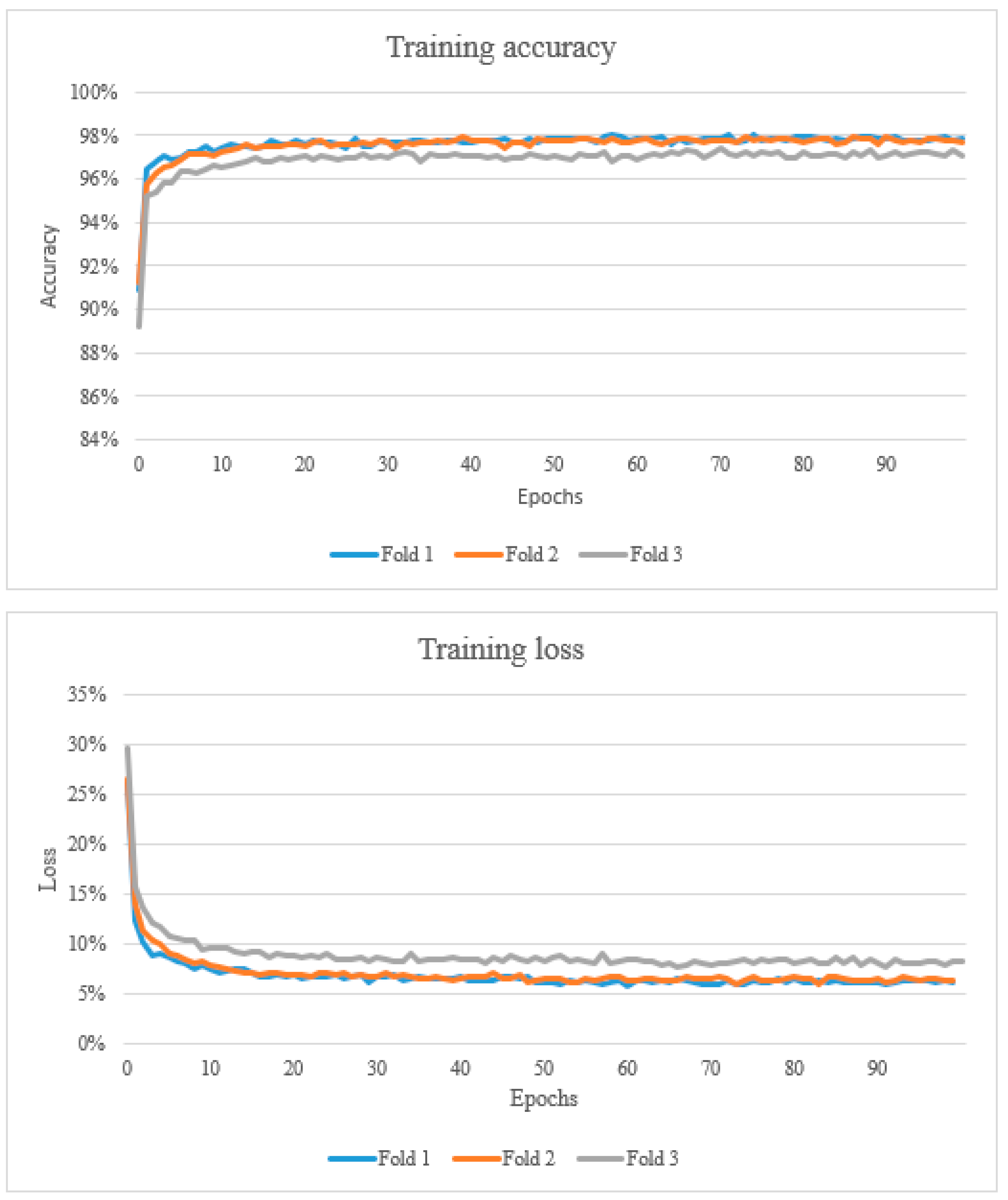
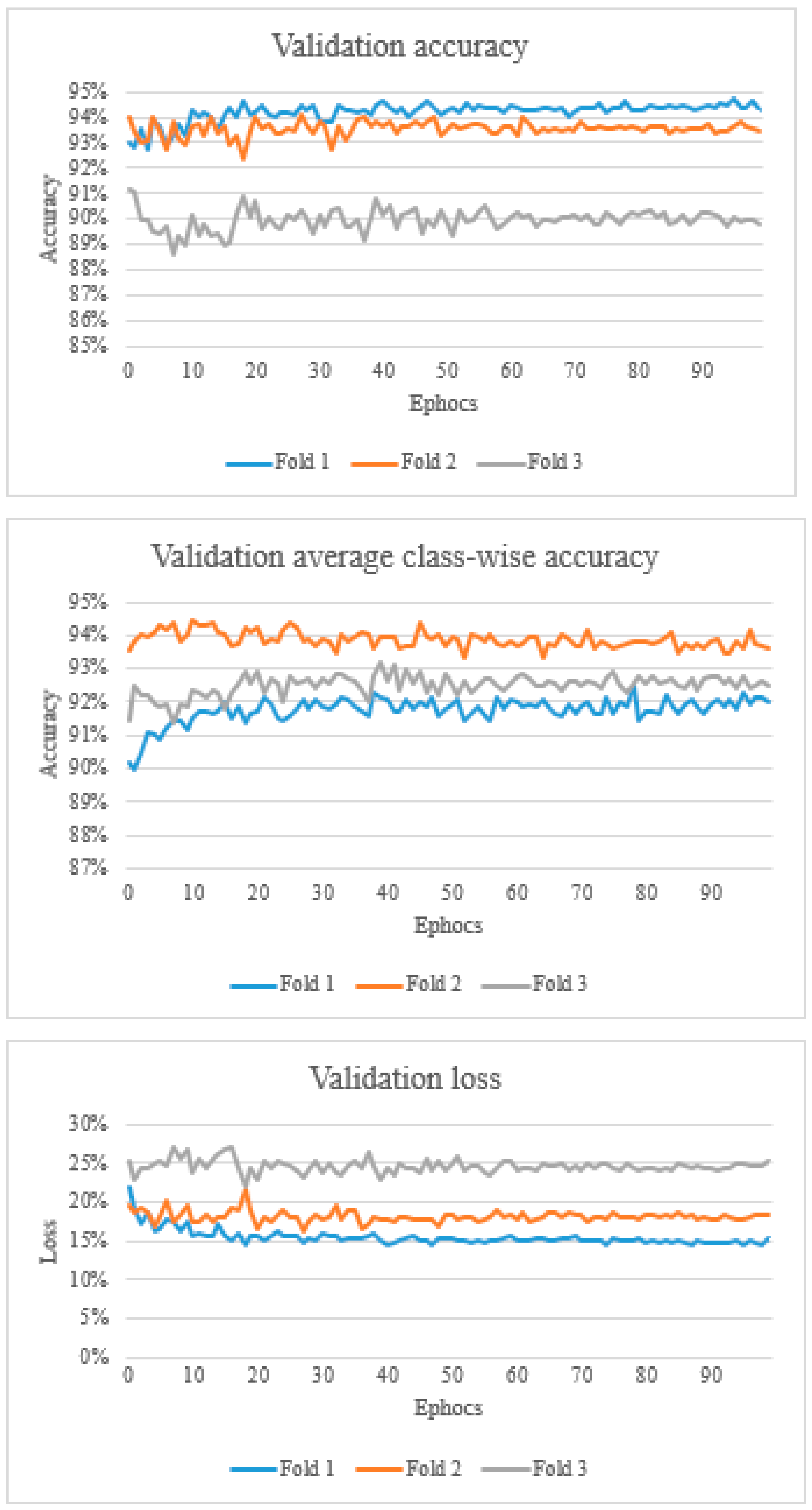
2.4. Training, Validation and Testing
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bostock, D.E.; Dye, M.T. Prognosis after surgical excision of canine fibrous connective tissue sarcoma. Vet. Pathol. 1980, 17, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Dernell, W.S.; Withrow, S.J.; Kuntz, C.A.; Powers, B.E. Principles of treatment for soft tissue sarcoma. Clin. Tech. Small Anim. Pract. 1988, 13, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Ehrhart, N. Soft-tissue sarcomas in dogs: A review. J. Am. Anim. Hosp. Assoc. 2005, 41, 241–246. [Google Scholar] [CrossRef]
- Mayer, M.N.; Larue, S.M. Soft tissue sarcomas in dogs. Can. Vet. J. 2005, 46, 040–1052. [Google Scholar]
- Dennis, M.M.; McSporran, K.D.; Bacon, N.J.; Schulman, F.Y.; Foster, R.A.; Powers, B.E. Prognostic factors for cutaneous and subcutaneous soft tissue sarcomas in dogs. Vet. Pathol. 2011, 48, 73–84. [Google Scholar] [CrossRef]
- Liptak, J.M.; Forrest, L.J. Soft tissue sarcomas. In Withrow and McEwen’s Small Animal Clinical Oncology, 5th ed.; Withrow, S.J., Vail, D.M., Page, R.L., Eds.; Elsevier: St. Louis, MO, USA, 2013. [Google Scholar]
- Roccabianca, P.; Schulman, Y.; Avallone, G.; Foster, R.; Scruggs, J.; Dittmer, K.; Kiupel, M. Surgical Pathology of Tumors of Domestic Animals: Volume 3: Tumors of Soft Tissue; Kiupel, M., Ed.; Davis Thompson Foundation: Gurnee, Illinois, 2020. [Google Scholar]
- Bray, J.P. Soft tissue sarcoma in the dog—Part 1: A current review. J. Small Anim. Pract. 2016, 57, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Coindre, J.M. Grading of soft tissue sarcomas: Review and update. Arch. Pathol. Lab. Med. 2006, 130, 1448–1453. [Google Scholar] [CrossRef]
- Yap, F.W.; Rasotto, R.; Priestnall, S.L.; Parsons, K.J.; Stewart, J. Intra- and inter-observer agreement in histological assessment of canine soft tissue sarcoma. Vet. Comp. Oncol. 2017, 15, 1553–1557. [Google Scholar] [CrossRef]
- Kuntz, C.A.; Dernell, W.S.; Powers, B.E.; Devitt, C.; Straw, R.C.; Withrow, S.J. Prognostic factors for surgical treatment of soft-tissue sarcomas in dogs: 75 cases (1986–1996). J. Am. Vet. Med. Assoc. 1997, 211, 1147–1151. [Google Scholar]
- McSporran, K.D. Histologic grade predicts recurrence for marginally excised canine subcutaneous soft tissue sarcomas. Vet. Pathol. 2009, 46, 928–933. [Google Scholar] [CrossRef]
- Swinson, D.E.; Jones, J.L.; Richardson, D.; Cox, G.; Edwards, J.G.; O’Byrne, K.J. Tumour necrosis is an independent prognostic marker in non-small cell lung cancer: Correlation with biological variables. Lung Cancer 2002, 37, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Sharma, H.; Zerbe, N.; Klempert, I.; Lohmann, S.; Lindequist, B.; Hellwich, O.; Hufnagl, P. Appearance-based necrosis detection using textural features and SVM with discriminative thresholding in histopathological whole slide images. In Proceedings of the IEEE 15th International Conference on Bioinformatics and Bioengineering (BIBE), Belgrade, Serbia, 2–4 November 2015; pp. 1–6. [Google Scholar]
- Kang, J.W.; Shin, S.H.; Choi, J.H.; Moon, K.C.; Koh, J.S.; Jung, C.; Park, Y.K.; Lee, K.B.; Chung, Y.G. Inter-and intra-observer reliability in histologic evaluation of necrosis rate induced by neo-adjuvant chemotherapy for osteosarcoma. Int. J. Clin. Exp. Pathol. 2017, 10, 359–367. [Google Scholar]
- Sharma, H.; Zerbe, N.; Klempert, I.; Hellwich, O.; Hufnagl, P. Deep convolutional neural networks for automatic classification of gastric carcinoma using whole slide images in digital histopathology. Comput. Med. Imaging Graph. 2017, 61, 2–13. [Google Scholar] [CrossRef]
- Fuchs, T.J.; Wild, P.J.; Moch, H.; Buhmann, J.M. Computational pathology analysis of tissue microarrays predict survival of renal clear cell carcinoma patients. In Medical Image Computing and Computer-Assisted Intervention, Proceedings of the 11th International Conference, New York, NY, USA, 6–10 September 2008; Springer: Berlin/Heidelberg, Germany, 2008; pp. 1–8. [Google Scholar]
- Yu, K.-H.; Zhang, C.; Berry, G.J.; Altman, R.B.; Ré, C.; Rubin, D.L.; Snyder, M. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat. Commun. 2016, 7, 12474. [Google Scholar] [CrossRef]
- Ertosun, M.G.; Rubin, D.L. Automated grading of gliomas using deep learning in digital pathology images: A modular approach with ensemble of convolutional neural networks. In AMIA Annual Symposium Proceedings; American Medical Informatics Association: Bethesda, MD, USA, 2015; Volume 2015, p. 1899. [Google Scholar]
- Petushi, S.; Garcia, F.U.; Haber, M.M.; Katsinis, C.; Tozeren, A. Large-scale computations on histology images reveal grade-differentiating parameters for breast cancer. BMC Med. Imaging 2006, 6, 14. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Zang, X.; Yang, L.; Huang, J.; Liang, F.; Rodriguez-Canales, J.; Wistuba, I.I.; Gazdar, A.; Xie, Y.; Xiao, G. Comprehensive computational pathological image analysis predicts lung cancer prognosis. J. Thorac. Oncol. 2017, 12, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Roa, A.; Basavanhally, A.; González, F.; Gilmore, H.; Feldman, M.; Ganesan, S.; Shih, N.; Tomaszewski, J.; Madabhushi, A. Automatic detection of invasive ductal carcinoma in whole slide images with convolutional neural networks. In Medical Imaging 2014: Digital Pathology, Proceedings of the SPIE Medical Imaging, San Diego, CA, USA, 15–20 February 2014; SPIE: Bellingham, WA, USA, 2014; p. 904103. [Google Scholar]
- Litjens, G.; Sánchez, C.I.; Timofeeva, N.; Hermsen, M.; Nagtegaal, I.; Kovacs, I.; Hulsbergen-van de Kaa, C.; Bult, P.; Van Ginneken, B.; Van Der Laak, J. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci. Rep. 2016, 6, 26286. [Google Scholar] [CrossRef]
- Cireşan, D.C.; Giusti, A.; Gambardella, L.M.; Schmidhuber, J. Mitosis detection in breast cancer histology images with deep neural networks. In Medical Image Computing and Computer-Assisted Intervention-MICCAI 2013, Proceedings of the 16th International Conference, Nagoya, Japan, 22–26 September 2016; Springer: Berlin/Heidelberg, Germany, 2013; pp. 411–418. [Google Scholar]
- Hou, L.; Samaras, D.; Kurc, T.M.; Gao, Y.; Davis, J.E.; Saltz, J.H. Patch-based Convolutional Neural Network for Whole Slide Tissue Image Classification. arXiv 2016, arXiv:1504.07947. [Google Scholar]
- Arunachalam, H.B.; Mishra, R.; Daescu, O.; Cederberg, K.; Rakheja, D.; Sengupta, A.; Leonard, D.; Hallac, R.; Leavey, P. Viable and necrotic tumor assessment from whole slide images of osteosarcoma using machine-learning and deep-learning models. PLoS ONE 2019, 14, e0210706. [Google Scholar] [CrossRef]
- Rai, T.; Morisi, A.; Bacci, B.; Bacon, N.J.; Dark, M.J.; Aboellail, T.; Thomas, S.A.; Bober, M.; La Ragione, R.; Wells, K. Deep learning for necrosis detection using canine perivascular wall tumour whole slide images. Sci. Rep. 2022, 12, 10634. [Google Scholar] [CrossRef]
- Rai, T.; Morisi, A.; Bacci, B.; Bacon, N.J.; Thomas, S.; La Ragione, R.; Bober, M.; Wells, K. Can imagenet feature maps be applied to small histopathological datasets for the classification of breast cancer metastatic tissue in whole slide images? Proc. SPIE 2019, 10956, 109560V. [Google Scholar]
- Rai, T.; Morisi, A.; Bacci, B.; Bacon, N.J.; Thomas, S.; La Ragione, R.; Bober, M.; Wells, K. An investigation of aggregated transfer learning for classification in digital pathology. Proc. SPIE 2019, 10956, 109560U. [Google Scholar]
- Talo, M. Automated Classification of Histopathology Images Using Transfer Learning. Artif. Intell. Med. 2019, 101, 101743. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Li, F.-F. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. In Proceedings of the 3rd International Conference for Learning Representations, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Veta, M.; van Diest, P.J.; Willems, S.M.; Wang, H.; Madabhushi, A.; Cruz-Roa, A.; Gonzalez, F.; Larsen, A.B.; Vestergaard, J.S.; Dahl, A.B.; et al. Assessment of algorithms for mitosis detection in breast cancer histopathology images. Med. Image Anal. 2015, 20, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Meuten, D.J.; Moore, F.M.; Donovan, T.A.; Bertram, C.A.; Klopfleisch, R.; Foster, R.A.; Smedley, R.C.; Dark, M.J.; Milovancev, M.; Stromberg, P.; et al. International Guidelines for Veterinary Tumor Pathology: A Call to Action. Vet. Pathol. 2021, 58, 766–794. [Google Scholar] [CrossRef] [PubMed]
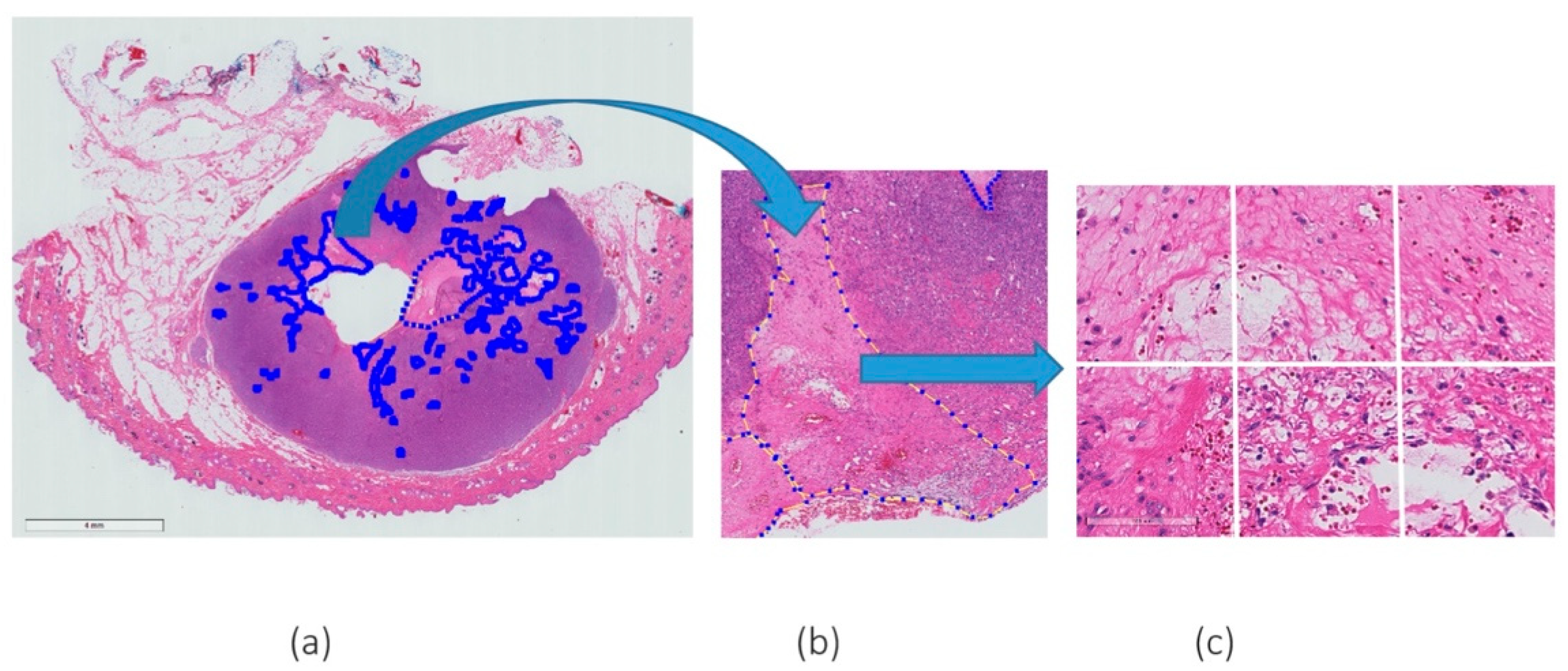

| Slide Code | Grade | Fold 1 | No. of Necrosis Patches 20× | No. of Negative Patches 20× |
|---|---|---|---|---|
| #1 | 1 | Validation | 0 | 2856 |
| #2 | 1 | Validation | 0 | 3379 |
| #3 | 2 | Validation | 0 | 2606 |
| #4 | 2 | Validation | 314 | 1417 |
| #5 | 3 | Validation | 86 | 2542 |
| #6 | 3 | Validation | 1560 | 2324 |
| Total | 1960 | 15,124 | ||
| #7 | 1 | Training | 0 | 800 |
| #8 | 1 | Training | 0 | 800 |
| #9 | 1 | Training | 0 | 800 |
| #10 | 2 | Training | 41 | 800 |
| #11 | 2 | Training | 0 | 800 |
| #12 | 2 | Training | 0 | 800 |
| #13 | 2 | Training | 0 | 800 |
| #14 | 2 | Training | 1696 | 800 |
| #15 | 2 | Training | 0 | 800 |
| #16 | 3 | Training | 57 | 800 |
| #17 | 3 | Training | 934 | 800 |
| #18 | 3 | Training | 210 | 800 |
| #19 | 3 | Training | 742 | 800 |
| #20 | 3 | Training | 144 | 800 |
| Total | 3824 | 11,200 | ||
| Slide Code | Grade | Fold 2 | No. of Necrosis Patches 20× | No. of Negative Patches 20× |
| #9 | 1 | Validation | 0 | 2132 |
| #8 | 1 | Validation | 0 | 2280 |
| #11 | 2 | Validation | 0 | 4944 |
| #19 | 3 | Validation | 742 | 2367 |
| #17 | 3 | Validation | 934 | 1332 |
| #20 | 3 | Validation | 144 | 1727 |
| #18 | 3 | Validation | 210 | 2280 |
| Total | 2030 | 17,062 | ||
| #1 | 1 | Training | 0 | 800 |
| #2 | 1 | Training | 0 | 800 |
| #7 | 1 | Training | 0 | 800 |
| #4 | 2 | Training | 314 | 800 |
| #10 | 2 | Training | 41 | 800 |
| #4 | 2 | Training | 0 | 800 |
| #12 | 2 | Training | 0 | 800 |
| #13 | 2 | Training | 0 | 800 |
| #14 | 2 | Training | 1696 | 800 |
| #15 | 2 | Training | 0 | 800 |
| #16 | 3 | Training | 57 | 800 |
| #6 | 3 | Training | 1560 | 800 |
| #5 | 3 | Training | 86 | 800 |
| Total | 3754 | 10,400 | ||
| Slide Code | Grade | Fold 3 | No. of Necrosis Patches 20× | No. of Negative Patches 20× |
| #7 | 1 | Validation | 0 | 2259 |
| #12 | 2 | Validation | 0 | 3562 |
| #13 | 2 | Validation | 0 | 2551 |
| #14 | 2 | Validation | 1696 | 2119 |
| #15 | 2 | Validation | 0 | 2936 |
| #10 | 2 | Validation | 41 | 2983 |
| #16 | 3 | Validation | 57 | 2379 |
| Total | 1794 | 18,789 | ||
| #1 | 1 | Training | 0 | 800 |
| #9 | 1 | Training | 0 | 800 |
| #2 | 1 | Training | 0 | 800 |
| #8 | 1 | Training | 0 | 800 |
| #4 | 2 | Training | 314 | 800 |
| #3 | 2 | Training | 0 | 800 |
| #20 | 3 | Training | 144 | 800 |
| #5 | 3 | Training | 86 | 800 |
| #18 | 3 | Training | 210 | 800 |
| #19 | 3 | Training | 742 | 800 |
| #17 | 3 | Training | 934 | 800 |
| #11 | 3 | Training | 0 | 800 |
| #6 | 2 | Training | 1560 | 800 |
| Total | 3990 | 10,400 |
| Slide Code | Grade | Positive | Total | Necrosis % |
|---|---|---|---|---|
| #21 | 1 | 0 | 4371 | 0.00 |
| #22 | 1 | 0 | 1611 | 0.00 |
| #23 | 1 | 0 | 2798 | 0.00 |
| #24 | 1 | 0 | 3040 | 0.00 |
| #25 | 2 | 14 | 1883 | 0.74 |
| #26 | 2 | 0 | 3368 | 0.00 |
| #27 | 2 | 20 | 1618 | 1.24 |
| #28 | 2 | 2 | 2714 | 0.07 |
| #29 | 3 | 302 | 3003 | 10.06 |
| #30 | 3 | 138 | 4528 | 3.05 |
| #31 | 3 | 369 | 3378 | 10.92 |
| #32 | 3 | 306 | 2890 | 10.59 |
| Slide Code | Sensitivity/Recall (%) | Precision (%) | Accuracy (%) | F1-Score (%) |
|---|---|---|---|---|
| Fold1_validation | 88.5 | 70.05 | 94.4 | 78.5 |
| Fold1_test | 93.4 | 30.0 | 92.7 | 45.4 |
| Fold2_validation | 94.3 | 63.2 | 93.6 | 75.7 |
| Fold2_test | 94.0 | 25.4 | 90.8 | 39.9 |
| Fold3_validation | 95.9 | 46.8 | 90.1 | 62.9 |
| Fold3_test | 94.6 | 22.4 | 89.1 | 36.2 |
| Fold 1 | |||
| Slide | Predicted Positive | Total | Necrosis % |
| #21 | 165 | 4371 | 3.77 |
| #22 | 343 | 1883 | 18.22 |
| #23 | 147 | 3040 | 4.84 |
| #24 | 44 | 3368 | 1.31 |
| #25 | 22 | 2798 | 0.79 |
| #26 | 45 | 1618 | 2.78 |
| #27 | 56 | 2714 | 2.06 |
| #28 | 56 | 1611 | 3.48 |
| #29 | 535 | 3003 | 17.82 |
| #30 | 905 | 4528 | 19.99 |
| #31 | 873 | 3378 | 25.84 |
| #32 | 395 | 2890 | 13.67 |
| Fold 2 | |||
| Slide | Predicted Positive | Total | Necrosis % |
| #21 | 192 | 4371 | 4.39 |
| #22 | 343 | 1883 | 18.22 |
| #23 | 270 | 3040 | 8.88 |
| #24 | 61 | 3368 | 1.81 |
| #25 | 31 | 2798 | 1.11 |
| #26 | 53 | 1618 | 3.28 |
| #27 | 55 | 2714 | 2.03 |
| #28 | 76 | 1611 | 4.72 |
| #29 | 537 | 3003 | 17.88 |
| #30 | 1153 | 4528 | 25.46 |
| #31 | 997 | 3378 | 29.51 |
| #32 | 498 | 2890 | 17.23 |
| Fold 3 | |||
| Slide | Predicted Positive | Total | Necrosis % |
| #21 | 388 | 4371 | 8.88 |
| #22 | 412 | 1883 | 21.88 |
| #23 | 474 | 3040 | 15.59 |
| #24 | 70 | 3368 | 2.08 |
| #25 | 30 | 2798 | 1.07 |
| #26 | 46 | 1618 | 2.84 |
| #27 | 74 | 2714 | 2.73 |
| #28 | 76 | 1611 | 4.72 |
| #29 | 621 | 3003 | 20.68 |
| #30 | 1130 | 4528 | 24.96 |
| #31 | 1172 | 3378 | 34.70 |
| #32 | 368 | 2890 | 12.73 |
| Slide | Fold 1 | Fold 2 | Fold 3 | Pathologists’ Annotations |
|---|---|---|---|---|
| #21 | 3.77 | 4.39 | 8.88 | 0.00 |
| #22 | 18.22 | 18.22 | 21.88 | 0.00 |
| #23 | 4.84 | 8.88 | 15.59 | 0.00 |
| #24 | 1.31 | 1.81 | 2.08 | 0.00 |
| #25 | 0.79 | 1.11 | 1.07 | 0.74 |
| #26 | 2.78 | 3.28 | 2.84 | 0.00 |
| #27 | 2.06 | 2.03 | 2.73 | 1.24 |
| #28 | 3.48 | 4.72 | 4.72 | 0.07 |
| #29 | 17.82 | 17.88 | 20.68 | 10.06 |
| #30 | 19.99 | 25.46 | 24.96 | 3.05 |
| #31 | 25.84 | 29.51 | 34.70 | 10.92 |
| #32 | 13.67 | 17.23 | 12.73 | 10.59 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morisi, A.; Rai, T.; Bacon, N.J.; Thomas, S.A.; Bober, M.; Wells, K.; Dark, M.J.; Aboellail, T.; Bacci, B.; La Ragione, R.M. Detection of Necrosis in Digitised Whole-Slide Images for Better Grading of Canine Soft-Tissue Sarcomas Using Machine-Learning. Vet. Sci. 2023, 10, 45. https://doi.org/10.3390/vetsci10010045
Morisi A, Rai T, Bacon NJ, Thomas SA, Bober M, Wells K, Dark MJ, Aboellail T, Bacci B, La Ragione RM. Detection of Necrosis in Digitised Whole-Slide Images for Better Grading of Canine Soft-Tissue Sarcomas Using Machine-Learning. Veterinary Sciences. 2023; 10(1):45. https://doi.org/10.3390/vetsci10010045
Chicago/Turabian StyleMorisi, Ambra, Taran Rai, Nicholas J. Bacon, Spencer A. Thomas, Miroslaw Bober, Kevin Wells, Michael J. Dark, Tawfik Aboellail, Barbara Bacci, and Roberto M. La Ragione. 2023. "Detection of Necrosis in Digitised Whole-Slide Images for Better Grading of Canine Soft-Tissue Sarcomas Using Machine-Learning" Veterinary Sciences 10, no. 1: 45. https://doi.org/10.3390/vetsci10010045
APA StyleMorisi, A., Rai, T., Bacon, N. J., Thomas, S. A., Bober, M., Wells, K., Dark, M. J., Aboellail, T., Bacci, B., & La Ragione, R. M. (2023). Detection of Necrosis in Digitised Whole-Slide Images for Better Grading of Canine Soft-Tissue Sarcomas Using Machine-Learning. Veterinary Sciences, 10(1), 45. https://doi.org/10.3390/vetsci10010045





