New Insights on Nucleotide Sequence Variants and mRNA Levels of Candidate Genes Assessing Resistance/Susceptibility to Mastitis in Holstein and Montbéliarde Dairy Cows
Abstract
Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Ethics Statement
2.2. Animals and Experimental Samples
2.3. DNA Extraction and Polymerase Chain Reaction (PCR)
2.4. DNA Sequencing and Polymorphism Detection
2.5. Total RNA Extraction, Reverse Transcription and Quantitative Real-Time PCR
2.6. Statistical Analysis
3. Results
3.1. Nucleotide Sequence Variants of Investigated Genes
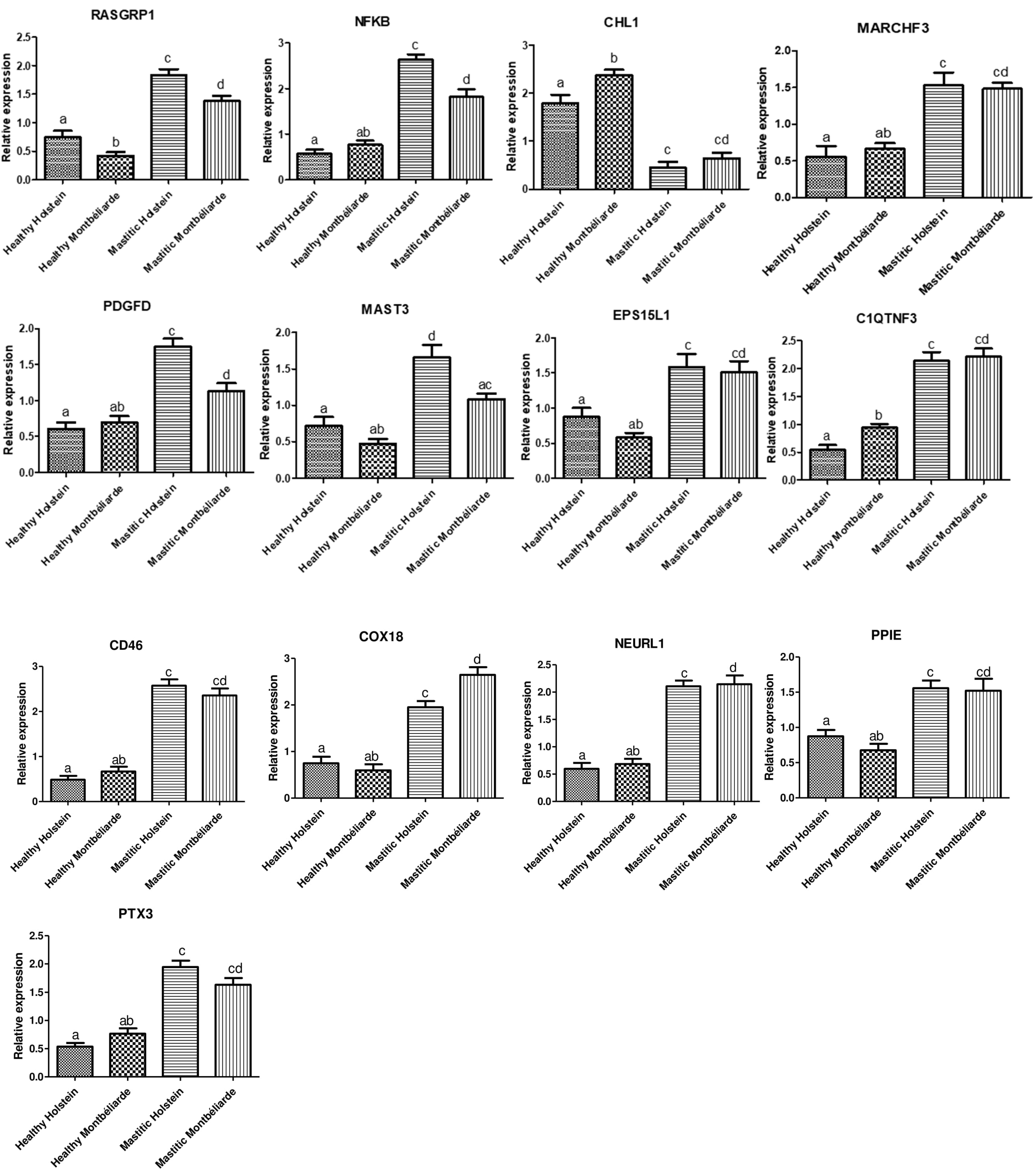
3.2. Gene Expression Pattern of Investigated Markers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kaneene, J.B.; Scott Hurd, H. The national animal health monitoring system in Michigan. III. Cost estimates of selected dairy cattle diseases. Prev. Vet. Med. 1990, 8, 127–140. [Google Scholar] [CrossRef]
- Seegers, H.; Fourichon, C.; Beaudeau, F. Production effects related to mastitis and mastitis economics in dairy cattle herds. Vet. Res. 2003, 34, 475–491. [Google Scholar] [CrossRef] [PubMed]
- Hofírek, B.; Haas, D. Categorization of mammary gland health, clinical forms of mastitis and their therapy. Sborník Ref. Odb. Semin. Mastitidy Skotu Čbs A VFU 2003, 1, 10–22. [Google Scholar]
- Sodeland, M.; Kent, M.P.; Olsen, H.G.; Opsal, M.A.; Svendsen, M.; Sehested, E.; Hayes, B.J.; Lien, S. Quantitative trait loci for clinical mastitis on chromosomes 2, 6, 14 and 20 in Norwegian red cattle. Anim. Genet. 2011, 42, 457–465. [Google Scholar] [CrossRef]
- Waller, K.P. Mammary gland immunology around parturition. Influence of stress, nutrition and genetics. Adv. Exp. Med. Biol. 2000, 480, 231–245. [Google Scholar]
- Wolfová, M.; Stípková, M.; Wolf, J. Incidence and economics of clinical mastitis in five Holstein herds in the Czech Republic. Prev. Vet. Med. 2006, 77, 48–64. [Google Scholar] [CrossRef]
- Svensson, C.; Hultgren, J. Associations between housing, management, and morbidity during rearing and subsequent first-lactation milk production of dairy cows in southwest Sweden. J. Dairy Sci. 2008, 91, 1510–1518. [Google Scholar] [CrossRef]
- Halasa, T.; Huijps, K.; Østerås, O.; Hogeveen, H. Economic effects of bovine mastitis and mastitis management: A review. Vet. Q. 2007, 29, 18–31. [Google Scholar] [CrossRef]
- Hogeveen, H.; Huijps, K.; Lam, T.J.G.M. Economic aspects of mastitis: New developments. N. Z. Vet. J. 2011, 59, 16–23. [Google Scholar] [CrossRef]
- Kvapilík, J. Mastitis of dairy cows and economic losses. Veterinářství 2014, 64, 946–955. [Google Scholar]
- Rollin, E.; Dhuyvetter, K.C.; Overton, M.W. The cost of clinical mastitis in the first 30 days of lactation: An economic modeling tool. Prev. Vet. Med. 2015, 122, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Rupp, R.; Boichard, D. Genetics of resistance to mastitis in dairy cattle. Vet. Res. 2003, 34, 671–688. [Google Scholar] [CrossRef] [PubMed]
- Tiezzi, F.; Parker-Gaddis, K.L.; Cole, J.B.; Clay, J.S.; Maltecca, C. A genome-wide association study for clinical mastitis in first parity US Holstein cows using single-step approach and genomic matrix re-weighting procedure. PLoS ONE 2015, 10, e0114919. [Google Scholar] [CrossRef] [PubMed]
- Bishop, S.C.; Woolliams, J.A. On the genetic interpretation of disease data. PLoS ONE 2010, 5, e8940. [Google Scholar] [CrossRef]
- Heringstad, B.; Klemetsdal, G.; Ruane, J. Selection for mastitis resistance in dairy cattle: A review with focus on the situation in the Nordic countries. Livest. Prod. Sci. 2000, 64, 95–106. [Google Scholar] [CrossRef]
- Carlén, E.; Strandberg, E.; Roth, A. Genetic parameters for clinical mastitis, somatic cell score, and production in the first three lactations of Swedish Holstein cows. J. Dairy Sci. 2004, 87, 3062–3070. [Google Scholar] [CrossRef]
- Meredith, B.K.; Berry, D.P.; Kearney, F.; Finlay, E.K.; Fahey, A.G.; Bradley, D.G.; Lynn, D.J. A genome-wide association study for somatic cell score using the Illumina high-density bovine beadchip identifies several novel QTL potentially related to mastitis susceptibility. Front. Genet. 2013, 4, 229. [Google Scholar] [CrossRef]
- Kühn, C.; Reinhardt, F.; Schwerin, M. Marker assisted selection of heifers improved milk somatic cell count compared to selection on conventional pedigree breeding values. Arch. Tierz. Dummerstorf. 2008, 51, 23–32. [Google Scholar] [CrossRef]
- Pighetti, G.M.; Elliott, A.A. Gene polymorphisms: The keys for marker assisted selection and unraveling core regulatory pathways for mastitis resistance. J. Mammary Gland Biol. Neoplasia. 2011, 16, 421–432. [Google Scholar] [CrossRef]
- Meier, S.; Arends, D.; Korkuć, P.; Neumann, G.B.; Brockmann, G.A. A genome-wide association study for clinical mastitis in the dual-purpose German Black Pied cattle breed. J. Dairy Sci. 2020, 103, 10289–10298. [Google Scholar] [CrossRef]
- Welderufael, B.G.; Løvendahl, P.; de Koning, D.J.; Janss, L.L.G.; Fikse, W.F. Genome wide association study for susceptibility to and recoverability from mastitis in Danish Holstein cows. Front. Genet. 2018, 24, 9–141. [Google Scholar] [CrossRef] [PubMed]
- Wagner, P.; Yin, T.; Brügemann, K.; Engel, P.; Weimann, C.; Schlez, K.; König, S. Genome wide associations for microscopic differential somatic cell count and specific mastitis pathogens in Holstein cows in compost-bedded pack and cubicle farming systems. Animals 2021, 11, 1839. [Google Scholar] [CrossRef] [PubMed]
- Kirsanova, E.; Heringstad, B.; Lewandowska-Sabat, A.; Olsaker, I. Identification of candidate genes affecting chronic subclinical mastitis in Norwegian Red cattle: Combining genome-wide association study, topologically associated domains and pathway enrichment analysis. Anim. Genet. 2020, 51, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, S.; Jagannadham, J.; Kumari, J.; Iquebal, M.A.; Gurjar, A.K.S.; Nayan, V.; Angadi, U.B.; Kumar, S.; Kumar, R.; Datta, T.K.; et al. Genome wide prediction, mapping and development of genomic resources of mastitis associated genes in water buffalo. Front. Vet. Sci. 2021, 8, 593871. [Google Scholar] [CrossRef] [PubMed]
- Kurz, J.P.; Yang, Z.; Weiss, R.B.; Wilson, D.J.; Rood, K.A.; Liu, G.E.; Wang, Z. A genome-wide association study for mastitis resistance in phenotypically well-characterized Holstein dairy cattle using a selective genotyping approach. Immunogenet 2019, 71, 35–47. [Google Scholar] [CrossRef]
- Boom, R.; Sol, C.J.; Salimans, M.M.; Jansen, C.L.; Wertheim-van Dillen, P.M.; Noordaa, J.V.D. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 1990, 28, 495–503. [Google Scholar] [CrossRef]
- Boesenberg-Smith, K.A.; Pessarakli, M.M.; Wolk, D.M. Assessment of DNA Yield and Purity: An Overlooked Detail of PCR Troubleshooting. Clin. Microbiol. Newsl. 2012, 34, 1–6. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Feuk, L.; Marshall, C.R.; Wintle, R.F.; Scherer, S.W. Structural variants: Changing the landscape of chromosomes and design of disease studies. Hum. Mol. Genet. 2006, 15, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Cartegni, L.; Chew, S.L.; Krainer, A.R. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat. Rev. Genet. 2002, 3, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Asadollahpour, N.H.; Ansari, M.S.; Edriss, M.A. Effect of LEPR, ABCG2 and SCD1 gene polymorphisms on reproductive traits in the Iranian Holstein cattle. Reprod. Domest. Anim. 2014, 49, 769–774. [Google Scholar] [CrossRef] [PubMed]
- Dusza, M.; Pokorska, J.; Makulska, J.; Kulaj, D.; Cupial, M. L-selectin gene polymorphism and its association with clinical mastitis, somatic cell score, and milk production in Polish Holstein-Friesian cattle. Czech J. Anim. Sci. 2018, 63, 256–262. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, S.; Cheng, Z.; Cooke, J.S.; Werling, D.; Wathes, D.C.; Pollott, G.E. Polymorphisms in the selectin gene cluster are associated with fertility and survival time in a population of Holstein Friesian cows. PLoS ONE 2017, 1812, 0175555. [Google Scholar] [CrossRef]
- Somasundaram, R.K.; Gupta, I.K.; Raja, N.; Periasamy, K.; Ramasamy, S. Polymorphism of Bovine Forebrain Embryonic Zinc Finger Like (FEZL) gene associated with resistance to mastitis in Indian cattle. Inter. J. Livest. Res. 2020, 10, 144–149. [Google Scholar] [CrossRef]
- Groeneveld, L.F.; Lenstra, A.J.; Eding, H.; Toro, A.M.; Scherf, B.; Pilling, D.; Negrini, R.; Finlay, K.E.; Jianlin, H.; Groeneveld, E.; et al. Genetic diversity in farm animals e a review. Anim. Genet. 2010, 41, 6–31. [Google Scholar] [CrossRef]
- Gautier, M.; Faraut, T.; Moazami-Goudarzi, K.; Navratil, V.; Foglio, M.; Grohs, C.; Boland, A.; Garnier, J.G.; Boichard, D.; Lathrop, G.M.; et al. Genetic and haplotypic structure in 14 European and African cattle breeds. Genetics. 2007, 177, 1059–1070. [Google Scholar] [CrossRef]
- McKay, D.S.; Schnabel, D.R.; Murdoch, M.B.; Matukumalli, K.L.; Aerts, J.; Coppieters, W.; Crews, D.; Neto, D.E.; Gil, A.C.; Gao, C.; et al. An assessment of population structure in eight breeds of cattle using a whole genome SNP panel. BMC Genet. 2008, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Socol, C.T.; Iacob, L.; Mihalca, I.; Criste, F.L. Molecular and population genetics tools for farm animal genetic resources conservation: Brief overview. J. Anim. Sci. Biotechnol. 2015, 48, 95–102. [Google Scholar]
- Svensson, E.M.; Anderung, C.; Baubliene, J.; Persson, P.; Malmström, H.; Smith, C.; Vretemark, M.; Daugnora, L.; Götherström, A. Tracing genetic change over time using nuclear SNPs in ancient and modern cattle. Anim. Genet. 2007, 38, 378–383. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.C.; Nap, J.P. Genetical genomics: Te added value from segregation. Trends Genet. 2001, 17, 388–391. [Google Scholar] [CrossRef] [PubMed]
- Fairfax, B.P.; Knight, J.C. Genetics of gene expression in immunity to infection. Curr. Opin. Immunol. 2014, 30, 63–71. [Google Scholar] [CrossRef]
- Cloney, R. Complex traits: Integrating gene variation and expression to understand complex traits. Nat. Rev. Genet. 2016, 17, 194. [Google Scholar] [CrossRef]
- Asadpour, R.; Zangiband, P.; Nofouzi, K.; Saberivand, A. Differential expression of antioxidant genes during clinical mastitis of cow caused by Staphylococcus aureus and Escherichia coli. Vet. Arh. 2021, 91, 451–458. [Google Scholar] [CrossRef]
- Darwish, A.; Ebissy, E.; Ateya, A.; El-Sayed, A. Single nucleotide polymorphisms, gene expression and serum profile of immune and antioxidant markers associated with postpartum disorders susceptibility in Barki sheep. Anim. Biotech. 2021, 1–13. [Google Scholar] [CrossRef]
- Bonnefont, C.M.D.; Toufeer, M.; Caubet, C.; Foulon, E.; Tasca, C.; Aurel, M.R.; Bergonier, D.; Boullier, S.; Robert-Granié, C.; Foucras, G.; et al. Transcriptomic analysis of milk somatic cells in mastitis resistant and susceptible sheep upon challenge with Staphylococcus epidermidis and Staphylococcus aureus. BMC Genom. 2011, 12, 208. [Google Scholar] [CrossRef]
- Brand, B.; Hartmann, A.; Repsilber, D.; Griesbeck-Zilch, B.; Wellnitz, O.; Kühn, C.; Ponsuksili, S.; Meyer, H.H.D.; Schwerin, M. Comparative expression profiling of E. coli and S. aureus inoculated primary mammary gland cells sampled from cows with different genetic predispositions for somatic cell score. Genet. Sel. Evol. 2011, 43, 24. [Google Scholar] [CrossRef][Green Version]
- Lewandowska-Sabat, A.M.; Boman, G.M.; Downing, A.; Talbot, R.; Storset, A.K.; Olsaker, I. The early phase transcriptome of bovine monocyte-derived macrophages infected with Staphylococcus aureus in vitro. BMC Genom. 2013, 14, 891. [Google Scholar] [CrossRef] [PubMed]
- Boutet, P.; Sulon, J.; Closset, R.; Detilleux, J.; Beckers, J.F.; Bureau, F.; Lekeux, P. Prolactin-induced activation of nuclear factor kappaB in bovine mammary epithelial cells: Role in chronic mastitis. J. Dairy Sci. 2007, 90, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Sun, J.; Rong, W.; Zhao, T.; Li, D.-H.; Ding, X.; Wu, L.-Y.; Wu, K.; Schachner, M.; Xiao, Z.-C.; et al. Loss of cell adhesion molecule CHL1 improves homeostatic adaptation and survival in hypoxic stress. Cell Death Dis. 2013, 4, e768. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.R.; Ning, L.; Zhou, F.H.; Sun, Q.; Meng, H.P.; Han, Z.; Liu, Y.; Huang, W.; Liu, S.; Li, X.H.; et al. Downregulation of Adhesion Molecule CHL1 in B Cells but Not T Cells of Patients with Major Depression and in the Brain of Mice with Chronic Stress. Neurotox. Res. 2020, 38, 914–928. [Google Scholar] [CrossRef]
- Fukuda, H.; Nakamura, N.; Hirose, S. MARCH-III is a novel component of endosomes with properties similar to those of MARCH-II. J. Biochem. 2006, 139, 137–145. [Google Scholar] [CrossRef]
- Labbe, C.; Goyette, P.; Lefebvre, C.; Stevens, C.; Green, T.; Tello-Ruiz, M.K.; Cao, Z.; Landry, A.L.; Stempak, J.; Annese, V.; et al. MAST3: A novel IBD risk factor that modulates TLR4 signaling. Genes Immun. 2008, 9, 602–612. [Google Scholar] [CrossRef]
- Uutela, M.; Wirzenius, M.; Paavonen, K.; Rajantie, I.; He, Y.; Karpanen, T.; Lohela, M.; Wiig, H.; Salven, P.; Pajusola, K.; et al. PDGF-D induces macrophage recruitment, increased interstitial pressure, and blood vessel maturation during angiogenesis. Blood. 2004, 104, 3198–3204. [Google Scholar] [CrossRef]
- Seiler, C.; Gebhart, N.; Zhang, Y.; Shinton, S.A.; Li, Y.-S.; Ross, N.L.; Liu, X.; Li, Q.; Bilbee, A.N.; Varshney, G.K.; et al. Mutagenesis screen identifies agtpbp1 and eps15L1 as essential for T lymphocyte development in Zebrafish. PLoS ONE 2015, 10, e0131908. [Google Scholar] [CrossRef]
- Rainard, P. The complement in milk and defense of the bovine mammary gland against infections. Vet. Res. 2003, 34, 647–670. [Google Scholar] [CrossRef]
- Wang, X.; Zhong, J.; Gao, Y.; Ju, Z.; Huang, J. A SNP in intron 8 of CD46 causes a novel transcript associated with mastitis in Holsteins. BMC Genom. 2014, 15, 1. [Google Scholar] [CrossRef]
- Bourens, M.; Barrientos, A. Human mitochondrial cytochrome c oxidase assembly factor COX18 acts transiently as a membrane insertase within the subunit 2 maturation module. J. Biol. Chem. 2017, 292, 7774–7783. [Google Scholar] [CrossRef] [PubMed]
- Iso-Touru, T.; Sahana, G.; Guldbrandtsen, B.; Lund, M.S.; Vilkki, J. Genome-wide association analysis of milk yield traits in Nordic Red Cattle using imputed whole genome sequence variants. BMC Genet. 2016, 17, 55. [Google Scholar] [CrossRef] [PubMed]
- Nath, P.R.; Isakov, N. Insights into peptidyl-prolyl cis-trans isomerase structure and function in immunocytes. Immunol. Lett. 2015, 163, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Cruciani, L.; Romero, R.; Vaisbuch, E.; Kusanovic, J.P.; Chaiworapongsa, T.; Mazaki-Tovi, S.; Mittal, P.; Ogge, G.; Gotsch, F.; Erez, O.; et al. Pentraxin 3 in amniotic fluid: A novel association with intra-amniotic infection and inflammation. J. Perinat. Med. 2010, 38, 161–171. [Google Scholar] [CrossRef]
- Lutzow, Y.C.S.; Donaldson, L.; Gray, C.P.; Vuocolo, T.; Pearson, R.D.; Reverter, A.; A Byrne, K.; A Sheehy, P.; Windon, R.; Tellam, R.L. Identification of immune genes and proteins involved in the response of bovine mammary tissue to Staphylococcus aureus infection. BMC Vet. Res. 2008, 4, 18. [Google Scholar] [CrossRef]
- Brenaut, P.; Lefèvre, L.; Rau, A.; Laloë, D.; Pisoni, G.; Moroni, P.; Bevilacqua, C.; Martin, P. Contribution of mammary epithelial cells to the immune response during early stages of a bacterial infection to Staphylococcus aureus. Vet. Res. 2014, 45, 16. [Google Scholar] [CrossRef]
- Garlanda, C.; Jaillon, S.; Doni, A.; Bottazzi, B.; Mantovani, A. PTX3, a humoral pattern recognition molecule at the interface between microbe and matrix recognition. Curr. Opin. Immunol. 2016, 38, 39–44. [Google Scholar] [CrossRef]
- Burvenich, C.; Van Merris, V.; Mehrzad, J.; Diez-Fraile, A.; Duchateau, L. Severity of E. coli Mastitis Is Mainly Determined by Cow Factors. Vet. Res. 2003, 34, 521–564. [Google Scholar] [CrossRef]
- Schukken, Y.H.; Günther, J.; Fitzpatrick, J.; Fontaine, M.; Goetze, L.; Holst, O.; Leigh, J.; Petzl, W.; Schuberth, H.-J.; Sipka, A.; et al. Host-Response Patterns of Intramammary Infections in Dairy Cows. Vet. Immunol. Immunopathol. 2011, 144, 270–289. [Google Scholar] [CrossRef]
- Miki, I.; Kusano, A.; Ohta, S.; Hanai, N.; Otoshi, M.; Masaki, S.; Sato, S.; Ohmori, K. Histamine Enhanced the TNF-Alpha-Induced Expression of E-Selectin and ICAM-1 on Vascular Endothelial Cells. Cell. Immunol. 1996, 171, 285–288. [Google Scholar] [CrossRef]
- Albrecht, E.A.; Chinnaiyan, A.M.; Varambally, S.; Kumar-Sinha, C.; Barrette, T.R.; Sarma, J.V.; Ward, P.A. C5a-Induced Gene Expression in Human Umbilical Vein Endothelial Cells. Am. J. Pathol. 2004, 164, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Paape, M.; Mehrzad, J.; Zhao, X.; Detilleux, J.; Burvenich, C. Defense of the Bovine Mammary Gland by Polymorphonuclear Neutrophil Leukocytes. J. Mammary Gland. Biol. Neoplasia 2002, 7, 109–121. [Google Scholar] [CrossRef] [PubMed]
| Gene | Forward | Reverse | Annealing Temperature (°C) | Length of PCR Product (bp) | Reference |
|---|---|---|---|---|---|
| RASGRP1 | 5′- ACTCATGGCTGCAGAGCAGTC-3′ | 5′- TCCATGGTGCTTGCCAGGCTG-3′ | 62 | 410 | Current study |
| NFKB | 5′- TCAGGTCAAACTCCAGAATGGC-3′ | 5′- GCCATTCTGGAGTTTGACCTGA-3′ | 58 | 396 | Current study |
| CHL1 | 5′- CGTGCAGATCGGCTGGGAGCT-3′ | 5′- ATTGTTAGATACAATTATCCGA-3′ | 60 | 547 | Current study |
| MARCHF3 | 5′- CTCTACGCGGCTGTCCGCCTC-3′ | 5′- GGTCCGCACTACTGTTGACAG-3′ | 64 | 455 | Current study |
| PDGFD | 5′- GCCAGCGAGTGCGGGCGCGCGT-3′ | 5′- GTAGTCATCAGACTTGAACGT-3′ | 62 | 531 | Current study |
| MAST3 | 5′- TCCTGTTACCGCTCCTTACCCA-3′ | 5′- CTTGTGGTTCAACAGAGAGTGAG-3′ | 62 | 650 | Current study |
| EPS15L1 | 5′- TCCATTATATGAGTCTTACTA-3′ | 5′- AACCATTGACAGGTAAGAGGCT-3′ | 58 | 383 | Current study |
| C1QTNF3 | 5′- CGAGGAGACCACGGCGGCCAGA-3′ | 5′- CCGGTCATGACATCAAAGAAGT-3′ | 60 | 526 | Current study |
| CD46 | 5′- CCGCTGAAGGCGCCGCTCCGC-3′ | 5′- ACAGCCCTCCTGGAGAGACGAC-3′ | 62 | 288 | Current study |
| COX18 | 5′- TGCGAGCGCGCGTGGTCTGTGA-3′ | 5′- TAGGTAAGTGAGCCTGGCAAC-3′ | 64 | 511 | Current study |
| NEURL1 | 5′- GTAACAACTTCTCCAGTATTC-3′ | 5′- CTCCTCAGGCAGCGCCTTGGCC-3′ | 58 | 476 | Current study |
| PPIE | 5′- GCAAGAGCAAGATGGCCACTAC-3′ | 5′- ACTTCTTCAACCAGTCATCATC-3′ | 60 | 324 | Current study |
| PTX3 | 5′- TCCAGCAATGCATATCTCTGTGA-3′ | 5′- TCATTGGTGTCACCGGATGCAC-3′ | 62 | 617 | Current study |
| Gene | Primer | Product Length (bp) | Annealing Temperature (°C) | Accession Number | Source |
|---|---|---|---|---|---|
| RASGRP1 | F5′-GAGAAGCTCCACGGAAACCA-3′ R5′-CAGAGGCACCATCATTCGGA-3′ | 137 | 60 | NM_001144078.1 | Current study |
| NFKB | F5-CAGATGGGCTACACTGAGGC-3′ R5′-TGCGGAAGGAGGTCTCTACA-3′ | 184 | 60 | NM_001076409.1 | Current study |
| CHL1 | F5′-CGGTTTCCTCGAAGGAAGGT-3′ R5′-GAAGGAGGCAGCCCAGAAAG-3′ | 172 | 59 | NM_001205541.3 | Current study |
| MARCHF3 | F5′-TGGAGACATGGTGTGCTTCC-3′ R5′-TCGAGCCGACTGCTAAAGTG-3′ | 105 | 58 | NM_001077941.1 | Current study |
| PDGFD | F5′-GGCTCTCGTTGACATCCAGT-3′ R5′-GTAAGTTCGGTTGCTGGTGG-3′ | 167 | 62 | NM_001083706.1 | Current study |
| MAST3 | F5′-CCTTACCCAGACTGGAGTGTC-3′ R5-CAGCCTCCTGCAGCAAATG-3′ | 211 | 60 | XM_024994781.1 | Current study |
| EPS15L1 | F5′-GAGTTCTCTGCCTTCCGTGC-3′ R5′-GGTGATGGTGTGAGGTTCCG-3′ | 144 | 59 | XM_024993963.1 | Current study |
| C1QTNF3 | F5′-ATAGAGCTCTGTTGACTGGCCG-3′ R5′-ACTCCATGCCAGTGTGTGTAA-3′ | 119 | 59 | NM_001101138.1 | Current study |
| CD46 | F5′-AGTTAGTGGCACACACTGGG-3′ R5′-CCACGTGCCTTACCCAAGAT-3′ | 161 | 60 | NM_001242563.2 | Current study |
| COX18 | F5′-ATGCGGAGGCTTGTTTCAGA-3′ R5′-CGGAGAGCGACAGACATGAA-3′ | 113 | 60 | NM_001082437.2 | Current study |
| NEURL1 | F5′-GGTAACAACTTCTCCAGTATTCCCA-3′ R5′-TTGTGGTGGCATCGGTGAGA-3′ | 131 | 58 | NM_001192253.3 | Current study |
| PPIE | F5-CTGACGTGTGACAAGACGGA-3′ R5′-TCCCCACAGTCGGAGATGAT-3′ | 149 | 59 | NM_001098161.1 | Current study |
| PTX3 | F5-GAACGTCGTCTCTCCAGCAA-3′ R5′-TGTCCCACTCGGAGTTCTCA-3′ | 191 | 60 | NM_001076259.2 | Current study |
| ß. actin | F5′-GCTCAGAGCAAGAGAGGCAT-3′ R5′-CACACGGAGCTCGTTGTAGA-3′ | 117 | 60 | AF191490.1 | Current study |
| Gene | SNPs | Healthy n = 90 | Mastitic n = 90 | Total | Type of Mutation | Amino Acid Number and Type | Chi Value | p Value | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Holstein n = 45 | Montbéliarde n = 45 | Holstein n = 45 | Montbéliarde n = 45 | |||||||
| RASGRP1 | C99T | 24 | - | - | - | 24/180 | Synonymous | 33 F | 20.02 | <0.0001 |
| T276C | - | 31 | - | - | 31/180 | 92 V | 25.86 | |||
| NFKB | C213A | - | - | 34 | - | 34/180 | Synonymous | 71 P | 28.36 | |
| CHL1 | A117T | - | 36 | - | - | 36/180 | Synonymous | 39 G | 30.03 | |
| MARCHF3 | C86T | - | 23 | - | - | 23/180 | Non-synonymous | 29 S to L | 19.19 | |
| T116G | - | 29 | - | - | 29/180 | 39 F to C | 24.19 | |||
| G216A | - | 22 | 15 | 37/180 | Synonymous | 72 S | 30.86 | |||
| PDGFD | G94A | - | 13 | 13- | 13/180 | Non-synonymous | 32 V to I | 10.84 | ||
| A140G | -39 | - | - | 39/180 | 47 D to G | 32.53 | ||||
| G232C | 29- | - | 29/180 | 78 E to Q | 24.19 | |||||
| C303A | 19 33 | - | 52/180 | Synonymous | 101 T | 43.38 | ||||
| MAST3 | T47C | -26 | - | 26/180 | Non-synonymous Non-synonymous | 16 V to A | 21.69 | |||
| A155G | - | 37 | - | 37/180 | 52 D to G | 30.86 | ||||
| A384C | 28- | - | 28/180 | Synonymous | 128 I | 23.36 | ||||
| EPS15L1 | C34T | 23 | 11 | - | 34/180 | Non-synonymous | 12 P to S | 28.36 | ||
| T79C | - | 19- | 19/180 | 27 S to P | 15.85 | |||||
| A202G | - | 26 | - | 26/180 | 68 T to A | 21.69 | ||||
| A250G | - | 31- | 31/180 | 84 T to A | 25.86 | |||||
| T280G | - | 23 | - | 23/180 | 94 S to A | 19.18 | ||||
| C1QTNF3 | G41A | - | -21 | 21/180 | Non-synonymous | 14 R to Q | 17.52 | |||
| CD46 | C27T | 17 | - | - | 17/180 | Synonymous | 9 P | 14.18 | ||
| G217A | - | 28- | 28/180 | Non-synonymous | 73 V to I | 23.36 | ||||
| C243T | 34 | 29 | - | 63/180 | Synonymous | 81 L | 52.55 | |||
| G131T | - | 36- | 36/180 | 44 R to L | 30.03 | |||||
| G272A | - | 18 | - | 18/180 | 91 R to H | 15.02 | ||||
| T339C | - | 33 | - | 33/180 | 113 G | 27.53 | ||||
| A466C | - | 27- | 27/180 | 156 T to P | 22.52 | |||||
| NEURL1 | G56A | - | 21- | 21/180 | Non-synonymous | 19 R to H | 17.51 | |||
| C108A | - | 19 | - | 19/180 | Synonymous | 36 S | 15.85 | |||
| C269T | 28 | - | - | 28/180 | Non-synonymous | 90 T to M | 23.36 | |||
| PPIE | T80C | - | 31- | 31/180 | Non-synonymous | 27 M to T | 25.86 | |||
| T134C | - | 29- | 29/180 | Non-synonymous | 45 M to T | 24.19 | ||||
| T287C | - | 15 | - | 15/180 | Non-synonymous | 96 L to P | 12.51 | |||
| PTX3 | A106G | - | 37 | - | 37/180 | Non-synonymous | 36 n to D | 30.86 | ||
| C189T | - | 21 | 21/180 | Synonymous | 63 H | 17.51 | ||||
| C364G | - | 18 | 18/180 | Non-synonymous | 122 P to A | 15.01 | ||||
| C488A | 32 | - | 32/180 | Non-synonymous | 163 A to E | 26.69 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Essa, B.; Al-Sharif, M.; Abdo, M.; Fericean, L.; Ateya, A. New Insights on Nucleotide Sequence Variants and mRNA Levels of Candidate Genes Assessing Resistance/Susceptibility to Mastitis in Holstein and Montbéliarde Dairy Cows. Vet. Sci. 2023, 10, 35. https://doi.org/10.3390/vetsci10010035
Essa B, Al-Sharif M, Abdo M, Fericean L, Ateya A. New Insights on Nucleotide Sequence Variants and mRNA Levels of Candidate Genes Assessing Resistance/Susceptibility to Mastitis in Holstein and Montbéliarde Dairy Cows. Veterinary Sciences. 2023; 10(1):35. https://doi.org/10.3390/vetsci10010035
Chicago/Turabian StyleEssa, Bothaina, Mona Al-Sharif, Mohamed Abdo, Liana Fericean, and Ahmed Ateya. 2023. "New Insights on Nucleotide Sequence Variants and mRNA Levels of Candidate Genes Assessing Resistance/Susceptibility to Mastitis in Holstein and Montbéliarde Dairy Cows" Veterinary Sciences 10, no. 1: 35. https://doi.org/10.3390/vetsci10010035
APA StyleEssa, B., Al-Sharif, M., Abdo, M., Fericean, L., & Ateya, A. (2023). New Insights on Nucleotide Sequence Variants and mRNA Levels of Candidate Genes Assessing Resistance/Susceptibility to Mastitis in Holstein and Montbéliarde Dairy Cows. Veterinary Sciences, 10(1), 35. https://doi.org/10.3390/vetsci10010035







