Abstract
Technologies, such as Wireless Sensor Networks (WSN) and Internet of Things (IoT), have captured the imagination of researchers, businesses, and general public, due to breakthroughs in embedded system development, sensing technologies, and ubiquitous connectivity in recent years. That resulted in the emergence of an enormous, difficult-to-navigate body of work related to WSN and IoT. In an ongoing research effort to highlight trends and developments in these technologies and to see whether they are actually deployed rather than subjects of theoretical research with presumed potential use cases, we gathered and codified a dataset of scientific publications from a five-year period from 2013 to 2017 involving actual sensor network deployments, which will serve as a basis for future in-depth analysis of the field. In the first iteration, 15,010 potentially relevant articles were identified in SCOPUS and Web of Science databases; after two iterations, 3059 actual sensor network deployments were extracted from those articles and classified in a consistent way according to different categories, such as type of nodes, field of application, communication types, etc. We publish the resulting dataset with the intent that its further analysis may identify prospective research fields and future trends in WSN and IoT.
Dataset License: CC0
1. Summary
As we are heading into the 21st century, digitalization trends in transportation [1,2], in-house logistics [3], education [4,5], agriculture [6], banking [7,8], and other fields are providing new and engaging ways for technology to improve our daily lives. This naturally leads to the emergence of applications of Wireless Sensor Networks (WSN) and Internet of Things (IoT) in large number of different domains. The WSN and IoT popularity is growing rapidly and, according to Grand View Research, the Narrow Band IoT (NB-IoT) market size will reach more than $6 billion by 2025 [9]. Yet, the majority of researchers still use simulation tools to validate their theories [10] rather than deploy actual devices; as a consequence, it is unclear to what extent the vast majority of the available WSN/IoT devices are actually used instead of theorized as being applicable and what design choices drive the selection of devices.
The aim of this work was to provide a comprehensive high level mapping of actual WSN and IoT deployments used by the research community to serve as a foundation for future in-depth analysis of related trends from the five-year period from 2013 to 2017. The presented dataset can be further used for various statistical and contextual analysis, as well as further extended to cover a broader time frame. As the complete marked data set is available, together with intermediate collection results, the authenticity of the data can be verified.
Altogether, 15,010 data articles were identified as potential candidates, from which after two iterations of screening 3059 actual sensor network deployments were extracted and codified according to multiple categories, as described in the next sections.
The data acquisition, analysis, and validation took around two years for a team of 12 volunteer researchers, of which eight provided significant value.
2. Data Description
The dataset contains data files that result from the data acquisition process as shown in Table 1 and described below in detail. The files are in one of three formats:

Table 1.
Data files in the dataset.
- .bib—BibTeX format containing entries representing published articles;
- .json—JSON format containing structured human readible data object entries; and,
- .txt—text files containing TAB delimited tabular data with a header row.
In the subsections below, the technical description of data entries with possible data types and values are described in detail. Verbatim data values in this description will be formatted, like this.
For the eager reader interested in the main resulting dataset, please refer to dataset (J) on page 7.
2.1. (A)—Identified Candidate Articles
This file contains 15,010 BibTeX entries, which have the following types: @article (7137), @book (74), @conference (1861), @incollection (67) and @inproceedings (5871).
Each entry in the file starts on a new line, and can continue over multiple lines. An example entry can be seen in Figure 1. The basic structure of an entry is @type{id,metadata}, where type is name of the document type e.g., article or book, id is a unique string in the document identifying that specific entry and metadata is a list of comma separated key/value pairs describing the entry. Not all entries contain the same metadata entries, but most have the following: abstract, author, doi, title, and year. Additionally, depending on the entry type, additional metadata, like page, volume, or url, could be present.
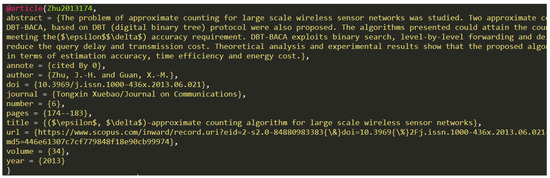
Figure 1.
Data entry example in dataset (A) and (B).
2.2. (B)—Screened Candidate Articles
This file contains 4915 BibTeX entries of the same format, as described in previous section, thus the related entry format is also shown in Figure 1. These entries represent candidate articles left from the (A) dataset after first step of screening and the file contains the following entry types: @article (2385, 33% left ater screening), @book (12, 16% left), @conference (569, 31% left), @incollection (12, 18% left), and @inproceedings (1937, 33% left).
2.3. (C)—Screening Statistics
This file is formatted as a table in a TAB delimited text file. It has 12 entries, each pertaining to one of the 12 volunteer researchers involved in the screening process.
Each row contains and entry (see Figure 2 for entry examples) that has the following three headers/columns with corresponding data types:
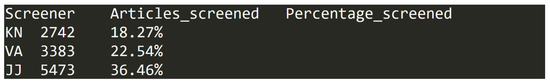
Figure 2.
Data entry examples in dataset (C).
- Screener—two letter code uniquely identifying each of the researchers. Example of data in column: KN;
- Articles_screened—number of articles processed by the corresponding researcher in the screening step. This is an integer value in range from 0 to 5473;
- Percentage_screened—the percentage of the total number of articles in dataset (A) that were processed by the researcher in the screening step. This number is formed as percentage value rounded to two decimal places and has values from 0.00% to 36.46%.
2.4. (D)—Screening Timeline
The file is formatted as a table in a TAB delimited text file. It has 17 entries, which each represent one of the 17 weeks, during which the screening process took place (for example entries, see Figure 3).
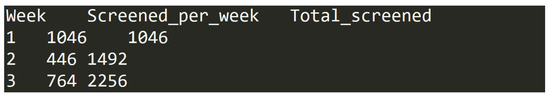
Figure 3.
Data entry examples in dataset (D).
Each row has the following three headers/columns with corresponding data types:
- Week—number of the week in screening process, represented by an integer value in range from 1 to 17;
- Screened_per_week—number of articles processed during the specific screening week by all researchers involved. This is an integer value in range from 50 to 2068;
- Total_screened—cumulative number of articles processed up to and including the specific screening week by all researchers involved. This is an integer value in range from 1046 to 15,010.
2.5. (E)—Eligibility Statistics
This file is formatted as a table in a TAB delimited text file. It has 12 entries, where each pertain to one of the 12 volunteer researchers involved in the eligibility checking process. Figure 4 shows example entries.
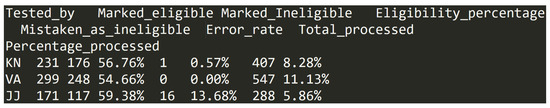
Figure 4.
Data entry examples in dataset (E).
Each row has the following eight headers/columns with corresponding data types:
- Tested_by—two letter code uniquely identifying each of the researchers. Example of data in column: KN;
- Marked_eligible—number of articles processed and marked as eligible by the corresponding researcher in the eligibility checking step. This is an integer value in range from 1 to 708;
- Marked_Ineligible—number of articles processed and marked as ineligible by the corresponding researcher in the eligibility checking step. This is an integer value in range from 0 to 475;
- Eligibility_percentage—the percentage of the total number of articles checked by the researcher in eligibility checking step that were marked as eligible. This number is formed as percentage value rounded to 2 decimal places and has values from 54.66% to 100.00%.
- Mistaken_as_ineligible—number of articles mistakenly marked as ineligible by the corresponding researcher in the eligibility checking step. This is an integer value in range from 0 to 20;
- Error_rate—the percentage of the total number of articles checked by the researcher in eligibility checking step that were mistakenly marked as ineligible. This number is formed as percentage value rounded to two decimal places and it has values from 0.00% to 33.33%. Additionally, one value is NaN or “not a number” representing value resulting from division by zero;
- Total_processed—number of articles processed by the corresponding researcher in the eligibility checking step. This is an integer value in range from 1 to 1183;
- Percentage_processed—the percentage of the total number of articles in dataset (B) that were processed by the researcher in the eligibility checking step. This number is formed as percentage value rounded to two decimal places and has values from 0.02% to 24.07%.
2.6. (F)—Candidate Articles Marked as Eligible
This file contains a JSON data object with 3017 entries, each representing a single article that is marked as eligible in the eligibility checking step. Figure 5 shows an example entry.
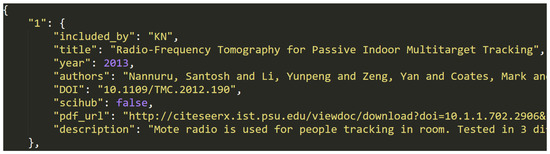
Figure 5.
Data entry example in dataset (F).
The object is structured, as follows: {entry1, entry2, ..., entry3017}. Each of the entries have the following structure: id:{key1:value1, ..., key8:value8}, where id is a unique string identifier of the entry (e.g., "42") and each key/value pair represents one of eight metadata entries from the Table 2 below. In some cases, where a specific metadata value was not available for an entry, the value can also be null.

Table 2.
Metadata format in dataset (F).
2.7. (G)—Candidate Articles Marked as Ineligible
This file contains a list of 1898 JSON data objects, which each represent a single article marked as ineligible in the eligibility checking step.
The list is structured, as follows: [entry1, entry2, ..., entry3017]. Each of the entries have the following structure: {key1:value1, ..., key8:value8}, where each key/value pair represents one of eight metadata entries from Table 3 below. In some cases, where a specific metadata value was not available for an entry, the value can also be null. Figure 6 shows an example entry .

Table 3.
Metadata format in dataset (G).
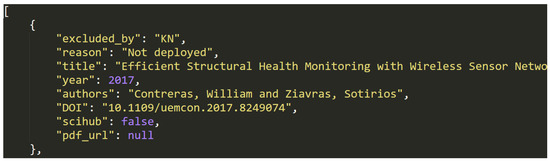
Figure 6.
Data entry example in dataset (G).
Because only articles that describe actual physical deployment of sensor network devices (more than one and networked) were included, several groups of articles were excluded, as ilustrated by the "reason" metadata field, which can take one of the following values (number of matching entries in the dataset in brackets):
- "Article not available" (438 entries)—we were not able to access full text of the article;
- "Theoretical" (160 entries)—the article described theoretical aspects not practical deployment;
- "Not deployed" (293 entries)—no deployment was described even though device might be developed;
- "Article not English" (88 entries)—article not available in English language;
- "Simulation" (485 entries)—experiment was simulated thus not using actual deployment;
- "No network" (183 entries)—non-networked devices (usually data loggers) or a single device deployed;
- "Review" (23 entries)—a review article of other deployment articles, excluded to avoid duplication; and,
- "Other" (166 entries)—some other reason for exclusion—usually not related to sensor networks at all.
2.8. (H)—Candidate Articles Mistaken as Ineligible
This file contains a list of 47 JSON data objects, which each represent a single article marked as ineligible by mistake, even though it was actually eligible, during the eligibility checking step.
The list is structured, as follows: [entry1, entry2, ..., entry47]. Each of the entries have the following structure: {key1:value1, key2:value2, key3:value3}, where each key/value pair represents one of three metadata entries from the Table 4, below. Figure 7 shows an example entry.

Table 4.
Metadata format in dataset (H).
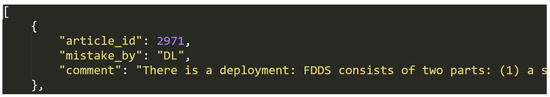
Figure 7.
Data entry example in dataset (H).
2.9. (I)—Timeline of Eligibility Check and Data Extraction Phase
This file is formatted as a table in a TAB delimited text file. It has 35 entries, which each represent one of the 35 weeks during which the eligiblity checking and data extraction phase took place. Figure 8 shows an example entry.
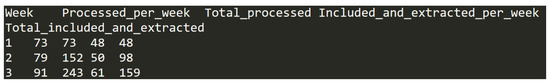
Figure 8.
Data entry example in dataset (I).
Each row has the following five headers/columns with coresponding data types:
- Week—the number of week for which statistics is given. This is an integer value in range from 1 to 35;
- Processed_per_week—the number of articles processed per week in the eligiblity checking and data extraction phase. This is an integer value in range from 56 to 302;
- Total_processed—the cumulative number of articles processed up to and including that week. This is an integer value in range from 73 to 4915;
- Included_and_extracted_per_week—the number of articles included and actually used for data extraction per week. This is an integer value in range from 33 to 186;
- Total_included_and_extracted—the cumulative number of articles included and actually used for data extraction up to and including that week. This is an integer value in the range from 48 to 2970.
2.10. (J)—Extracted Codified Data
This file contains a JSON data object with 3059 entries, which each represent a single deployment from the previously identified articles and containing extracted codified data that are related to this deployment.
The object is structured, as follows: {entry1, entry2, ..., entry3059}. Each of the entries have the following structure: id:{key1:value1, ..., key12:value12} where id is a unique string identifier of the entry (e.g., "42") and each key/value pair represents one of 12 metadata entries from the Table 5 below. In some cases, where a specific metadata value was not available for an entry, the value can also be null. Figure 9 shows an example entry.

Table 5.
Metadata format in dataset (J).

Figure 9.
Data entry example in dataset (J).
In the context of these data, a device is considered to be a “rich” device instead of ordianry sensor network device, if it is an interactive computer like system with some multimedia capabilities, e.g., smartphone, personal computer, Raspberry PI, etc.
In addition to the overall description of the sensor network deployment itself, such as type of connection of sensor nodes and technology readiness level of the deployment, as described in the article, an additional group of metadata was extracted related to the potential future goal network that the reserach is building towards. Although a major part of the deployments is driven by technology development (1234 entries) not application and don’t have such a goal network, for those deployments that have some practical application in mind (1825 entries), the following metadata object is stored under the key "goal_network": {key1:value1, ..., key4:value4}. In this object for each deployment, four keys with these possible values and number of entries in dataset are provided:
- "field"—The target field of application with one of the following values:
- "Health & wellbeing", including patient, frail, and elderly monitoring systems, sports performance, and general health and wellbeing of both body and mind (349 entries);
- "Education" including systems meant for educational purpouses and serious games (10 entries);
- "Entertainment" including computer games, AR/VR systems, broadcasting, sporting and public events, gambling and other entertainment (17 entries);
- "Safety" including anti-theft, security, privacy enhancing, reliability improving, emergency response and military applications and tracking people and objects for these applications (163 entries);
- "Agriculture" including systems related to farming, crop growing, farm and domesticated animal monitoring, precision agriculture (229 entries);
- "Environment" monitoring of environment both in wild life and city, including weather, pollution, wild life, forest fires, aquatic life, volcanic activity, flooding, earthquakes etc. (297 entries);
- "Communications" general communications like power lines, water and gas pipes, energy consumption monitoring, internet, telephony, radio etc. (51 entries);
- "Transport" inlcuding intelligent transport systems (ITS) smart mobility, logistics and goods tracking, smart road infrastructure etc. (123 entries);
- "Infrastructure" general infrastructure, such as tunnels, bridges, dams, ports, smart homes and buildings etc. (413 entries);
- "Industry" anything related to industrial processes, production and business in general like coal mine monitoring, production automation, quality control, process monitoring etc. (143 entries);
- "Research" not related to other fields, but to support future research—better resaerch tools and protocols, testbeds etc. (20 entries); and,
- "Multiple" the deployed network will serve multiple of the previously described fields (10 entries).
- "scale"—The target deployment scale of the sensor network with one of the following values (from smallest to larges):
- "Single actor" including such single entities as a person (e.g., body area network), animal, vehicle, or robot (345 entries);
- "Room" include such relatively small territories as rooms, garages, small yards (131 entries);
- "Building" include larger areas with separate zones, like houses, private gardens, shops, hospitals (530 entries);
- "Property" include even larger zones capable of containing multiple buildings, like city blocks, farms, small private forest or orchard (447 entries);
- "Region" include areas of city or self-government scale like a rural area, forest, lake, river, city or suburbs (317 entries);
- "Country" include objects of scale relative to countries, like national road grid, large agricultural or forest areas, smaller seas (27 entries);
- "Global" include networks of scale not limited to a single country, such as oceans, jungle or space (24 entries); and,
- null—no scale information of target deployment provided or it is not clearly defined (four entries).
- "subject"—The main target subject meant to be monitored by the goal network with one of the following values:
- "Environment" includes all types of environmental phenomena, like weather, forests, bodies of water, habitats, etc. (728 entries);
- "Equipment" includes all types of inanimate objects, including industrial equipment, buildings, vehicles or robots as systems not actors in environment, dams, walls etc. (498 entries);
- "Opposing actor" include all types of actors in environment, which do not want to be monitored, thus including security and spying applications, tracking and monitoring of perpetrators or military opponents, pest control etc. (126 entries);
- "Friendly actor" includes actors that do not mind to be tracked or monitored for some purpose, like domestic or wild animals (tagging), elderly or frail, people in general if compliant (456 entries);
- "SELF" includes cases where the sensor network monitors itself—location of nodes, communication quality etc. (one entry); and,
- "Mixed"—this includes target deployments with multiple subjects from the previously stated values (16 entries).
- "interactivity"—The interactivity of the goal sensor network with the following values:
- "Passive" includes passive monitoring nodes and data gathering for decision making outside the system or for general statistics purposes (1448 entries);
- "Interactive" includes sensor networks providing some kind of feedback, control or interactivity within the loop or confines of the system, like automated irrigation systems, real time alarms etc. (375 entries); and,
- null no specific interactivity of target deployment is provided or clearly defined in the article (two entries).
2.11. (K)—Statistics of Extraction Process
This file is formatted as a table in a TAB delimited text file. It has 12 entries, each pertaining to one of the 12 volunteer researchers that are involved in the data extraction process. Example entries shown in Figure 10.
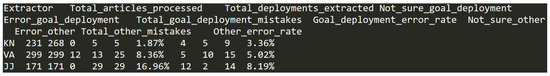
Figure 10.
Data entry examples in dataset (K).
Each row has the following 11 headers/columns with coresponding data types:
- Extractor—two letter code uniquely identifying each of the researchers. Example of data in column: KN;
- Total_articles_processed—number of articles processed by the corresponding researcher in the data extraction step. This is an integer value in range from 1 to 708;
- Total_deployments_extracted—number of actual sensor network deployments extracted from these articles by the corresponding researcher in the data extraction step. This is an integer value in range from 1 to 708;
- Not_sure_goal_deployment—number of articles in which the extractor was not sure about the goal deployment of the sensor network and required peer input to get the final value. This is an integer value in range from 0 to 12;
- Error_goal_deployment—number of articles in which the extractor mistakenly marked a wrong goal deployment value, which was later corrected in validation stage. This is an integer value in range from 0 to 79;
- Total_goal_deployment_mistakes—sum of two previous values representing the total amount of errors related to the goal deployment made by the specific extractor. This is an integer value in range from 0 to 87;
- Goal_deployment_error_rate—the percentage of the total number of deployments processed by the researcher in data extraction stage that contained some sort of error related to goal deployment data extraction. This number is formed as percentage value rounded to 2 decimal places and has values from 0.00% to 30.00%;
- Not_sure_other—number of articles in which the extractor was not sure about the some other metadata value not related to goal deployment and required peer input to get the final value. This is an integer value in range from 0 to 60;
- Error_other—number of articles in which the extractor mistakenly marked a wrong metadata value not related to goal deployment, which was later corrected in validation stage. This is an integer value in range from 0 to 10;
- Total_other_mistakes—sum of two previous values representing the total amount of errors not related to the goal deployment made by the specific extractor. This is an integer value in range from 0 to 66; and,
- Other_error_rate—the percentage of the total number of deployments processed by the researcher in data extraction stage that contained some sort of error not related to goal deployment data extraction. This number is formed as percentage value rounded to two decimal places and has values from 0.00% to 50.00%.
2.12. (L)—Candidate Articles Mistaken as Eligible
This file contains JSON data object with 15 entries, each representing a single article that is marked as eligible by mistake, even though it was actually ineligible, discovered during the data extraction step.
The object is structured, as follows: {entry1, entry2, ..., entry15}. Each of the entries have the following structure: id:{key1:value1, key2:value2, key3:value3}, where id is a unique string identifier of the article and each key/value pair represents one of 3 metadata entries from the Table 6 below. Figure 11 shows an example entry.

Table 6.
Metadata format in dataset (L).
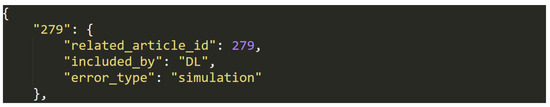
Figure 11.
Data entry example in dataset (L).
2.13. (M)—Overall Timeline of Dataset Creation
This file is formatted as a table in a TAB delimited text file. It has 14 entries, each pertaining to a milestone date in the progress of dataset creation and has no column headers. Figure 12 shows example entries.

Figure 12.
Data entry examples in dataset (M).
Each row has the following 2 columns with coresponding data types:
- Date in format of yyyy-mm-dd with values in the range from 2018-06-12 to 2020-07-02; and,
- Milestone event description in the form of a text string.
2.14. (N)—Readme File
This file contains a short human readable description of the data in this dataset in the form of a Markdown document.
2.15. (O)—Notebook Folder
In this folder, several Jupyter notebook files are stored for easy loading of and access to the data files. These contain example Python 3 code for opening the files and extracting the data within.
3. Methods
To acquire this dataset, the scope of the problem was first defined, as follows: to gather and codify all scientific peer reviewed publications describing original practical sensor network deployments from a five-year period from 2013 to 2017. The scope was narrowed for practical purposes, as follows:
- only publications in English language were considered;
- only publications that could be accessed by the research team without use of additional funds were considered;
- to be considered a network, the deployment had to have at least two actually deployed sensor devices;
- devices did not have to be wireless, to be considered sensor network—also wired, acoustic, or other networks were considered;
- only research doing the deployment themselves was considered—no use of ready datasets from other deployments was included;
- no simulated experiments were included;
- the timeframe was selected as 2013–2017, because the data acquisition was started in the middle of 2018, and only full years were chosen for comparability; and,
- to avoid duplicates only original deployments were included instead of review articles.
Based on this scope, a systematic literature review methodology was devised and followed consisting of the following steps (note that in dataset, the files related to these steps are enumerated starting from 0 not 1):
- Candidate article acquisition
- Screening (exclusion)
- Screening (inclusion/eligibility)
- Codification and data extraction
- Verification
3.1. Candidate Article Acquisition
Due to their popularity and wide access in the institutions represented by the authors, two main indexing databases were selected for querying articles: SCOPUS and Web of Science.
For each of these databases, a query with the same information based on the scope defined above was prepared:
- SCOPUS: KEY (sensor network OR sensor networks) AND TITLE-ABS-KEY (test* OR experiment* OR deploy*) AND NOT TITLE-ABS-KEY (review) AND NOT TITLE-ABS-KEY (simulat*) AND (LIMIT-TO (PUBYEAR,2017) OR LIMIT-TO (PUBYEAR,2016) OR LIMIT-TO (PUBYEAR,2015) OR LIMIT-TO (PUBYEAR,2014) OR LIMIT-TO (PUBYEAR,2013))
- Web of Science: TS = (“sensor network” OR “sensor networks”) AND TS = (test* OR experiment* OR deploy*) NOT TI =“review” NOT TS = simulat* with additional parameters: Indexes = SCI-EXPANDED, SSCI, A&HCI, CPCI-S, CPCI-SSH, BKCI-S, BKCI-SSH, ESCI, CCR-EXPANDED, IC Timespan = 2013–2017
The querying was done on 12 June 2018 and it yielded the following results:
- SCOPUS: 11,536 total articles identified of which 4814 were not found in Web of Science database;
- Web of Science: 10,204 total articles identified of which 3636 were not found in SCOPUS database;
- After checking for duplicates 15,010 unique candidate articles were identified of which 6560 articles were found in both databases. Duplicates were checked both automatically while using features that were provided by Mendeley software and manually by title/author/year combination.
The resulting dataset was saved as BibTeX file (see dataset (A)) and imported in Mendeley software for collaborative screening for exclusion.
3.2. First Screening Iteration-Exclusion
During this stage, the research team was instructed to exclude articles conservatively—only exclude those that definitely match the exclusion citeria and leave all others for a more thorough examination in the next stage.
The exclusion criteria was defined, as follows: The article does not feature a real life deployment of a sensor network or is not in English language.
The team of volunteers participating in the screening process were all provided access to a shared Mendeley group with the dataset of 15,010 articles and given the following instructions:
- spend no more than 10 min on a single article;
- only look at article title and abstract for exclusion;
- when processing an article mark it as “read” (a gray/green circle mark in Mendelay);
- if the article matches exclusion citeria move onto the next article;
- otherwise, include it for the next stage by marking it with favourite/star icon in Mendeley;
- regularly synchronize progress and follow randomized article slots based on alphabetic order of article titles to avoid collisions of multiple reviewers; and,
- in the case of doubt, articles could be tagged for second opinion by another reviewer.
One researcher took lead of it and re-evaluated the first 100 articles processed by all other researchers, and discussed any differences or problems, in order to ensure consistent understanding of the exclusion process. Weekly discussions on progress, problematic articles, etc. were held.
The screening for exclusion took place from 13 June 2018 until 8 October 2018 with the weekly progress that is shown in Figure 13. Subsequently, validation stage started, during which the randomized sample or articles was double checked by other researchers and 142 articles identified as requiring second opinion were discussed and marked appropriately. The validation of screening/exclusion phase ended on 3 November 2018 with 4915 articles left for the next stage (thus 10,095 articles were excluded in this phase).
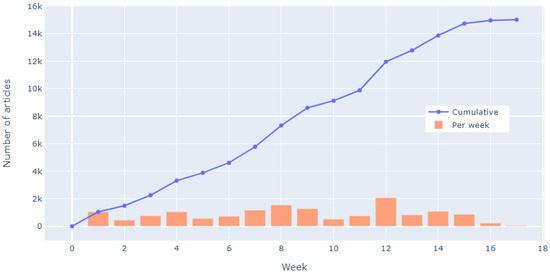
Figure 13.
Weekly progress of first screening phase.
3.3. Second Screening Iteration-Inclusion/Eligibility
After the first stage of screening, the second screening iteration phase started. This phase required opening and reading the full text of the articles, thus, for time conservation, it was done in parallel with the next phase—codification and data extraction (see next section).
First, from 4 November 2018 till 13 Janaury 2019, an instruction for full text eligibility validation was developed together with data codification and data extraction methodology. An online spreadsheet was developed with the 4915 articles from the previous screening phase, with columns for the required data as dropboxes.
Subsequently, from 14 Janaury 2019 till 15 September 2019 data inclusion/eligibility and codified data extraction stage took place—the weekly progress can be seen in Figure 14. The main steps in this stage for all researchers involved were, as follows:
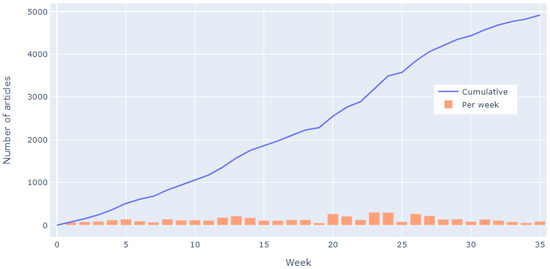
Figure 14.
Weekly progress of second screening phase.
- mark the row corresponding to the selected article with unique identifier of the researcher, so that no one else accidentally takes the same article for analysis;
- locate the full text of the article—if it is not available in English language from any source (indexing pages, preprint publishing pages, author pages, Researchgate, Sci-hub, general google search, etc.), then exclude the article from data extraction, otherwise move to the next step;
- read the article to identify any sensor network deployments in it. If there are no deployments, then the article must be excluded. If there are several deployments, insert new lines in the table, thus describing each deployment separately;
- do not include any articles that should have been excluded in the previous stage (review articles, articles without actual deployments or using old data from previous deployments, or even deployments with single sensor device or multiple devices, which have no sensors or network between them;
- for each included row, leave a comment on which/how many actual sensor network deployments are there—these deployment rows in the spreadhseet table are then filled by the same researcher as part of the next phase (see next Section).
During the second screening stage, 2970 articles were first included and codified. Subsequently, on 17 September 2019 a verification phase of excluded articles was begun, and involved both randomized reviews, as well as multiple reviews of any article marked as uncertain by the original researcher. After this phase ended on 2 December 2019, an additional 47 articles were found in the mistakenly excluded article list and included, thus leading to 3017 total articles eligible for extraction.
From the excluded articles, the reasons for exclusion from most frequent to least frequent were: (1) article describes simulation not actual deployment—485 articles; (2) article full text not available—438 articles; (3) sensor network only described, but not actually deployed—293 articles; (4) only separate sensor devices with no network/local data logging—183 articles; (5) theorethical article with no practical experiments—160 articles; (6) article not available in English—88 articles; (7) article uses existing data gathered from a previous deployment or public data set—62 articles; and, (8) Article is a review article of other deployments—23 articles. Additionally, 166 articles were excluded due to other reasons, that didn’t correspond to one of the above mentioned categories (e.g., nothing to do with sensor devices or disqualified due to multiple categories).
Until 5 Janaury 2020, all of the deployments in these articles were identified and codified and a thoroguh validation phase of codified data was carried out during the process in which 15 articles were identified as mistakenly included for codification leaving only 3002 articles.
The total number of identified sensor network deployments in these articles was 3059.
3.4. Data Codification and Extraction
For all of the 3059 deployments the researchers involved had to extract two codified groups of data:
- details on the actual sensor network deployment described in the article; and,
- if exists—the goal deployment towards which this research is aimed in the future.
The specific codification values are described in detail in the data description of dataset (J), as shown in Section 2.10. In addition to these values, all of the researchers were allowed to provide null value if the article did not mention or describe the specific value of interest and OTHER value if the researcher did not think that the value could fit in any previously defined category. Additionally on every field the researchers could leave comments asking for second opinion or leaving discussion points about the codification system.
As with the exclusion stage, the data extraction stage also contained coordination between the researchers involved—the first 10 codification efforts by each of the researchers were double checked by one researcher, so that everyone had a common understanding. All of the questions and unclear values were discussed weekly for clarifications.
Finally the codified data was verified—all of the comments were manually processed, outlier values, null values, and OTHER values were double checked by other researchers, in order to verify that something was not missed by the original reader of the article. Additionally, random validation of codified entries occured.
The errors during validation were labeled and counted for each of the researchers involved (as can be seen in datasets (E), (H), (K), and (L). The deployments that were checked by researchers who were outliers (with low amount of articles processed or high amount of specific errors) were re-checked by other researchers.
Finally, on 29 May 2020, the dataset was completed and preparation started for publishing the data set. Data set was cleaned up, formatted, and submitted to an open access Git repository on 2 July 2020. Afterwards, the text of this publication was prepared together with Jupyter Notebook examples on use of these data.
3.5. Data Quality
In addition to random validation and checking for errors, as described in the previous steps, additional checks on the data set were done to ensure quality of the data.
First, the number of article candidates from each year were compared to see if there is a bias for specific years (e.g., older articles). Each year the number of articles was around the mean 3002, with deviation of less than 4.5%.
Subsequently, the screening phase results were analyzed to test for bias related to year. In all years, 30% to 35% of articles survived the first screening/exclusion phase, with no observable bias towards any particular year (see Figure 15).
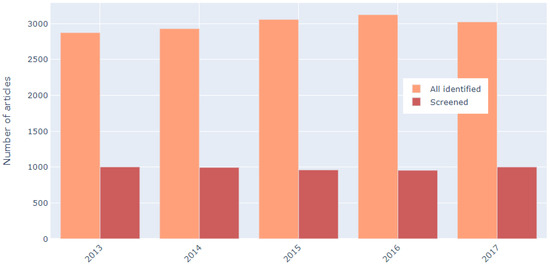
Figure 15.
Number of articles initially identified per year and corresponding number of articles included in first screening phase.
The approximate 1/3 inclusion rate also held true for the three most represented categories of articles: @article with 33.42%, @conference with 30.57% and @inproceedings with 32.99% inclusion rates. The two less represented groups @book and @incollection each had less than 75 instances in the first dataset and, thus, even though their inclusion rate differed from the expected (16.22% and 17.91%, respectively) this is most likely due to the small number of articles in these categories not an inherent bias towards them in the screening process.
Another potential source of bias is the differences in researchers doing the screening, so all of the involved researchers were analyzed. Most had a similar approval ratio of articles (articles marked eligible over all articles processed) and similar low error percentage from total articles processed. As seen in Figure 16, there are three main outliers: RZ who has 100% approval ratio, which is due to the fact that this researcher only processed one article in this stage, SO whose approval ratio is closer to 70% instead of 60%, like others, which is also due to the low number of articles processed (less than 40), and finally RB, who had around 15% error rate in comparison to other researchers who had error rate below 5%. This is also due to the low number of processed articles (7). All other researchers in this phase processed several hundreds of articles and their statistics and error rates were very similar, showing that the efforts to reduce bias that were introduced by individuals were successful.
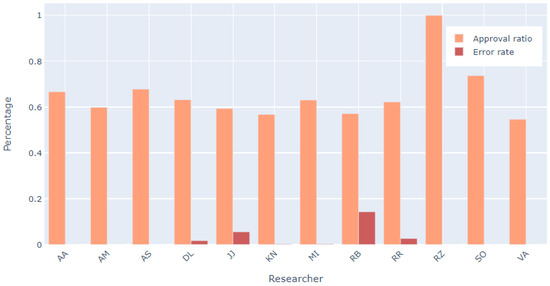
Figure 16.
For each researcher—the ratio of articles marked as eligible and their detected error percentage in eligibility screening phase.
Overall, wherever a potential source for bias was detected due to a low number of articles being processed by a researcher, their work was re-validated by at least one other researcher to guarantee high data quality.
4. User Notes
The data set was primarily meant for easy processing while using programming tools, such as Python/Jupyter Notebooks, thus it is machine readible first.
The data is made freely accessible to everybody, although we would appreciate credit if at all possible. To the best of our knowledge, this is the only data set of its kind and currently only covers years 2013 to 2017.
The data is published as a frozen mirror at https://doi.org/10.5281/zenodo.4048214.
For user convenience live version can be accessed as a Git repository:
In this way, you will get all of the files described in Section 2.
For examples on loading and processing this data using Python, you can access the folder Notebooks where Jupyter notebook files with examples on data exploration are stored.
The data structure and examples promote the easy expandability of the dataset—for example, researchers interested in the impact or citation count of the articles containing identified deployments, can use Python libraries such as scholarly (for Google Scholar), wos (for Web of Science), or pyscopus (for SCOPUS) to automatically acquire this additional information—see example in notebook Explore_extraction_step.ipynb.
Author Contributions
Conceptualization, J.J. and K.N.; methodology, K.N.; software, K.N.; validation, J.J., A.M., V.A. and K.N.; formal analysis, K.N.; investigation, J.J., A.M., V.A., A.S., D.L., R.R., M.I. and K.N.; data curation, K.N.; writing–original draft preparation, J.J.; writing–review and editing, K.N., M.I. and R.R.; visualization, K.N.; supervision, K.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research is co-financed by ERDF funds under the project No. 1.1.1.1/18/A/183 “iTrEMP: Intelligent transport and emergency management platform”.
Acknowledgments
In addition to the authors of this article also these people provided their work to the acquisition and processing of this data: Rihards Balass, Reinholds Zviedris, Armands Ancans and Sandra Ose.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Abbreviations
The following abbreviations are used in this manuscript:
| WSN | Wireless Sensor Network |
| IoT | Internet of Things |
| TAB | Tabulator character |
| JSON | JavaScript Object Notation |
References
- Noussan, M.; Hafner, M.; Tagliapietra, S. Digitalization Trends. In The Future of Transport Between Digitalization and Decarbonization; Springer: Berlin/Heidelberg, Germany, 2020; pp. 51–70. [Google Scholar]
- Noussan, M.; Tagliapietra, S. The effect of digitalization in the energy consumption of passenger transport: An analysis of future scenarios for Europe. J. Clean. Prod. 2020, 258, 120926. [Google Scholar] [CrossRef]
- Winkler, H.; Zinsmeister, L. Trends in digitalization of intralogistics and the critical success factors of its implementation. Braz. J. Oper. Prod. Manag. 2019, 16, 537–549. [Google Scholar] [CrossRef]
- Dorofeeva, A.A.; Nyurenberger, L.B. Trends in digitalization of education and training for industry 4.0 in the Russian Federation. In IOP Conference Series: Materials Science and Engineering; IOP Publishing: Bristol, UK, 2019; Volume 537, p. 042070. [Google Scholar]
- Li, F.; Yang, J.; Wang, J.; Li, S.; Zheng, L. Integration of digitization trends in learning factories. Procedia Manuf. 2019, 31, 343–348. [Google Scholar] [CrossRef]
- Kosareva, O.A.; Eliseev, M.N.; Cheglov, V.P.; Stolyarova, A.N.; Aleksina, S.B. Global trends of digitalization of agriculture as the basis of innovative development of the agro-industrial complex of Russia. Eurasian J. Biosci. 2019, 13, 1675–1681. [Google Scholar]
- Rodin, B.; Ganiev, R.; Orazov, S. «Fintech» in digitalization of banking services. In Proceedings of the International Scientific and Practical Conference on Digital Economy (ISCDE 2019), Chelyabinsk, Russia, 7–8 November 2019; Atlantis Press: Paris, France, 2019. [Google Scholar]
- Evdokimova, Y.; Shinkareva, O.; Bondarenko, A. Digital banks: Development trends. In Proceedings of the 2nd International Scientific Conference on New Industrialization: Global, National, Regional Dimension (SICNI 2018), Ekaterinburg, Russia, 4–5 December 2018; Atlantis Press: Paris, France, 2019. [Google Scholar]
- NB-IoT Market Size Worth $6.02 Billion by 2025. Available online: https://www.bloomberg.com/press-releases/2019-07-23/nb-iot-market-size-worth-6-02-billion-by-2025-cagr-34-9-grand-view-research-inc (accessed on 16 June 2020).
- Lima, L.E.; Kimura, B.Y.L.; Rosset, V. Experimental environments for the internet of things: A review. IEEE Sens. J. 2019, 19, 3203–3211. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).