Abstract
We present CaosDB, a Research Data Management System (RDMS) designed to ensure seamless integration of inhomogeneous data sources and repositories of legacy data in a FAIR way. Its primary purpose is the management of data from biomedical sciences, both from simulations and experiments during the complete research data lifecycle. An RDMS for this domain faces particular challenges: research data arise in huge amounts, from a wide variety of sources, and traverse a highly branched path of further processing. To be accepted by its users, an RDMS must be built around workflows of the scientists and practices and thus support changes in workflow and data structure. Nevertheless, it should encourage and support the development and observation of standards and furthermore facilitate the automation of data acquisition and processing with specialized software. The storage data model of an RDMS must reflect these complexities with appropriate semantics and ontologies while offering simple methods for finding, retrieving, and understanding relevant data. We show how CaosDB responds to these challenges and give an overview of its data model, the CaosDB Server and its easy-to-learn CaosDB Query Language. We briefly discuss the status of the implementation, how we currently use CaosDB, and how we plan to use and extend it.
1. Introduction
Despite the technological advances over the last decades, the scientific community still faces the problem of storing and accessing scientific data in a structured and future-proof manner [1,2,3,4]. Although principles for good scientific data management have since been formulated under the acronym FAIR [5] and are now widely recognized in the community, real-life obstacles tend to prevent their wide-spread adoption. Especially in cross-disciplinary environments, the interaction between different user groups, e.g., numerical scientists conducting simulation studies and experimenters working in the laboratory, often leads to highly inhomogeneous approaches to data management. For such heterogeneous data, inefficiencies become inevitable when different kinds of data have to be combined in a joint research project or when data has to be accessed by scientists who were not involved in the recording and storage procedure. In the worst case, this can lead to data being de facto inaccessible after their creators can no longer be reached.
The ongoing issues are rooted in some ubiquitous properties of scientific environments themselves [6,7].
- Scientists use specific or customized tools, software and data formats with good reason. A research data management system (RDMS) must be built around their workflows and practices and be open for change. It furthermore should assist in integrating data, even if stored in propriertary formats. Enforcing one of the individual systems cannot work in a heterogeneous scientific environment.
- If the system imposes too many restrictions on individual scientists they are likely to be unwilling or even unable to use it. Additionally, the required time and financial cost for a RDMS must be adequate for possibly small research groups [2,7].
- If the RDMS requires too much extra work for learning and understanding, it is likely that the individual scientist will be unable to use it efficiently or just be unwilling to use it at all. This holds in particular for the construction of queries which should retrieve data according to powerful criteria while being simple and intuitive at the same time. For this reason it is unlikely that a scientist without computer science background will be able to use Structured Query Language (SQL) or SPARQL efficiently [8,9,10].
- The system should strongly encourage to develop and use standards for workflows and data models without being overly restrictive. Users of the database can only profit from the system when data is organized sufficiently structured to enable everyone to search and retrieve data easily and to understand the structure of the data intuitively. At the same time the database has to be prepared for constantly evolving data models and standards [2].
- File systems and other types of storage usually organize data into some kind of hierarchy which can be folders or projects. In scientific environments this can raise issues, especially when data belongs to multiple projects or is part of cooperations.
Our open source software CaosDB is a research data management system built on top of robust data management technologies (MySQL, HTTP, XML, file storages) that is specifically suited for scientific research. Our approach is guided by two central principles:
- Simple and intuitive access to complex and changing data models
- Reuse and integration of existing workflows and technologies
2. Results
2.1. Requirements
Based on the considerations described in the previous section, we define the following requirements for a data management system to address the mentioned issues. For reference, we use the abbreviations from the original FAIR publication [5] to link our statements to the four principles:
- Findable (F1–F4)
- Accessible (A1–A2)
- Interoperable (I1–I3)
- Reusable (R1–R1.3)
Architecture. The system must be built in a client/server architecture for separating the high-performance workload on the database and filesystem from the lightweight clients. Create/Read/Update/Delete (CRUD) transactions on the server side must be ACID1 compliant in order to keep the structure consistent at any time. The communication Application Programming Interface (API) must be built around a transparent human-readable protocol with RESTful2 identifiers [11]. This API can then be used by libraries and clients that can be integrated into existing data management workflows. These requirements are needed to comply with A1–A1.2 of the FAIR principles.
Access control, file system. Heterogenous scientific environments require fine-grained access-control on object level (A1.2). In order to seamlessly integrate into existing data acquisition and data analysis workflows the system must be able to incorporate an existing file system with its grown folder structure. Separation between file system storage and (meta)data storage factilitates data management in compliance with A2.
Query language. One of the most important requirements is the query language which has to fulfill several properties that guarantee that heterogenous data in big amounts can be searched and retrieved easily. The logic behind the query language can also have a major impact on the data models used. To spell this out more precisely, the data model (for implementing F1–F3) and the query language (F4) must support:
- Entities with subtyping
- User-defined n-ary relationships and properties
- Integration of files and directories as entities
- Native support for primitive data types which include several numeric data types with their physical units and uncertainties, standard compliant date and time values, booleans, strings, and undefined values
- Compound data types for lists, sets, tuples, and dictionaries
This general nature of the data model enables data management in compliance with I1–I3, R1, R1.2, and R1.3.
Extensibility. The system must be able to adapt to new software and hardware requirements. Furthermore, the system must be flexible enough to adapt to continously changing scientific workflows. The simplest way to ensure this extensibility is to implement a server-side API for extensions and plug-ins.
Although we highlighted, that our requirements are feasible for implementing data management in accordance with the FAIR guiding principles, we acknowledge that data management standards might evolve in the future. Our requirements therefore have a strong focus on extensibility.
2.2. Implementation
CaosDB [12] is our in-house solution for fulfilling these conditions, to our knowledge it is currently the only existing software to satisfy the mentioned requirements. CaosDB is an object-oriented database with a powerful query language based on English natural language and a flexible and adaptive data model. For example, a typical query could look like this:
SELECT flavour, rating, ingredients FROM Experiment WHICH HAS A room_temperature > 26C AND WHICH IS REFERENCED BY ExperimentSeries WHICH HAS A name LIKE *ice cream testing*
It also integrates efficient management of large data files directly into the core functionality to accomodate specific requirements by the scientific users:
- offers a flexible data model
- can be seamlessly integrated into existing workflows
- allows search for values of specific fields (not just full text search), with automatic unit conversion and search for (back)references of linked objects.
2.3. Architecture
The software design follows a server/client architecture. The CaosDB server handles all CRUD requests, implements consistency checks, and translates the requests into SQL commands which are redirected to the MySQL backend. It furthermore provides a transparent layer for interactions with the file system. The server frontend is written entirely in Java and is accessed using a RESTful API over HTTP with XML messages. The frontend also serves a web user interface (WebUI, shown in Figure 1) written in XSLT, HTML, and JavaScript that can be used for browsing data and maintenance operations.

Figure 1.
Screenshot of the WebUI (graphical web user interface) of CaosDB showing a search query that returned a fictitious experiment.
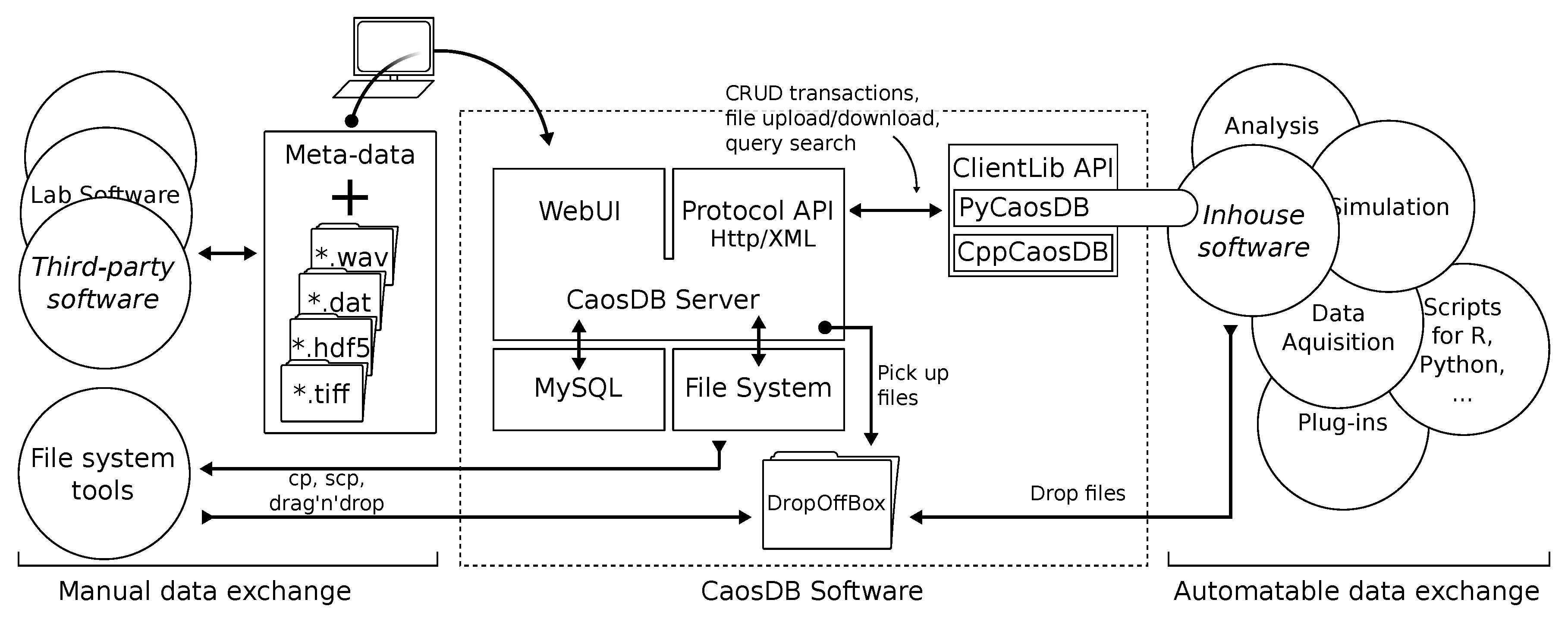
The server is complemented by client libraries for Python and C++ that encapsulate the XML API for usage in scripting, data acquisition (manual and automated via file system crawlers) and data analysis tools. Figure 2 gives a schematic overview of the software architecture.

Figure 2.
An overview of the components of the CaosDB project and its relations to contexts of application. The CaosDB server encapsulates access to the MySQL back end and the local file system. It implements the protocol Application Programming Interface (API) for CRUD transactions (creation, retrieval, update, deletion), file exchange, and query search. The client libraries (ClientLib API) can communicate with the server via the protocol API and provide interfaces in several programming languages for automatable data exchange with data acquisition software and analysis tools. Data files can be imported into CaosDB using the filesystem-based DropOffBox. The WebUI for convenient database access is directly integrated into the core application. These also facilitate the manual data exchange with non-customizable third-party tools and data sources.
2.4. Data Model
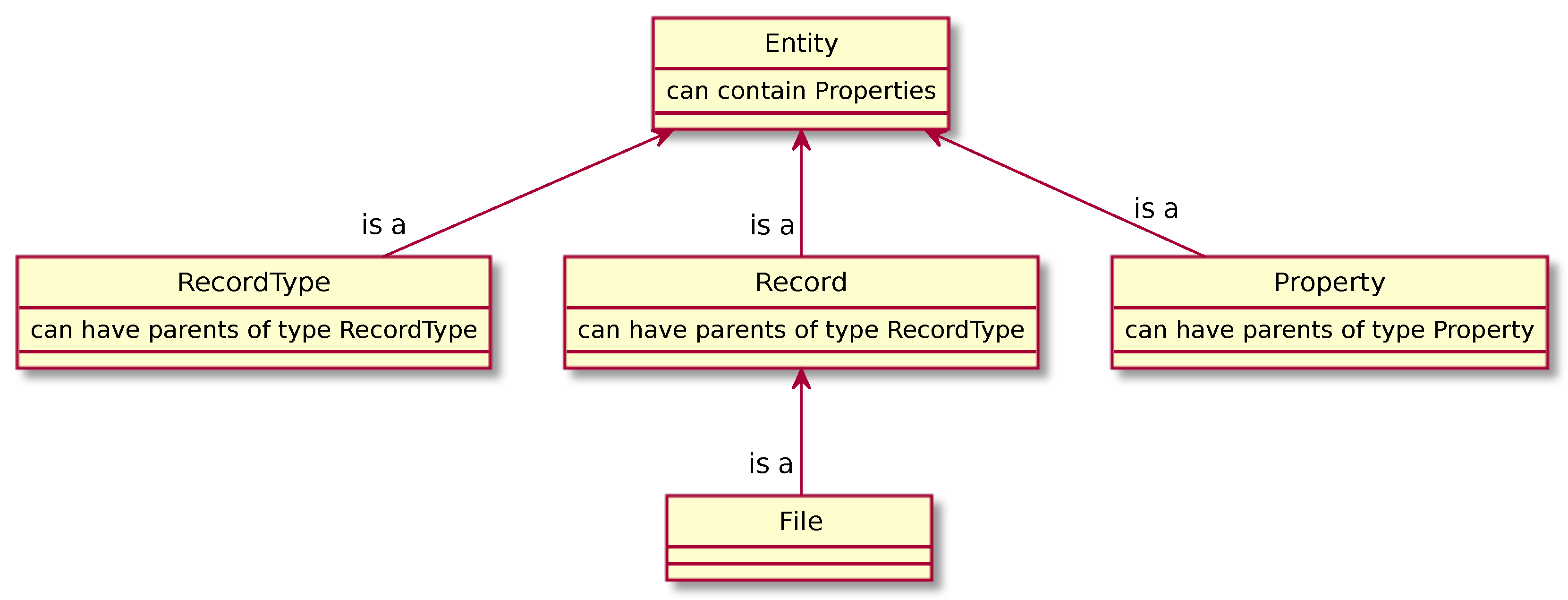
CaosDB has a general purpose object-oriented data model, depicted in Figure 3, which is not tied to any particular scientific field or structure of data. It has a base object called Entity. Entities are either Record Types, Records, or Abstract Properties and every Entity has a unique, server-generated Id.

Figure 3.
An overview over the generic data model in CaosDB.
Record Types and Abstract Properties are used to define the ontology for a particular domain in which the RDMS is used. Records are used to store the actual data and therefore represent individuals or particular things, e.g., a particular experiment, a particular time series, etc. Record Types define classes or types of things, e.g., persons, experiments, time series, etc. Records can be viewed as members of the class defined by its Record Type. These classes can contain Abstract Properties which define key-value relationships for properties of the things along with the expected data type and possibly the default unit, a default value, or a range of permitted values. As files on the back-end file system are a major focus of this database management system, there is a special entity File that encapsulates typical file properties like path, size and checksum. Entities can be related via binary, directed, transitive is-a relations which model both subtyping and instantiation, depending on the relata. These relations construct a directed graph of the Entities. If A is-a B we call A the child of B and B the parent of A. No adamant restrictions are imposed on the relate of the is-a relation and thus, Entities can be children of multiple Entities.
Each Entity has a list of Entity Properties, or in short just Properties. An Entity Property is not an Entity of its own, but a triple of an Abstract Property, a value or Null, and an Importance. The values can be numericals, strings, dates, any other valid value that fits into one of several built-in data types, or, most notably, references to other Entities. The importance is either obligatory, recommended, suggested, or fix. A valid child of an Entity implicitly inherits its parent’s Properties according to their Importance, which means that it is obliged, recommended, or only suggested to have a Property with the same Abstract Property (or any subtype thereof). As opposed to Properties with other priorities, Fixed Properties have no effect on the Entity’s children. During the creation or update of Entities, the importances of the parents are being checked by the Server. Missing obligatory Properties invalidate the transaction and result in an error, by default. Missing Properties, when they are recommended, result in a warning, but the transaction is considered valid. Entities with missing suggested Properties are silently accepted as valid.
This novel approach to ontology standardization is inspired by the operators from deontic logics, the logics of obligation and permission [13]. It is designed to guide the users without restricting them too heavily and ensures that they do not insert their data wrongly by accident. Furthermore, it helps them to find the most relevant or best fitting Properties for their Entity based on the supertype(s).
CaosDB thus facilitates the definition and observation of standards for data storage.
2.5. Query Language
Existing data management technologies already provide very comprensive and expressive query languages. Two prominent examples are SQL and SPARQL. SPARQL is a language for querying RDF triple stores and therefore also suited for complex queries of semantic data models.
However, SPARQL statements for simple requests often result in long and complex statements [8,9,10] which motivated the need for a simpler but similarly expressive query language also suited for scientists without computer science background.
We would like to illustrate this with the following example:
Suppose we would like to retrieve all datasets from experiments that were conducted in 2017 at a room temperature of 293.15 K. This simple request would result in a highly complex SPARQL statement. The filter for the dates alone would read as:
(?date >= xsd:date("2017-01-01") && ?date < xsd:date("2018-01-01"))
The implementation of a temperature filter covering unit conversion would even rely on external unit conversion extensions.In contrast to SPARQL, the CaosDB Query Language (CQL) which we implemented in CaosDB allows for a much simpler expression for the whole request:
Find Experiment with date in 2017 and room temperature=293.15K
CQL is translated into SQL statements by the CaosDB server. These statements are then passed on to the MySQL backend which carries out the actual request.
CQL is designed to express simple questions with simple queries resembling English. Its syntax is illustrated in Figure 4 using EBNF3. The language is case-insensitive, but for clarity some terms are explicitly spelled in upper or mixed case here.

Figure 4.
The first levels of the CaosDB Query Language (CQL) syntax in Extended Backus-Naur Form (EBNF). This is only a schematic overview and does not include the syntactic sugar or white spaces. However, it should be noted that the top level of this syntax is not too complex and has only very few keywords. Yet even simple queries are very powerful, mainly due to the transitivity of the is-a relation. In case of errors, the server responds with a detailed listing of syntactical errors which the clients then use to inform the user.
The first term (query prefix in Figure 4) in a CQL expression is the desired return type of the query:
- A query starting with Count returns a non-negative integer.
- A query starting with Find returns a list of entities.
- A query starting with Select returns a table containing the values of selected Properties.
This is optionally followed by an entity type which restricts the query to specific entities. The most important information searched for is probably the entity name which specifies the actual “thing” searched for. This term makes use of the object-oriented structure of the database and—in addition to searching for all entities having a specific name—also returns subtypes and Records being of that type. In a CQL expression, entity name is followed by a list of filters which are connected by filter separators. Filters can address any possible Property of an Entity and restrict the values to ranges or particular values, use a range of comparison operators, and even search with wildcards or regular expressions. Furthermore, relations between Entities can be expressed precisely. Filters can be combined with logical operators like And, Or, and Not. The query processor is able to interpret and convert physical units. This unique feature simplifies working with scientific data and sets CQL apart from SQL and various modern query languages for RDF(S), OWL or graph data.
We will illustrate the basic concepts by giving some typical examples:
COUNT Experiment with date in 2017will return the number of experiments from 2017. In this query, Experiment is typically the name of a Record Type with a possibly large number of subtypes and instances. All Entities which have the name Experiment or have a parent with this name are filtered for those which have a Property with the name date and a date value in the year 2017. CQL filters can also express the equivalence of complex SQL joins in an easily understandable syntax:
FIND Person which is referenced as an Author by an Article which has aTitle like *terminating ventricular fibrillation*
In this example, Person is a Record Type. Article is another Record Type having an Author and a Title as Properties. The statement would therefore return all Records, if they are a Person, that are assigned as values of an Author Property of a Record of type Article with a specific title. Since the returned objects are themselves Records of Record Type Person, they have Properties, presumably a name, affiliation(s), possibly an ORCiD, an email-address or some other contact information.
Another special feature are Select queries which follow an SQL-like syntax and represent their results as a table, e.g., the result of
SELECT first name, family name from person with date of birth > 2000will appear as an HTML table in the WebUI (downloadable as a tsv table), with three columns—id, first name, and family name. This feature is intended to provide one of the interfaces between CaosDB and existing scientific workflows.
CQL is inspired by SQL and therefore probably feels familiar to users with knowledge of prevalent database management systems. It should be clear from the aforementioned examples that the query language is structured, precise and powerful, but nevertheless resembles English sentences. This makes it easier to learn for users without SQL experience.
2.6. User Management and Access Control
CaosDB provides a fine-grained role-based access control system with access control lists. It is possible to define the permissions for insertion, update, retrieval and deletion of Entities, single Properties, and is-a relations, as well as the access to the transaction log and the user management.
CaosDB has a built-in user database where users can sign up or be registered by administrators. Furthermore, users can login with the credentials of their user accounts from PAM (Pluggable Authentication Modules). Access roles—which are relevant for the authorization—can be assigned to clients based on various criteria including their authentication status, the Unix groups of the user—if PAM is used—and connection details, like IP address and others. This makes it possible to share subsets of the data base with collaborators and even a greater audience of anonymous users.
3. Discussion
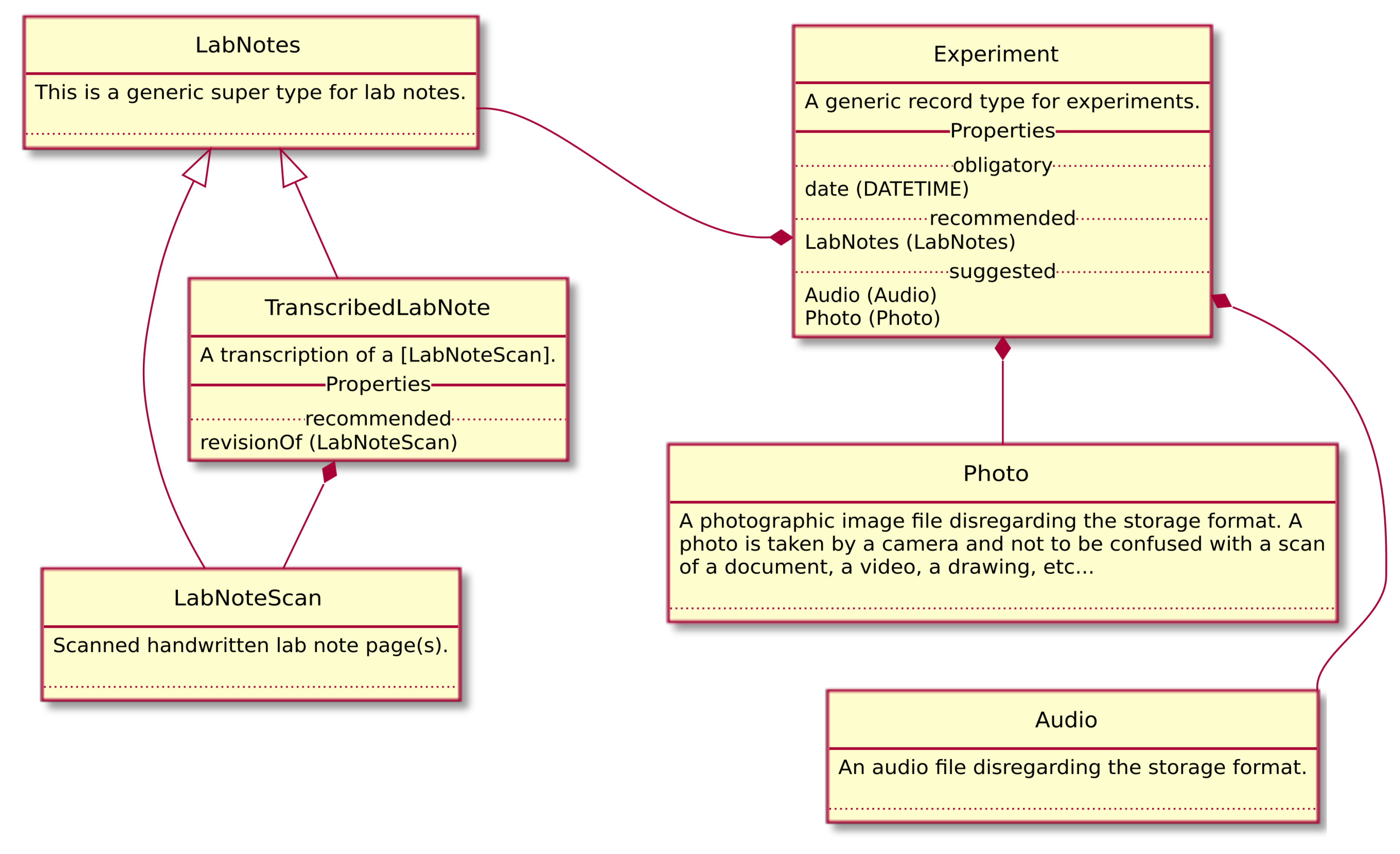
CaosDB is currently in productive use and handles around 40TiB of experimental data from biomedical physics in 250,000 Files along with detailed meta data contained in about 320 Record Types and 95,000 Records. Data file types include video recordings from optical imaging, electrophysiological time series, scanned lab notes, and image files. A small excerpt of the data model of this instance can be seen in Figure 5. Furthermore, data and parameters from simulations and information about source code is stored along with analysis results of experimental and numerical data. The analysis results are thereby linked to the data from which they stem. Many file types are automatically parsed and integrated as parts of Records into our data model. The hash sums computed and stored for every file allow for comprehensive consistency checks. One advantage of our strict separation between file system and data model is that the system can be directly used on top of the established file system structure without the need to move or modify any existing file.

Figure 5.
A Unified Modeling Language (UML) diagram of a small excerpt of the data model we currently use for experimental data. Experiment is a Record Type with Properties date, LabNotes, Audio and Photo. date is a simple Property of data type DATETIME. The other Properties reference the corresponding Record Types LabNotes, Audio and Photo respectively. TranscribedLabNote is a Record Type with parent LabNotes with a Property named revisionOf that has the data type LabNoteScan. LabNoteScan is as well a Record Type with parent LabNotes.
For data analysis mainly the CaosDB Python client is used, which can directly query and retrieve the relevant data and use it for more specific analyses. The Python client can also be used in more complex data analysis scenarios: for example, data can be retrieved from within a Jupyter notebook using the Python client library. During this interactive data analysis, results and figures are created which can be directly sent back to the CaosDB server and connected with their source data. Finally, the whole Jupyter notebook can be committed as a file object to CaosDB for documenting the data analysis workflow.
The power of CQL leads to much more complex SQL queries in the background than what users would typically enter manually, and subsequently to perceived slower responses. Nevertheless, it already proves to be faster than using simple file operations for finding specific data.
Our current efforts to improve the data model focus on connecting experimental data to intermediate and final results of data analysis, and the integration of data from cardiac simulations.
The software is publicly available at https://gitlab.gwdg.de/bmp-caosdb under the GNU Affero General Public License [14]. A snapshot of the software version described here can be accessed at http://dx.doi.org/10.17617/3.1s [12]. For setting up the system from scratch, a typical Linux server setup (e.g., Debian or Ubuntu with MySQL/MariaDB) is required. The server software is written in Java, some components need compilation with a C++ compiler (e.g., gcc). Detailed installation instructions are provided along with the sources. Packages for typical Linux distributions will be available soon to ease the installation process. Creating an own data model is straightforward and can be done using the WebUI or the Python client. Challenges in designing a data model can differ among workgroups.
Future Work
The evaluation of CaosDB in different scientific environements is currently ongoing, specifically in the fields of geosciences and medical research. The flexibility of the data model allowed for an effortless integration of data from these different scientific areas. Of special interest during these evaluations is the adoption of CQL by people without computer science background, as well as an overall feedback on usability.
One focus of the future development is in query language processing which is still subject to algorithmic optimizations. Here, two specific projects are the improvement of caching facilities and the optimization of the execution order of sub queries.
4. Conclusions
In this article we presented our approach to improve research data management in heterogeneous scientific environments. We presented our perspective on the current situation and proposed a list of requirements an RDMS must fulfill and further described how our research data management system lives up to these requirements.
The most important differences to existing solutions include a smart data model framework which enforces the development of standards while allowing for enough flexibility to adapt to rapidly changing scientific workflows. Furthermore, the powerful and intuitive query language enables quick access to data and simplifies data retrieval and data analysis.
We conclude that our database management system could provide a solution for ongoing issues with research data management in heterogeneous environments and promote the development of standards of data storage and retrieval. It will thereby improve the FAIRness of research data management.
Author Contributions
Conceptualization, T.F., A.S., D.H., H.t.W., U.P. and S.L.; Funding acquisition, U.P. and S.L.; Software, T.F., A.S., D.H. and H.t.W.; Writing—original draft, T.F., A.S., D.H., H.t.W. and U.P.; Writing—review & editing, A.S., D.H., U.P. and S.L.
Funding
We acknowledge support from the German Federal Ministry of Education and Research (BMBF) (project FKZ 031A147, GO-Bio), the German Research Foundation (DFG) (Collaborative Research Centers SFB 1002 Project C3 and SFB 937 Project A18) and the German Center for Cardiovascular Research (DZHK e.V.).
Acknowledgments
We would like to thank the contributors to the software listed in the file caosdb/HUMANS.md and especially Philip Bittihn and Johannes Schröder-Schetelig for their valuable ideas on the design of CaosDB.
Conflicts of Interest
A company for providing commercial support (IndiScale GmbH i.G.) for CaosDB was founded by T.F., D.H., A.S., H.t.W.
Abbreviations
The following abbreviations are used in this manuscript:
| RDMS | Research Data Management System |
| FAIR | Findable, Accessible, Interoperable and Reusable |
| REST | Representational State Transfer |
| CRUD | Create/Read/Update/Delete |
| ACID | Atomicity, Consistency, Isolation, Durability |
| API | Application Programming Interface |
| XML | Extensible Markup Language |
| HTTP | Hypertext Transfer Protocol |
| XSLT | Extensible Stylesheet Language Transformations |
| HTML | Hypertext Markup Language |
| SQL | Structured Query Language |
| UI | User Interface |
| CQL | CaosDB Query Language |
| GNU | GNU’s not Unix |
| PAM | Pluggable Authentication Modules |
| IP | Internet Protocol |
| EBNF | Extended Backus-Naur Form |
| RDF | Resource Description Framework |
| OWL | Web Ontology Language |
| UML | Unified Modeling Language |
| SPARQL | SPARQL Protocol and RDF Query Language |
References
- Nelson, E.K.; Piehler, B.; Eckels, J.; Rauch, A.; Bellew, M.; Hussey, P.; Ramsay, S.; Nathe, C.; Lum, K.; Krouse, K.; et al. LabKey Server: An open source platform for scientific data integration, analysis and collaboration. BMC Bioinform. 2011, 12, 71. [Google Scholar] [CrossRef] [PubMed]
- Anderson, N.R.; Lee, E.S.; Brockenbrough, J.S.; Minie, M.E.; Fuller, S.; Brinkley, J.; Tarczy-Hornoch, P. Issues in Biomedical Research Data Management and Analysis: Needs and Barriers. J. Am. Med. Inform. Assoc. 2007, 14, 478–488. [Google Scholar] [CrossRef] [PubMed]
- Wruck, W.; Peuker, M.; Regenbrecht, C.R. Data management strategies for multinational large-scale systems biology projects. Brief. Bioinform. 2014, 15, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Rocha da Silva, J.; Aguiar Castro, J.; Ribeiro, C.; Correia Lopes, J. Dendro: Collaborative Research Data. In The Semantic Web: ESWC 2014 Satellite Events; Presutti, V., Blomqvist, E., Troncy, R., Sack, H., Papadakis, I., Tordai, A., Eds.; Springer International Publishing: Cham, Switzerland, 2014; pp. 483–487. [Google Scholar]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef] [PubMed]
- Marcus, D.S.; Olsen, T.; Ramaratnam, M.; Buckner, R.L. XNAT: A software framework for managing neuroimaging laboratory data. In Proceedings of the 12th Annual Meeting of the Organization for Human Brain Mapping, Florence, Italy, 11–15 June 2006. [Google Scholar]
- Kanza, S.; Willoughby, C.; Gibbins, N.; Whitby, R.; Frey, J.G.; Erjavec, J.; Zupančič, K.; Hren, M.; Kovač, K. Electronic lab notebooks: Can they replace paper? J. Cheminform. 2017, 9, 31. [Google Scholar] [CrossRef] [PubMed]
- Schweiger, D.; Trajanoski, Z.; Pabinger, S. SPARQLGraph: A web-based platform for graphically querying biological Semantic Web databases. BMC Bioinform. 2014, 15, 279. [Google Scholar] [CrossRef] [PubMed]
- Haag, F.; Lohmann, S.; Siek, S.; Ertl, T. QueryVOWL: A Visual Query Notation for Linked Data. In The Semantic Web: ESWC 2015 Satellite Events; Gandon, F., Guéret, C., Villata, S., Breslin, J., Faron-Zucker, C., Zimmermann, A., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 387–402. [Google Scholar]
- Chochiang, K.; Grabmann, B.; Betbeder, M.L.; Lapayre, J.C.; Sa-ngiamsak, C. OntoQuer: A Tool for Building SPARQL Query Automatically Applying with Our Ontologies. J. Softw. 2017, 12, 145–152. [Google Scholar]
- Fielding, R.T. Architectural Styles and the Design of Network-Based Software Architectures. Ph.D. Thesis, University of California, Irvine, CA, USA, 2000. [Google Scholar]
- Fitschen, T.; Hornung, D.; Schlemmer, A.; tom Wörden, H. CaosDB. 2018. Available online: http://dx.doi.org/10.17617/3.1s (accessed on 12 October 2018).
- Føllesdal, D.; Hilpinen, R. Deontic Logic: An Introduction. In Deontic Logic: Introductory and Systematic Readings; Hilpinen, R., Ed.; Springer: Dordrecht, The Netherlands, 1971; pp. 1–35. [Google Scholar]
- Free Software Foundation. GNU Affero General Public License; Free Software Foundation, Inc.: Boston, MA, USA, 2018. [Google Scholar]
| 1 | Atomicity, Consistency, Isolation, Durability. |
| 2 | Representational State Transfer. |
| 3 | Extended Backus-Naur Form. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).