Graph Theoretic and Pearson Correlation-Based Discovery of Network Biomarkers for Cancer
Abstract
1. Introduction
2. Results
2.1. Topology of Gene Co-Expression Networks
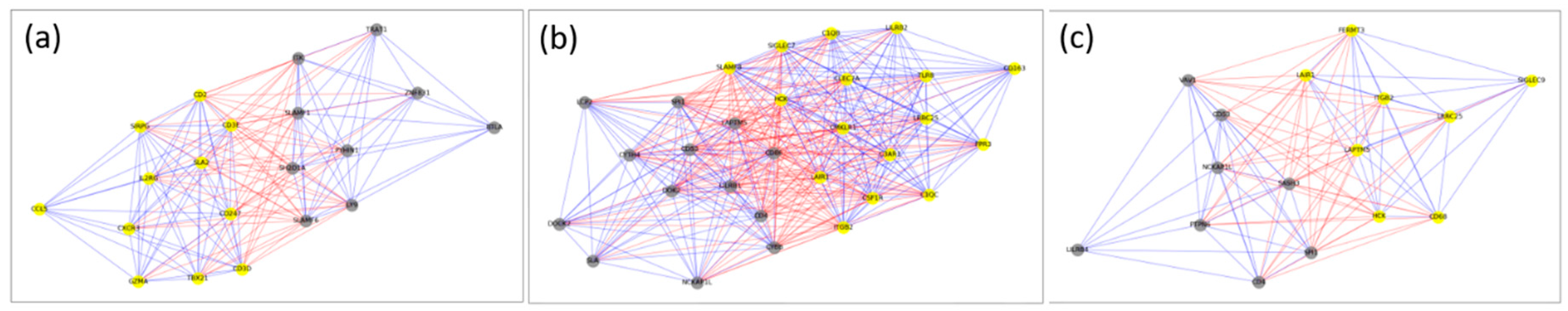
2.2. Cliques and Maximal Clique-Like Modules
2.3. 2-Clique-1-Bipartite Modules
2.4. 3-Clique-2-Bipartite Modules
3. Discussion
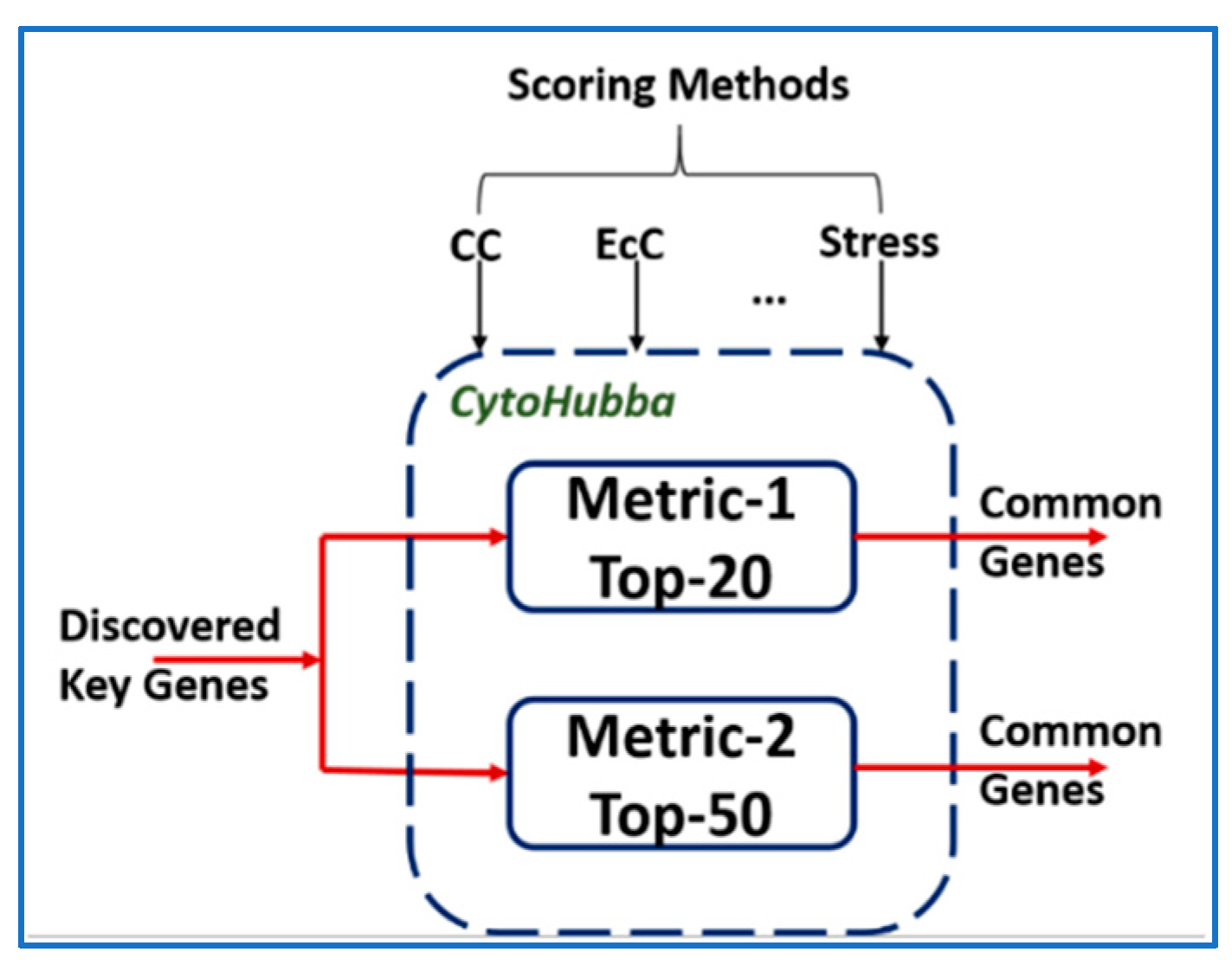
3.1. Validation Using CytoHubba
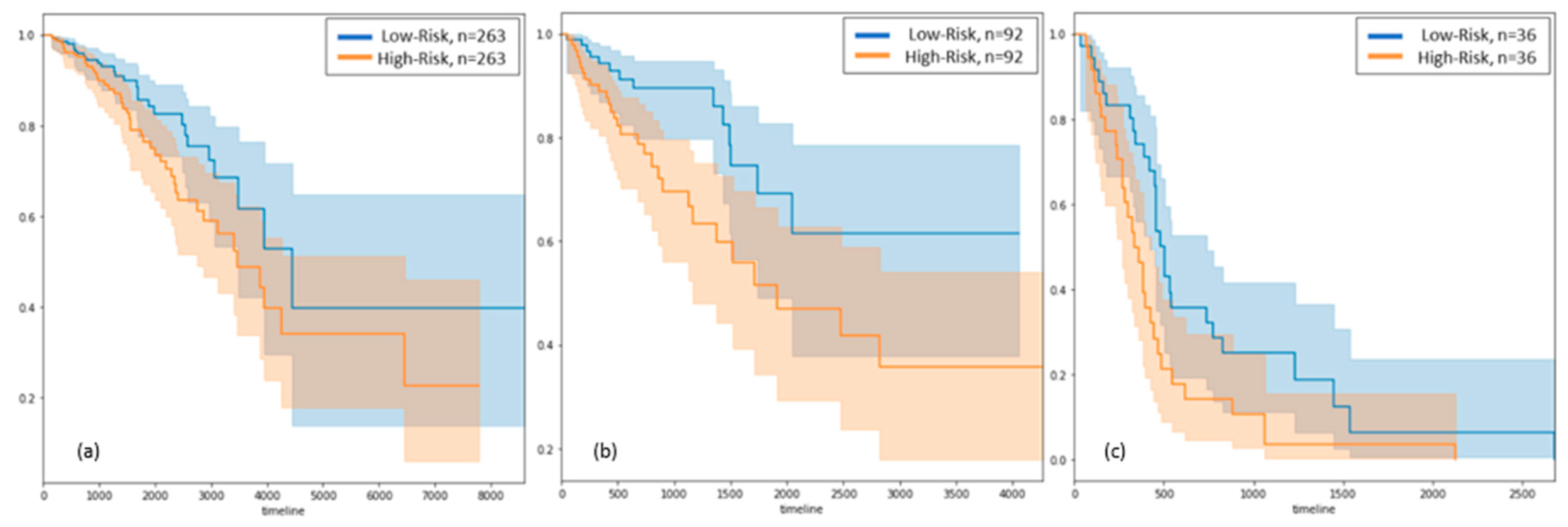
3.2. Survival Analysis
3.3. Pathway and Gene Ontology Enrichment of Key Genes
3.4. Future Direction
4. Materials and Methods
4.1. Dataset Preparation
4.2. Method to Identify Clique and Clique-Bipartite-Like Modules
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ideker, T.; Sharan, R. Protein networks in disease. Genome Res. 2008, 18, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, S.; Mehta, M.; Kuo, D.; Sung, M.-K.; Chuang, R.; Jaehnig, E.J.; Bodenmiller, B.; Licon, K.; Copeland, W.; Shales, M.; et al. Rewiring of Genetic Networks in Response to DNA Damage. Science 2010, 330, 1385–1389. [Google Scholar] [CrossRef] [PubMed]
- Haugen, A.C.; Kelley, R.; Collins, J.B.; Tucker, C.J.; Deng, C.; Afshari, C.A.; Brown, J.M.; Ideker, T.; Van Houten, B. Integrating phenotypic and expression profiles to map arsenic-response networks. Genome Boil. 2004, 5, R95. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Tu, Z.; Deng, M.; Sun, F.; Chen, T. Diffusion Kernel-Based Logistic Regression Models for Protein Function Prediction. OMICS A J. Integr. Boil. 2006, 10, 40–55. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Suhail, Y.; Lin, Y.; Boeke, J.D.; Bader, J.S. Finding friends and enemies in an enemies-only network: A graph diffusion kernel for predicting novel genetic interactions and co-complex membership from yeast genetic interactions. Genome Res. 2008, 18, 1991–2004. [Google Scholar] [CrossRef] [PubMed]
- Ananda, M.M.; Hu, J. NetLoc: Network based protein localization prediction using protein-protein interaction and co-expression networks. In Proceedings of the 2010 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Hong Kong, China, 18–21 December 2010; pp. 142–148. [Google Scholar]
- Mondal, A.; Lin, J.-R.; Hu, J. Network based subcellular localization prediction for multi-label proteins. In Proceedings of the 2011 IEEE International Conference on Bioinformatics and Biomedicine Workshops (BIBMW), Atlanta, GA, USA, 12–15 November 2011. [Google Scholar]
- Mondal, A.M.; Hu, J. Protein Localization by Integrating Multiple Protein Correlation Networks. Proceedings of The 2012 International Conference on Bioinformatics & Computational Biology (BIOCOMP’12), Las Vegas, NV, USA, 16–19 July 2012; pp. 82–88. [Google Scholar]
- Lin, J.-R.; Mondal, A.M.; Liu, R.; Hu, J. Minimalist ensemble algorithms for genome-wide protein localization prediction. BMC Bioinform. 2012, 13, 157. [Google Scholar] [CrossRef]
- Mondal, A.; Hu, J. Scored Protein-Protein Interaction to Predict Subcellular Localizations for Yeast Using Diffusion Kernel. In International Conference on Pattern Recognition and Machine Intelligence; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Mondal, A.; Hu, J. Network based prediction of protein localisation using diffusion kernel. Int. J. Data Min. Bioinform. 2014, 9, 386–400. [Google Scholar] [CrossRef]
- Faisal, F.E.; Milenkovic, T. Dynamic networks reveal key players in aging. Bioinformatics 2014, 30, 1721–1729. [Google Scholar] [CrossRef]
- Kevin, C.; Andrews, A.; Ananda, M. Protein Subnetwork Biomarkers for Yeast Using Brute Force Method. In Proceedings of the International Conference on Bioinformatics & Computational Biology (BIOCOMP), Las Vagas, NV, USA, 22–25 July 2013; pp. 218–223. [Google Scholar]
- Timalsina, P.; Charles, K.; Mondal, A.M. STRING PPI Score to Characterize Protein Subnetwork Biomarkers for Human Diseases and Pathways. In Proceedings of the 2014 IEEE International Conference on Bioinformatics and Bioengineering, Boca Raton, FL, USA, 10–12 November 2014; pp. 251–256. [Google Scholar]
- Maharjan, M.; Tanvir, R.B.; Chowdhury, K.; Mondal, A.M. Determination of Biomarkers for Diagnosis of Lung Cancer Using Cytoscape-based GO and Pathway Analysis. In Proceedings of the 20th International Conference on Bioinformatics & Computational Biology (BIOCOMP’19), Las Vegas, NV, USA, 29 July–01 Aug 2019. (Accepted). [Google Scholar]
- Eisen, M.B.; Spellman, P.T.; Brown, P.O.; Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 1998, 95, 14863–14868. [Google Scholar] [CrossRef]
- Wolfe, C.J.; Kohane, I.S.; Butte, A.J. Systematic survey reveals general applicability of ‘guilt-by-association’ within gene coexpression networks. BMC Bioinform. 2005, 6, 227. [Google Scholar] [CrossRef]
- Butte, A.J.; Kohane, I.S. Mutual information relevance networks: Functional genomic clustering using pairwise entropy measurements. Pac. Symp. Biocomput. 2000, 418–429. [Google Scholar]
- Zhang, B.; Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 17. [Google Scholar] [CrossRef]
- Tang, J.; Lu, M.; Cui, Q.; Zhang, D.; Kong, D.; Liao, X.; Ren, J.; Gong, Y.; Wu, G. Overexpression of ASPM, CDC20, and TTK Confer a Poorer Prognosis in Breast Cancer Identified by Gene Co-expression Network Analysis. Front. Oncol. 2019, 9, 310. [Google Scholar] [CrossRef]
- Lalremmawia, H.; Tiwary, B.K. Identification of Molecular Biomarkers for Ovarian Cancer using Computational Approaches. Carcinogenesis 2019. [Google Scholar] [CrossRef]
- Maertens, A.M.; Tran, V.; Kleensang, A.; Hartung, T. Weighted Gene Correlation Network Analysis (WGCNA) Reveals Novel Transcription Factors Associated With Bisphenol A Dose-Response. Front. Genet. 2018, 9, 508. [Google Scholar] [CrossRef]
- Shi, H.; Zhang, L.; Qu, Y.; Hou, L.; Wang, L.; Zheng, M. Prognostic genes of breast cancer revealed by gene co-expression network analysis. Oncol. Lett. 2017, 14, 4535–4542. [Google Scholar] [CrossRef][Green Version]
- Liu, X.; Hu, A.-X.; Zhao, J.-L.; Chen, F. Identification of Key Gene Modules in Human Osteosarcoma by Co-Expression Analysis Weighted Gene Co-Expression Network Analysis (WGCNA). J. Cell. Biochem. 2017, 118, 3953–3959. [Google Scholar] [CrossRef]
- Zhang, C.; Sun, Q. Weighted gene co-expression network analysis of gene modules for the prognosis of esophageal cancer. J. Huazhong Univ. Sci. Technol. [Med. Sci.] 2017, 37, 319–325. [Google Scholar] [CrossRef]
- Liu, R.; Zhang, W.; Liu, Z.; Zhou, H. Associating transcriptional modules with colon cancer survival through weighted gene co-expression network analysis. BMC Genom. 2017, 18, 361. [Google Scholar] [CrossRef]
- Liu, Z.-P.; Gao, R. Detecting pathway biomarkers of diabetic progression with differential entropy. J. Biomed. Inform. 2018, 82, 143–153. [Google Scholar] [CrossRef]
- Guan, L.; Luo, Q.; Liang, N.; Liu, H. A prognostic prediction system for hepatocellular carcinoma based on gene co-expression network. Exp. Ther. Med. 2019, 17, 4506–4516. [Google Scholar] [CrossRef]
- Shi, Z.; Derow, C.K.; Zhang, B. Co-expression module analysis reveals biological processes, genomic gain, and regulatory mechanisms associated with breast cancer progression. BMC Syst. Biol. 2010, 4, 74. [Google Scholar] [CrossRef]
- Perkins, A.D.; Langston, M.A. Threshold selection in gene co-expression networks using spectral graph theory techniques. BMC Bioinform. 2009, 10, S4. [Google Scholar] [CrossRef]
- Zhang, D.-Q.; Zhou, C.; Chen, S.-Z.; Yang, Y.; Shi, B. Identification of hub genes and pathways associated with bladder cancer based on co-expression network analysis. Oncol. Lett. 2017, 14, 1115–1122. [Google Scholar] [CrossRef]
- Mondal, A.M.; Schultz, C.A.; Sheppard, M.; Carson, J.; Tanvir, R.B.; Aqila, T. Graph Theoretic Concepts as the Building Blocks for Disease Initiation and Progression at Protein Network Level: Identification and Challenges. In Proceedings of the 2018 IEEE International Conference on Bioinformatics and Biomedicine, BIBM, Madrid, Spain, 3–6 December 2018. [Google Scholar]
- Hagberg, A.A.; Schult, D.A.; Swart, P.J. Exploring network structure, dynamics, and function using NetworkX. In Proceedings of the 7th Python in Science Conference (SciPy), Pasadena, CA, USA, 19–24 August 2008; pp. 11–15. [Google Scholar]
- Chin, C.-H.; Chen, S.-H.; Wu, H.-H.; Ho, C.-W.; Ko, M.-T.; Lin, C.-Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. 4), S11. [Google Scholar] [CrossRef]
- Mauger, E.A.; Wolfe, R.A.; Port, F.K. Transient effects in the cox proportional hazards regression model. Stat. Med. 1995, 14, 1553–1565. [Google Scholar] [CrossRef]
- Sotiriou, C.; Wirapati, P.; Loi, S.; Harris, A.; Fox, S.; Smeds, J.; Nordgren, H.; Farmer, P.; Praz, V.; Haibe-Kains, B.; et al. Gene Expression Profiling in Breast Cancer: Understanding the Molecular Basis of Histologic Grade to Improve Prognosis. J. Natl. Cancer Inst. 2006, 98, 262–272. [Google Scholar] [CrossRef]
- Wu, G.; Dawson, E.; Duong, A.; Haw, R.; Stein, L. ReactomeFIViz: The Reactome FI Cytoscape app for pathway and network-based data analysis. F1000Research 2014, 3, 146. [Google Scholar] [CrossRef]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of Gene Ontology categories in Biological Networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef]
- Monette, A.; Bergeron, D.; Ben Amor, A.; Meunier, L.; Caron, C.; Mes-Masson, A.-M.; Kchir, N.; Hamzaoui, K.; Jurisica, I.; Lapointe, R. Immune-enrichment of non-small cell lung cancer baseline biopsies for multiplex profiling define prognostic immune checkpoint combinations for patient stratification. J. Immunother. Cancer 2019, 7, 86. [Google Scholar] [CrossRef]
- Erazo-Oliveras, A.; Fuentes, N.R.; Wright, R.C.; Chapkin, R.S. Functional link between plasma membrane spatiotemporal dynamics, cancer biology, and dietary membrane-altering agents. Cancer Metastasis Rev. 2018, 37, 519–544. [Google Scholar] [CrossRef]
- Vasaikar, S.V.; Straub, P.; Wang, J.; Zhang, B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2017, 46, D956–D963. [Google Scholar] [CrossRef]

| Cancer Name | # Of Genes | # Of Edges | Max Degree | Min Degree | Avg Degree |
|---|---|---|---|---|---|
| BRCA | 380 | 1034 | 39 | 1 | 5.4 |
| COAD | 607 | 3651 | 75 | 1 | 12.0 |
| GBM | 506 | 1243 | 49 | 1 | 4.9 |
| Cancer | List of Genes in Maximal Clique-Like Modules |
|---|---|
| BRCA | CD2, CD247, CD3D, CD3E, CD5, CD96, CXCR3, IL2RG, LCK, LY9, PTPN7, SH2D1A, SIRPG, SIT1, SLA2, SLAMF1, SLAMF6, TBX21, UBASH3A |
| COAD | C1QB, C1QC, C3AR1, CD300A, CD4, CD53, CD86, CLEC7A, CSF1R, CYBB, CYTH4, DOK2, FCER1G, FPR3, HAVCR2, HCK, ITGB2, LAIR1, LAPTM5, LILRB1, LILRB4, LRRC25, MS4A4A, PDCD1LG2, SIGLEC7, SIGLEC9, SLAMF8, SPI1, TFEC, TYROBP |
| GBM | ALOX5, CD4, FERMT3, HCK, ITGB2, LAIR1, LAPTM5, NCKAP1L, PTPN6, SASH3, SPI1, STXBP2, VAV1, WAS |
| BRCA | COAD | GBM | |
|---|---|---|---|
| Clique1 | CCL5, CD2, CD247, CD3D, CD3E, CXCR3, GZMA, IL2RG, SIRPG, SLA2, TBX21 | C1QB, C1QC, C3AR1, CD163, CLEC7A, CMKLR1, CSF1R, FPR3, HCK, ITGB2, LAIR1, LILRB2, LRRC25, SIGLEC7, SLAMF8, TLR8 | CD68, FERMT3, HCK, ITGB2, LAIR1, LAPTM5, LRRC25, SIGLEC9 |
| Clique2 | BTLA, ITK, LY9, PYHIN1, SH2D1A, SLAMF1, SLAMF6, TRAT1, ZNF831 | CD4, CD53, CD86, CYBB, CYTH4, DOCK2, DOK2, LAPTM5, LCP2, LILRB1, NCKAP1L, SLA, SPI1 | CD4, CD53, LILRB4, NCKAP1L, PTPN6, SASH3, SPI1, VAV1 |
| Clique-A | Clique-B | Clique-C | Connections A-B | Connections B-C | |
|---|---|---|---|---|---|
| BRCA-Module1 | 13 | 7 | 4 | 56 | 13 |
| BRCA-Module2 | 11 | 7 | 4 | 35 | 10 |
| BRCA-Module3 | 8 | 6 | 6 | 8 | 18 |
| COAD-Module1 | 16 | 14 | 7 | 85 | 53 |
| COAD-Module2 | 16 | 14 | 6 | 111 | 51 |
| COAD-Module3 | 16 | 12 | 7 | 69 | 40 |
| GBM-Module1 | 9 | 9 | 5 | 30 | 19 |
| GBM-Module2 | 9 | 7 | 6 | 22 | 23 |
| GBM-Module3 | 9 | 7 | 4 | 36 | 14 |
| List of Genes | |
|---|---|
| BRCA-Modules | ACAP1, CCL5, CD2, CD247, CD3D, CD3E, CD3G, CD5, CD53, CD96, CXCR3, CXCR6, DOCK2, EVI2B, FYB, GZMA, GZMM, IKZF1, IL2RG, ITK, LCP2, LY9, NCKAP1L, PLEK, PRF1, PRKCB, PTPRC, PTPRCAP, PYHIN1, S1PR4, SH2D1A, SIRPG, SIT1, SLA2, SLAMF1, SLAMF6, SPN, TBC1D10C, TBX21, THEMIS, TRAT1, UBASH3A, ZAP70, ZNF831 |
| COAD-Modules | APBB1IP, ARHGAP30, ARHGAP9, BTK, C3AR1, CD163, CD4, CD53, CD84, CD86, CLEC7A, CSF1R, CYBB, CYTH4, DOCK10, DOCK2, FPR3, HAVCR2, HCK, HCLS1, IKZF1, IL10RA, ITGAL, ITGB2, ITK, KLHL6, LAIR1, LAPTM5, LILRB1, LILRB4, LRRC25, MAP4K1, MNDA, MYO1G, NCKAP1L, PIK3R5, PTPRC, RASAL3, SASH3, SIGLEC7, SIGLEC9, SIRPB2, SLA, SLAMF8, SPI1, TBC1D10C, TRAF3IP3, WAS |
| GBM-Modules | ARHGAP30, ARL11, C1QA, C1QB, C1QC, CD33, CD4, CD53, CD68, CD86, CSF1R, DOCK2, DOCK8, FCER1G, FCGR3A, FERMT3, HCK, IKZF1, ITGB2, LAIR1, LAPTM5, MYO1F, NCF4, NCKAP1L, PLCG2, SASH3, SPI1, STXBP2, SYK, TYROBP, VAMP8, VAV1 |
| Dataset. | Key Genes | Keys Genes Common with | |
|---|---|---|---|
| Metric-1 | Metric-2 | ||
| BRCA | 47 | 26 | 45 |
| COAD | 61 | 23 | 53 |
| GBM | 38 | 25 | 36 |
| Cancer | No of Genes | No of Samples | Reduced no of Genes |
|---|---|---|---|
| Breast invasive carcinoma (BRCA) | 20,155 | 1093 | 16,011 |
| Colorectal adenocarcinoma (COAD) | 19,828 | 379 | 15,769 |
| Glioblastoma multiforme (GBM) | 19,660 | 153 | 16,186 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tanvir, R.B.; Aqila, T.; Maharjan, M.; Mamun, A.A.; Mondal, A.M. Graph Theoretic and Pearson Correlation-Based Discovery of Network Biomarkers for Cancer. Data 2019, 4, 81. https://doi.org/10.3390/data4020081
Tanvir RB, Aqila T, Maharjan M, Mamun AA, Mondal AM. Graph Theoretic and Pearson Correlation-Based Discovery of Network Biomarkers for Cancer. Data. 2019; 4(2):81. https://doi.org/10.3390/data4020081
Chicago/Turabian StyleTanvir, Raihanul Bari, Tasmia Aqila, Mona Maharjan, Abdullah Al Mamun, and Ananda Mohan Mondal. 2019. "Graph Theoretic and Pearson Correlation-Based Discovery of Network Biomarkers for Cancer" Data 4, no. 2: 81. https://doi.org/10.3390/data4020081
APA StyleTanvir, R. B., Aqila, T., Maharjan, M., Mamun, A. A., & Mondal, A. M. (2019). Graph Theoretic and Pearson Correlation-Based Discovery of Network Biomarkers for Cancer. Data, 4(2), 81. https://doi.org/10.3390/data4020081





