Transcriptomic Response of L. monocytogenes to Co-Culture with S. cerevisiae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Sample Preparation
2.2. Sampling and Microbiological Analyses
2.3. Gene Transcription Assay
2.4. Statistical Analysis
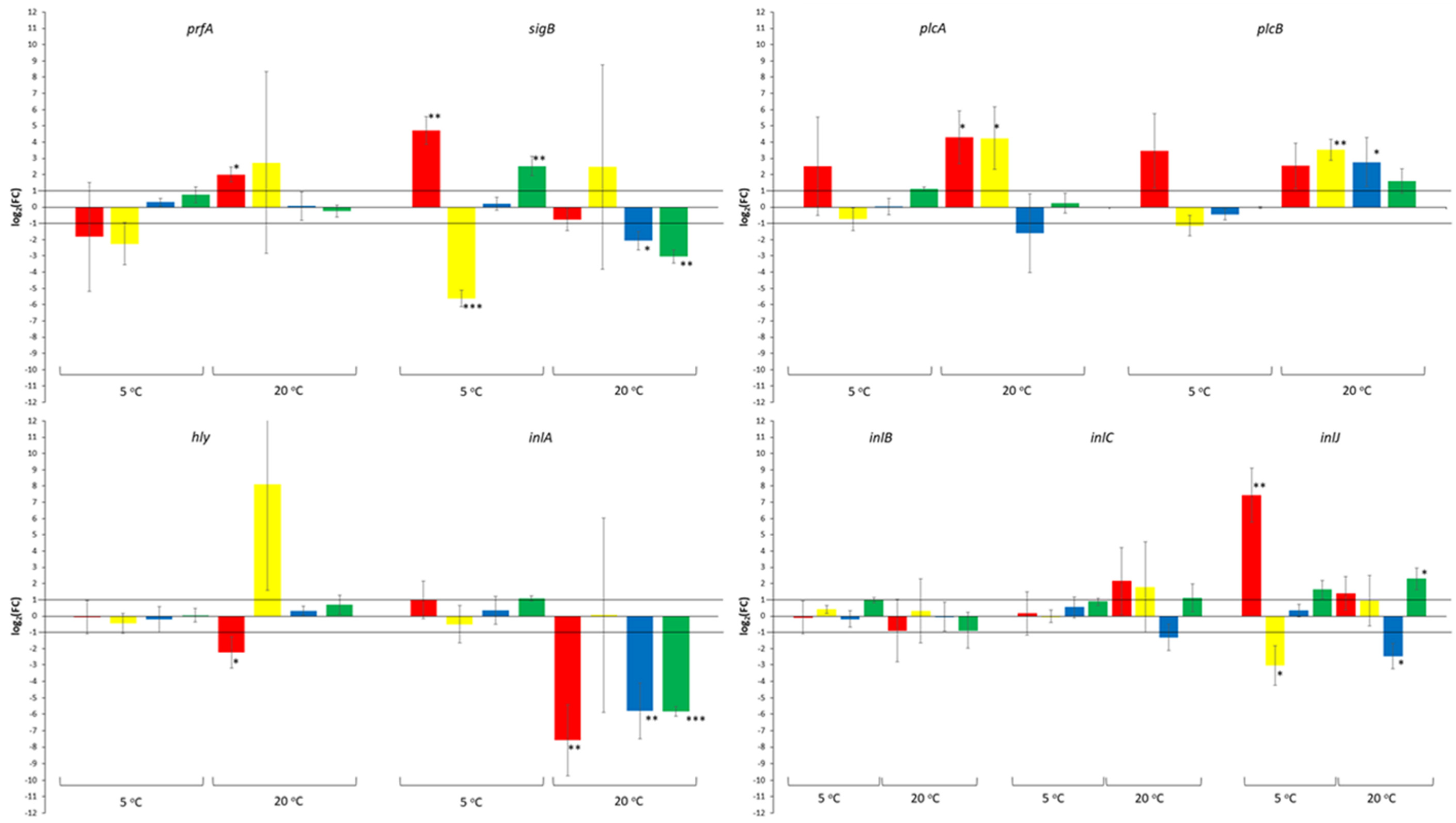
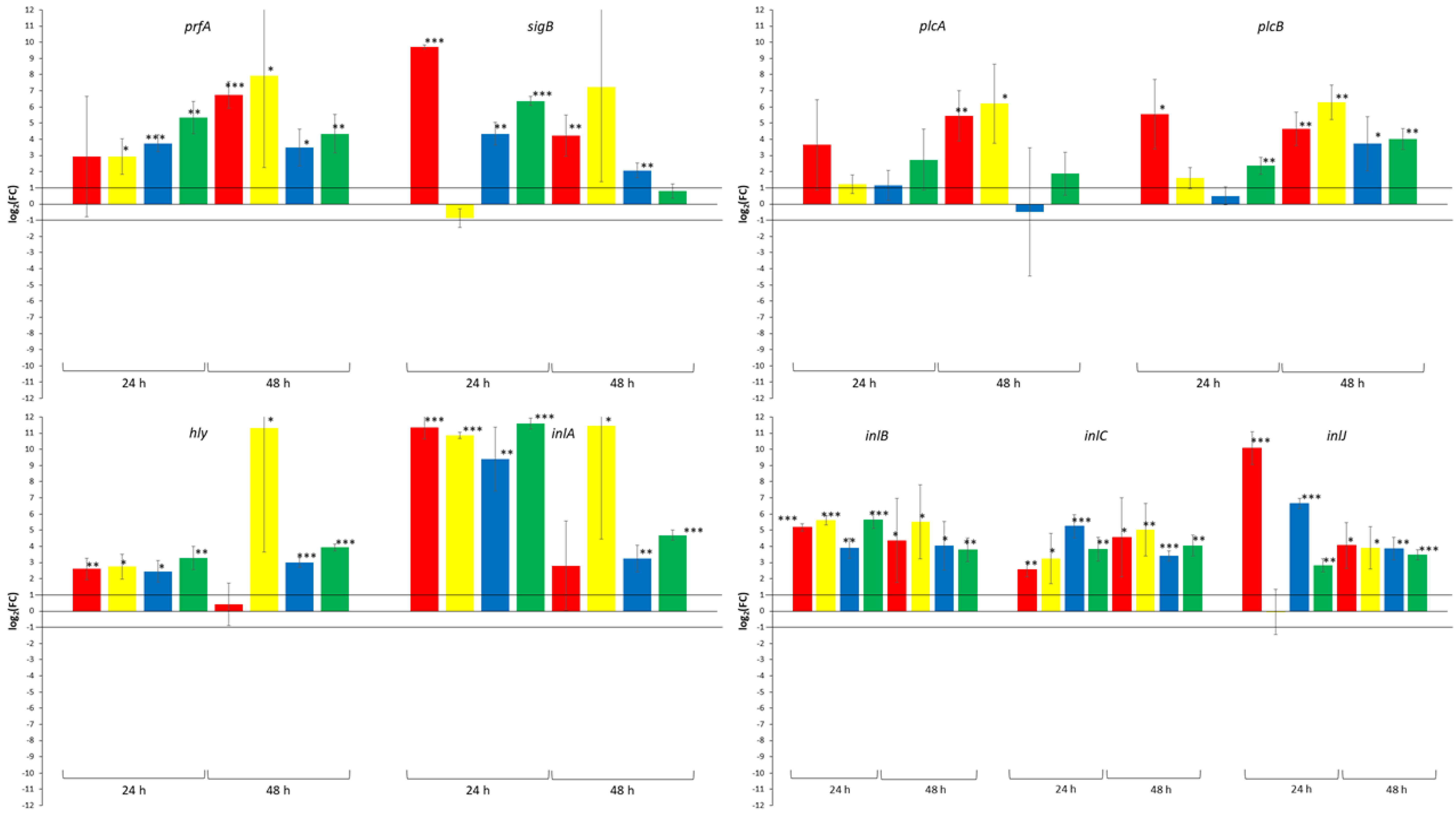
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- EFSA (European Food Safety Authority); ECDC (European Centre for Disease Prevention and Control). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 2018, 16, e05500. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). Surveillance for Foodborne Disease Outbreaks, 2017, Annual Report; Department of Health and Human Services, CDC: Atlanta, GA, USA, 2019. [Google Scholar]
- Conte, M.P.; Petrone, G.; Di Biase, A.; Ammendolia, M.G.; Superti, F.; Seganti, L. Acid tolerance in Listeria monocytogenes influences invasiveness of enterocyte-like cells and macro-phagelike cells. Microb. Pathog. 2000, 29, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Garner, M.R.; James, K.E.; Callahan, M.C.; Wiedmann, M.; Boor, K.J. Exposure to salt and organic acids increases the ability of Listeria monocytogenes to invade Caco-2 cells but de-creases its ability to survive gastric stress. Appl. Environ. Microbiol. 2006, 72, 5384–5395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Werbrouck, H.; Vermeulen, A.; Van Coillie, E.; Messens, W.; Herman, L.; Devlieghere, F.; Uyttendaele, M. Influence of acid stress on survival, expression of virulence genes and invasion capacity into Caco-2 cells of Listeria monocytogenes strains of different origins. Int. J. Food Microbiol. 2009, 134, 140–146. [Google Scholar] [CrossRef]
- Duodu, S.; Holst-Jensen, A.; Skjerdal, T.; Cappelier, J.-M.; Pilet, M.-F.; Loncarevic, S. Influence of storage temperature on gene expression and virulence potential of Listeria monocytogenes strains grown in a salmon matrix. Food Microbiol. 2010, 27, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Rieu, A.; Guzzo, J.; Piveteau, P. Sensitivity to acetic acid, ability to colonize abiotic surfaces and virulence potential of Listeria monocytogenes EGD-e after incubation on parsley leaves. J. Appl. Microbiol. 2010, 108, 560–570. [Google Scholar] [CrossRef] [PubMed]
- Wałecka, E.; Molenda, J.; Karpíšková, R.; Bania, J. Effect of osmotic stress and culture density on invasiveness of Listeria monocytogenes strains. Int. J. Food Microbiol. 2011, 144, 440–445. [Google Scholar] [CrossRef] [PubMed]
- Pricope-Ciolacu, L.; Nicolau, A.I.; Wagner, M.; Rychli, K. The effect of milk components and storage conditions on the virulence of Listeria monocytogenes as determined by a Caco-2 cell assay. Int. J. Food Microbiol. 2013, 166, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Colas-Meda, P.; Vinas, I.; Oliveira, M.; Anguera, M.; Serrano, J.C.E.; Abadias, M. Exposure to minimally processed pear and melon during shelf life could modify the pathogenic potential of Listeria monocytogenes. Food Microbiol. 2017, 62, 275–281. [Google Scholar] [CrossRef] [Green Version]
- Alves, A.; Magalhães, R.; Brandão, T.R.S.; Pimentel, L.; Rodríguez-Alcalá, L.M.; Teixeira, P.; Ferreira, V. Impact of exposure to cold and cold-osmotic stresses on virulence-associated characteristics of Listeria monocytogenes strains. Food Microbiol. 2020, 87, 103351. [Google Scholar] [CrossRef]
- Wałecka-Zacharska, E.; Korkus, J.; Skowron, K.; Wietlicka-Piszcz, M.; Kosek-Paszkowska, K.; Bania, J. Effect of temperatures used in food storage on duration of heat stress induced invasiveness of L. monocytogenes. Microorganisms 2019, 7, 467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brackett, R.E. Shelf stability and safety of fresh produce as influenced by sanitation and disinfection. J. Food Prot. 1992, 55, 808–814. [Google Scholar] [CrossRef] [PubMed]
- Samelis, J.; Metaxopoulos, J.; Vlassi, M.; Pappa, A. Stability and safety of traditional Greek salami—a microbiological ecology study. Int. J. Food Microbiol. 1998, 44, 69–82. [Google Scholar] [CrossRef]
- Del Campo, J.; Carlin, F.; Nguyen-The, C. Effects of epiphytic Enterobacteriaceae and pseudomonads on the growth of Listeria monocytogenes in model media. J. Food Prot. 2001, 64, 721–724. [Google Scholar] [CrossRef] [PubMed]
- Gómez, D.; Iguácel, L.P.; Rota, M.C.; Carramiñana, J.J.; Ariño, A.; Yangüela, J. Occurrence of Listeria monocytogenes in ready-to-eat meat products and meat processing plants in Spain. Foods 2015, 4, 271–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falardeau, J.; Trmčić, A.; Wang, S. The occurrence, growth, and biocontrol of Listeria monocytogenes in fresh and surface-ripened soft and semisoft cheeses. Compr. Rev. Food Sci. Food Saf. 2021, 20, 4019–4048. [Google Scholar] [CrossRef]
- Tan, Q.; Xu, H.; Chen, T.; Li, P.; Aguilar, Z.P.; Xu, D.; Ming, X.; Xu, F.; Wei, H. Differential expression of virulence and stress fitness genes during interaction between Listeria monocytogenes and Bifidobacterium longum. Biosci. Biotechnol. Biochem. 2012, 76, 699–704. [Google Scholar] [CrossRef] [Green Version]
- Tirumalai, P.S. Metabolic gene expression shift by Listeria monocytogenes in coculture biofilms. Can. J. Microbiol. 2015, 61, 327–334. [Google Scholar] [CrossRef]
- Dutra, V.; Silva, A.C.; Cabrita, P.; Peres, C.; Malcata, X.; Brito, L. Lactobacillus plantarum LB95 impairs the virulence potential of Gram-positive and Gram-negative food-borne pathogens in HT-29 and Vero cell cultures. J. Med Microbiol. 2016, 65, 28–35. [Google Scholar] [CrossRef]
- Collazo, C.; Abadias, M.; Colás-Medà, P.; Iglesias, M.B.; Granado-Serrano, A.B.; Serrano, J.; Viñas, I. Effect of Pseudomonas graminis strain CPA-7 on the ability of Listeria monocytogenes and Salmonella enterica subsp. enterica to colonize Caco-2 cells after pre-incubation on fresh-cut pear. Int. J. Food Microbiol. 2017, 262, 55–62. [Google Scholar] [CrossRef] [Green Version]
- Iglesias, M.; Viñas, I.; Colás-Medà, P.; Collazo, C.; Serrano, J.; Abadias, M. Adhesion and invasion of Listeria monocytogenes and interaction with Lactobacillus rhamnosus GG after habituation on fresh-cut pear. J. Funct. Foods 2017, 34, 453–460. [Google Scholar] [CrossRef]
- Montiel, R.; Quesille-Villalobos, A.; Alessandria, V.; Medina, M.; Cocolin, L.S.; Rantsiou, K. Antilisterial Effect and influence on Listeria monocytogenes gene expression of enterocin or Enterococcus faecalis in sliced dry-cured ham stored at 7 °C. J. Food Prot. 2019, 82, 1598–1606. [Google Scholar] [CrossRef]
- Lappa, I.; Dionysopoulou, A.M.; Paramithiotis, S.; Georgiadou, M.; Drosinos, E.H. Dual transcriptional profile of Aspergillus flavus during co-culture with Listeria monocytogenes and AFB1 production: A pathogen: Pathogen interaction. Pathogens 2019, 8, 198. [Google Scholar] [CrossRef] [Green Version]
- Zilelidou, E.A.; Milina, V.; Paramithiotis, S.; Zoumpopoulou, G.; Poimenidou, S.V.; Mavrogonatou, E.; Kletsas, D.; Papadimitriou, K.; Tsakalidou, E.; Skandamis, P.N. Differential modulation of Listeria monocytogenes fitness, in vitro virulence, and transcription of virulence-associated genes in response to the presence of different microorganisms. Appl. Environ. Microbiol. 2020, 86, 01165-20. [Google Scholar] [CrossRef]
- Kotova, I.B.; Cherdyntseva, T.A.; Netrusov, A.I. Russian kefir grains microbial composition and its changes during production process. Adv. Exp. Med. Biol. 2016, 932, 93–121. [Google Scholar] [CrossRef]
- Gao, W.; Zhang, L. Genotypic diversity of bacteria and yeasts isolated from Tibetan kefir. Int. J. Food Sci. Technol. 2018, 53, 1535–1540. [Google Scholar] [CrossRef]
- Akabanda, F.; Owusu-Kwarteng, J.; Tano-Debrah, K.; Glover, R.L.; Nielsen, D.S.; Jespersen, L. Taxonomic and molecular characterization of lactic acid bacteria and yeasts in nunu, a Ghanaian fermented milk product. Food Microbiol. 2013, 34, 277–283. [Google Scholar] [CrossRef]
- Bayili, G.R.; Johansen, P.G.; Nielsen, D.S.; Sawadogo, H.; Ouedraogo, G.A.; Diawara, B.; Jespersen, L. Identification of the predominant microbiota during production of lait caillé, a spontaneously fermented milk product made in Burkina Faso. World J. Microbiol. Biotechnol. 2019, 35, 100. [Google Scholar] [CrossRef]
- Gulmez, M.; Guven, A. Survival of Escherichia coli O157:H7, Listeria monocytogenes 4b and Yersinia enterocolitica O3 in different yogurt and kefir combinations as prefermentation contaminant. J. Appl. Microbiol. 2003, 95, 631–636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dias, P.A.; da Silva, D.T.; Tejada, T.S.; Leal, M.C.G.M.; da Conceicao, R.C.S.; Timm, C.D. Survival of pathogenic microorganisms in kefir. Rev. Inst. Adolfo Lutz 2012, 71, 182–186. [Google Scholar]
- Owusu-Kwarteng, J.; Wuni, A.; Akabanda, F.; Jespersen, L. Prevalence and characteristics of Listeria monocytogenes isolates in raw milk, heated milk and Nunu, a spontaneously fermented milk beverage, in Ghana. Beverages 2018, 4, 40. [Google Scholar] [CrossRef] [Green Version]
- Hadjilouka, A.; Andritsos, N.; Paramithiotis, S.; Mataragas, M.; Drosinos, E.H. Listeria monocytogenes serotype prevalence and biodiversity in diverse food products. J. Food Prot. 2014, 77, 2115–2120. [Google Scholar] [CrossRef] [PubMed]
- Hadjilouka, A.; Paramithiotis, S.; Drosinos, E.H. Genetic analysis of the Listeria Pathogenicity Island 1 of Listeria monocytogenes 1/2a and 4b isolates. Curr. Microbiol. 2018, 75, 857–865. [Google Scholar] [CrossRef]
- Bonatsou, S.; Paramithiotis, S.; Panagou, E.Z. Evolution of yeast consortia during the fermentation of Kalamata natural black olives upon two initial acidification treatments. Front. Microbiol. 2018, 8, 2673. [Google Scholar] [CrossRef]
- Hadjilouka, A.; Mavrogiannis, G.; Mallouchos, A.; Paramithiotis, S.; Mataragas, M.; Drosinos, E.H. Effect of lemongrass essential oil on Listeria monocytogenes gene expression. LWT 2017, 77, 510–516. [Google Scholar] [CrossRef]
- Hadjilouka, A.; Gkolfakis, P.; Patlaka, A.; Grounta, A.; Vourli, G.; Paramithiotis, S.; Touloumi, G.; Triantafyllou, K.; Drosinos, E.H. In vitro gene transcription of Listeria monocytogenes after exposure to human gastric and duodenal aspirates. J. Food Prot. 2019, 83, 89–100. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization—Applied to bladder- and colon-cancer data-sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [Green Version]
- Syrokou, M.; Tziompra, S.; Psychogiou, E.-E.; Mpisti, S.-D.; Paramithiotis, S.; Bosnea, L.; Mataragas, M.; Skandamis, P.; Drosinos, E. Technological and safety attributes of lactic acid bacteria and yeasts isolated from spontaneously fermented Greek wheat sourdoughs. Microorganisms 2021, 9, 671. [Google Scholar] [CrossRef]
- Rios-Covian, D.; Nogacka, A.; Salazar, N.; Hernández-Barranco, A.M.; Cuesta, I.; Gueimonde, M.; Gavilán, C.G.R. Bifidobacterium breve IPLA20005 affects in vitro the expression of hly and luxS genes, related to the virulence of Listeria monocytogenes Lm23. Can. J. Microbiol. 2018, 64, 215–221. [Google Scholar] [CrossRef] [Green Version]
- Goerges, S.; Aigner, U.; Silakowski, B.; Scherer, S. Inhibition of Listeria monocytogenes by food-borne yeasts. Appl. Environ. Microbiol. 2006, 72, 313–318. [Google Scholar] [CrossRef] [Green Version]
- Silva, T.L.; Reto, M.; Sol, M.; Peito, A.; Peres, C.; Malcata, F. Characterization of yeasts from Portuguese brined olives, with a focus on their potentially probiotic behavior. LWT 2011, 44, 1349–1354. [Google Scholar] [CrossRef]
- Alía, A.; Córdoba, J.J.; Rodríguez, A.; García, C.; Andrade, M.J. Evaluation of the efficacy of Debaryomyces hansenii as protective culture for controlling Listeria monocytogenes in sliced dry-cured ham. LWT 2020, 119, 108886. [Google Scholar] [CrossRef]
- Ozmen Togay, S.; Capece, A.; Siesto, G.; Aksu, H.; Sandikci Altunatmaz, S.; Yilmaz Aksu, F.; Romano, P.; Karagul Yuceer, Y. Molecular characterization of yeasts isolated from traditional Turkish cheeses. Food Sci. Technol. (Camp.) 2020, 40, 871–876. [Google Scholar] [CrossRef]
- Kim, Y.J.; Yu, H.H.; Song, Y.J.; Park, Y.J.; Lee, N.-K.; Paik, H.-D. Anti-biofilm effect of the cell-free supernatant of probiotic Saccharomyces cerevisiae against Listeria monocytogenes. Food Control. 2021, 121, 107667. [Google Scholar] [CrossRef]
- Radoshevich, L.; Cossart, P. Listeria monocytogenes: Towards a complete picture of its physiology and pathogenesis. Nat. Rev. Genet. 2018, 16, 32–46. [Google Scholar] [CrossRef]
- Gaballa, A.; Oropeza, V.G.; Wiedmann, M.; Boor, K.J. Cross Talk between SigB and PrfA in Listeria monocytogenes facilitates transitions between extra- and intracellular environments. Microbiol. Mol. Biol. Rev. 2019, 83, 00034-19. [Google Scholar] [CrossRef] [PubMed]
- Leimeister-Wächter, M.; Domann, E.; Chakraborty, T. The expression of virulence genes in Listeria monocytogenes is thermoregulated. J. Bacteriol. 1992, 174, 947–952. [Google Scholar] [CrossRef] [Green Version]
- Phelps, C.; Vadia, S.; Arnett, E.; Tan, Y.; Zhang, X.; Pathak-Sharma, S.; Gavrilin, M.A.; Seveau, S. Relative roles of Listeriolysin O, InlA, and InlB in Listeria monocytogenes uptake by host cells. Infect. Immun. 2018, 86, e00555-18. [Google Scholar] [CrossRef] [Green Version]
- Sabet, C.; Lecuit, M.; Cabanes, D.; Cossart, P.; Bierne, H. LPXTG protein InlJ, a newly identified internalin involved in Listeria monocytogenes virulence. Infect. Immun. 2005, 73, 6912–6922. [Google Scholar] [CrossRef] [Green Version]
- Dramsi, S.; Biswas, I.; Maguin, E.; Braun, L.; Mastroeni, P.; Cossart, P. Entry of Listeria monocytogenes into hepatocytes requires expression of InIB, a surface protein of the internalin multigene family. Mol. Microbiol. 1995, 16, 251–261. [Google Scholar] [CrossRef]
- Engelbrecht, F.; Chun, S.K.; Ochs, C.; Hess, J.; Lottspeich, F.; Goebel, W.; Sokolovic, Z. A new PrfA-regulated gene of Listeria monocytogenes encoding a small, secreted protein which belongs to the family of internalins. Mol. Microbiol. 1996, 21, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Dussurget, O. New insights into determinants of Listeria monocytogenes virulence. Int. Rev. Cell Mol. Biol. 2008, 270, 1–38. [Google Scholar] [CrossRef] [PubMed]

| L. monocytogenes LQC 15257 | S. cerevisiae Y32 | S. cerevisiae Y34 | S. cerevisiae Y37 | |
|---|---|---|---|---|
| Initial inoculum | 7.12 (0.20) | 4.10 (0.15) | 3.95 (0.24) | 4.20 (0.22) |
| Mono-culture | ||||
| 5 °C 24 h | 7.61 (0.27) a | 4.12 (0.20) a | 4.27 (0.23) a | 4.20 (0.20) a |
| 5 °C 48 h | 7.72 (0.18) a | 4.35 (0.15) a | 4.52 (0.20) a | 4.40 (0.34) a |
| 20 °C 24 h | 9.29 (0.21) a | 5.68 (0.21) a | 5.49 (0.18) a | 5.60 (0.25) a |
| 20 °C 48 h | 9.00 (0.17) a | 6.12 (0.24) b | 5.95 (0.10) b | 6.10 (0.11) b |
| Co-culture | ||||
| 5 °C 24 h | 7.75 (0.19) a | 4.20 (0.10) a | - | - |
| 7.70 (0.31) a | - | 4.36 (0.21) a | - | |
| 7.71 (0.12) a | - | - | 4.25 (0.15) a | |
| 5 °C 48 h | 7.77 (0.07) a | 4.31 (0.12) a | - | - |
| 7.59 (0.14) a | - | 4.40 (0.08) a | - | |
| 7.57 (0.18) a | - | - | 4.35 (0.14) a | |
| 20 °C 24 h | 9.24 (0.12) a | 5.57 (0.20) a | - | - |
| 9.17 (0.14) a | - | 5.57 (0.15) a | - | |
| 9.10 (0.15) a | - | - | 5.69 (0.19) a | |
| 20 °C 48 h | 9.07 (0.30) a | 6.10 (0.15) b | - | - |
| 9.14 (0.21) a | - | 5.97 (0.12) b | - | |
| 9.11 (0.15) a | - | - | 6.12 (0.17) b |
| log2(FC) | Effect of Yeast Strains 1 | Effect of Incubation Time 2 | Effect of Incubation Temperature 3 |
|---|---|---|---|
| prfA | |||
| <−1 | 2 (16.7) | 0 (0.0) | 0 (0.0) |
| −1 to 1 | 10 (83.3) | 7 (87.5) | 1 (12.5) |
| >1 | 0 (0.0) | 1 (12.5) | 7 (87.5) |
| sigB | |||
| <−1 | 2 (16.7) | 3 (37.5) | 0 (0.0) |
| −1 to 1 | 7 (33.3) | 3 (37.5) | 3 (37.5) |
| >1 | 3 (25.0) | 2 (25.0) | 5 (62.5) |
| plcA | |||
| <−1 | 2 (16.7) | 0 (0.0) | 0 (0.0) |
| −1 to 1 | 10 (83.3) | 6 (75.0) | 6 (75.0) |
| >1 | 0 (0.0) | 2 (25.0) | 2 (25.0) |
| plcB | |||
| <−1 | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| −1 to 1 | 10 (83.3) | 6 (75.0) | 2 (25.0) |
| >1 | 2 (16.7) | 2 (25.0) | 6 (75.0) |
| hly | |||
| <−1 | 0 (0.0) | 1 (12.5) | 0 (0.0) |
| −1 to 1 | 10 (83.3) | 7 (87.5) | 1 (12.5) |
| >1 | 2 (16.7) | 0 (0.0) | 7 (87.5) |
| inlA | |||
| <−1 | 0 (0.0) | 3 (37.5) | 0 (0.0) |
| −1 to 1 | 12 (100.0) | 5 (62.5) | 1 (12.5) |
| >1 | 0 (0.0) | 0 (0.0) | 7 (87.5) |
| inlB | |||
| <−1 | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| −1 to 1 | 12 (100.0) | 8 (100.0) | 0 (0.0) |
| >1 | 0 (0.0) | 0 (0.0) | 8 (100.0) |
| inlC | |||
| <−1 | 1 (8.3) | 0 (0.0) | 0 (0.0) |
| −1 to 1 | 10 (83.3) | 8 (100.0) | 0 (0.0) |
| >1 | 1 (8.3) | 0 (0.0) | 8 (100.0) |
| inlJ | |||
| <−1 | 0 (0.0) | 2 (25.0) | 0 (0.0) |
| −1 to 1 | 8 (66.6) | 4 (50.0) | 1 (12.5) |
| >1 | 4 (33.3) | 2 (25.0) | 7 (87.5) |
| Total | |||
| <−1 | 7 (6.5) | 9 (12.5) | 0 (0.0) |
| −1 to 1 | 89 (82.5) | 54 (75.0) | 15 (20.8) |
| >1 | 12 (11.0) | 9 (12.5) | 57 (79.2) |
| prfA | sigB | plcA | plcB | hly | inlA | inlB | inlC | ||
|---|---|---|---|---|---|---|---|---|---|
| sigB | r | 0.4792 | |||||||
| p | 0.0099 | ||||||||
| plcA | r | 0.6961 | 0.6497 | ||||||
| p | 0.0000 | 0.0002 | |||||||
| plcB | r | 0.6065 | 0.7028 | 0.7614 | |||||
| p | 0.0006 | 0.0000 | 0.0000 | ||||||
| hly | r | 0.4862 | 0.2925 | 0.2265 | 0.3533 | ||||
| p | 0.0087 | 0.1309 | 0.2465 | 0.0652 | |||||
| inlA | r | 0.5698 | 0.5640 | 0.2897 | 0.3161 | 0.6024 | |||
| p | 0.0015 | 0.0018 | 0.1349 | 0.1013 | 0.0007 | ||||
| inlB | r | 0.8461 | 0.4665 | 0.4898 | 0.5449 | 0.4646 | 0.8140 | ||
| p | 0.0000 | 0.0123 | 0.0081 | 0.0027 | 0.0128 | 0.0000 | |||
| inlC | r | 0.9004 | 0.3930 | 0.6235 | 0.4746 | 0.3879 | 0.5914 | 0.8211 | |
| p | 0.0000 | 0.0385 | 0.0004 | 0.0107 | 0.0414 | 0.0009 | 0.0000 | ||
| inlJ | r | 0.1837 | 0.7920 | 0.4719 | 0.6052 | −0.0284 | 0.3145 | 0.2403 | 0.3003 |
| p | 0.3495 | 0.0000 | 0.0112 | 0.0006 | 0.8860 | 0.1031 | 0.2180 | 0.1205 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paramithiotis, S.; Katidi, A.; Drosinos, E.H. Transcriptomic Response of L. monocytogenes to Co-Culture with S. cerevisiae. Beverages 2021, 7, 55. https://doi.org/10.3390/beverages7030055
Paramithiotis S, Katidi A, Drosinos EH. Transcriptomic Response of L. monocytogenes to Co-Culture with S. cerevisiae. Beverages. 2021; 7(3):55. https://doi.org/10.3390/beverages7030055
Chicago/Turabian StyleParamithiotis, Spiros, Alexandra Katidi, and Eleftherios H. Drosinos. 2021. "Transcriptomic Response of L. monocytogenes to Co-Culture with S. cerevisiae" Beverages 7, no. 3: 55. https://doi.org/10.3390/beverages7030055
APA StyleParamithiotis, S., Katidi, A., & Drosinos, E. H. (2021). Transcriptomic Response of L. monocytogenes to Co-Culture with S. cerevisiae. Beverages, 7(3), 55. https://doi.org/10.3390/beverages7030055








