Sugary Kefir: Microbial Identification and Biotechnological Properties
Abstract
1. Introduction
2. Materials and Methods
2.1. Origin and Types of Sugar Kefir Grains
2.2. Fermentation Tests
2.3. Yeasts and Lactic Acid Bacteria Enumeration and Isolation
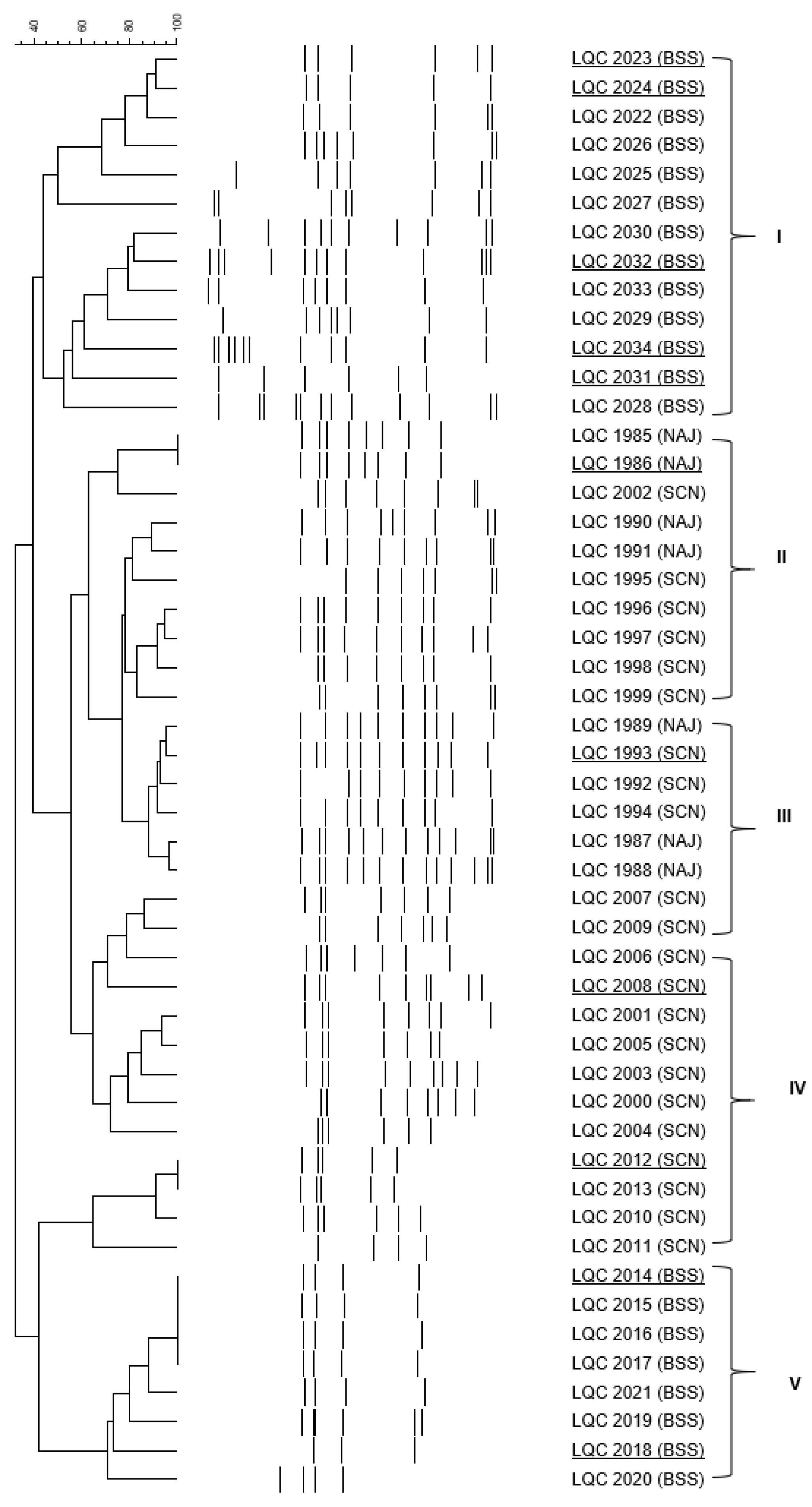
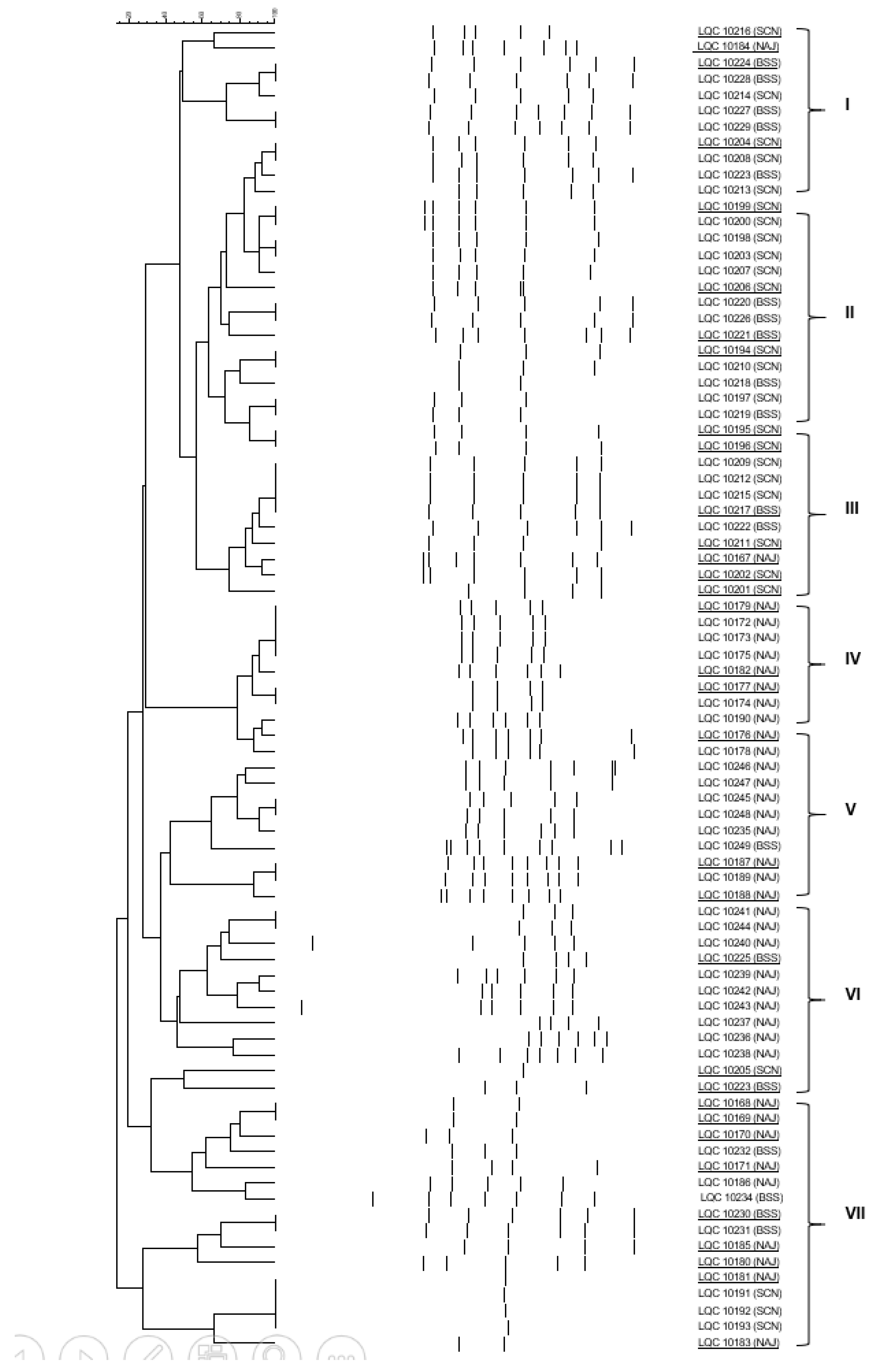
2.4. Clustering and Identification of the Isolates
2.5. Assessment of Technological Properties
2.5.1. Proteolytic Activity
2.5.2. Lipolytic Activity
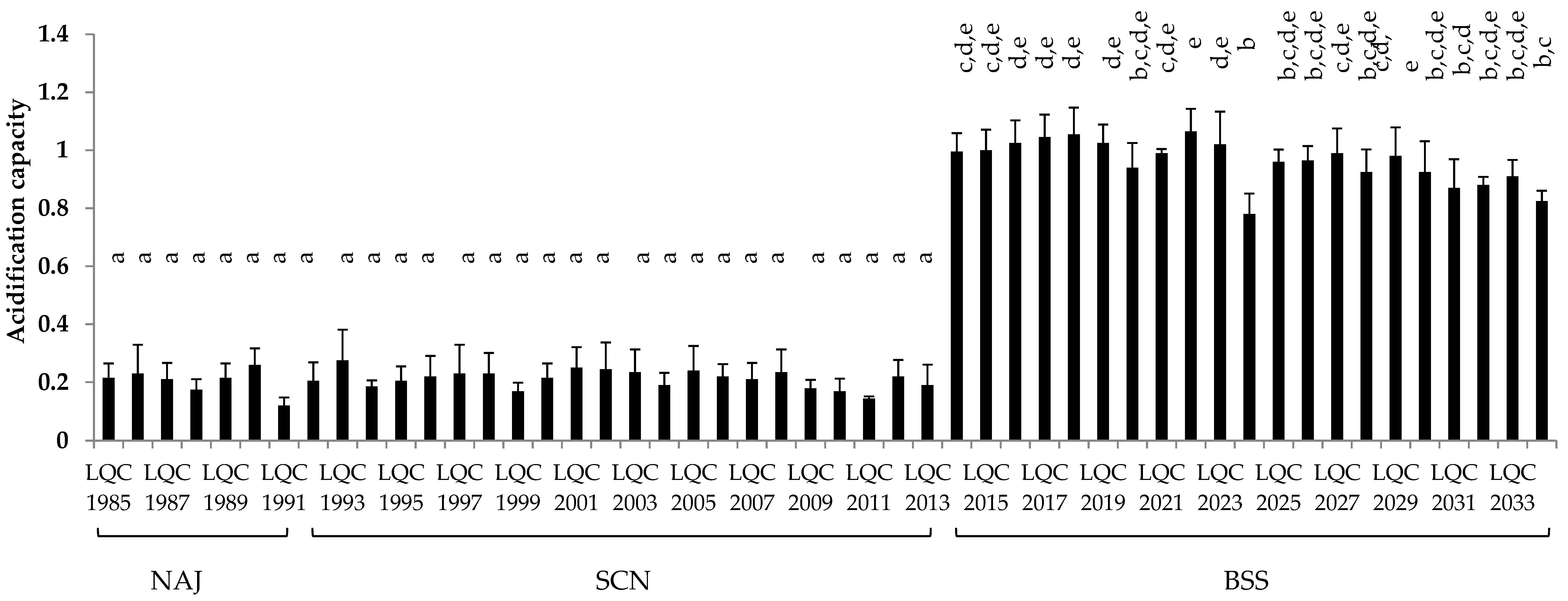
2.5.3. Acidification Capacity
2.5.4. Antimicrobial Activity
2.6. Analytical Methods
3. Results
3.1. Fermentation Tests
3.2. Yeast and Bacterial Enumeration in Kefir Grains
3.3. Assessment of Technological Properties
3.3.1. Proteolytic Activity
3.3.2. Lipolytic Activity
3.3.3. Acidification Capacity
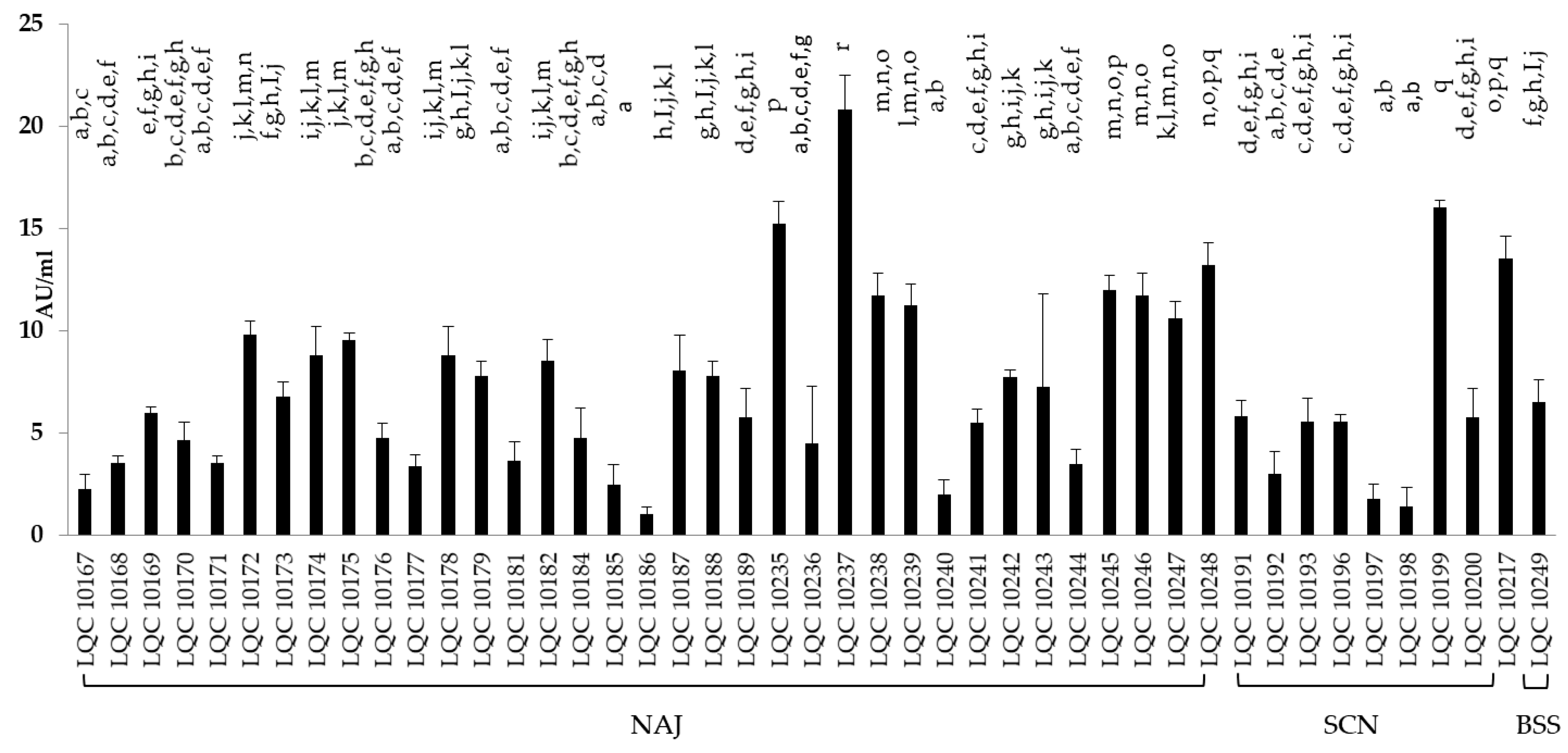
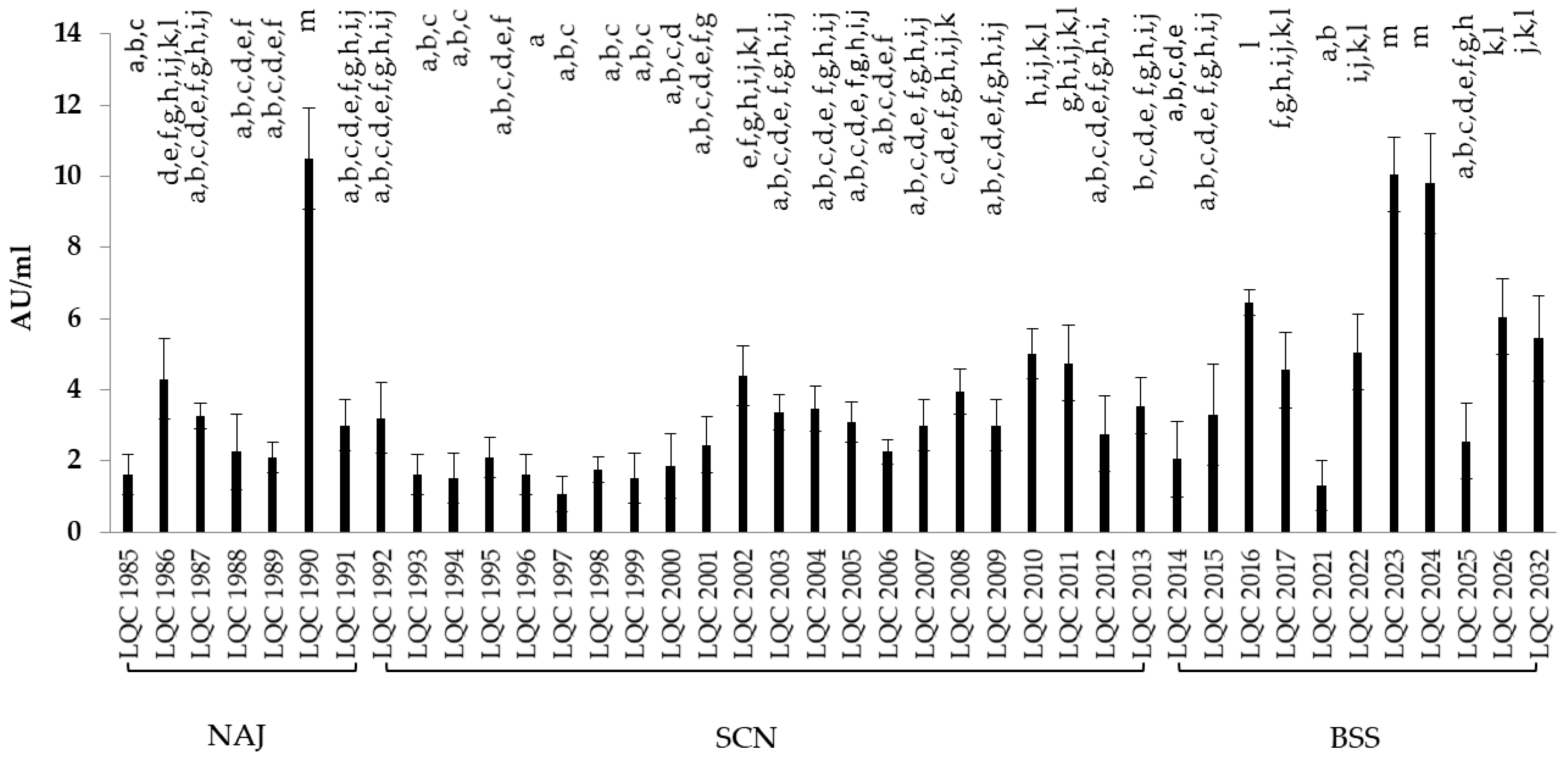
3.3.4. Antimicrobial Activity
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Prado, M.R.; Blandón, L.M.; Vandenberghe, L.P.S.; Rodrigues, C.; Castro, G.R.; Thomaz-Soccol, V.; Soccol, C.R. Milk kefir: Composition, microbial cultures, biological activities, and related products. Front. Microbiol. 2015, 6, 422. [Google Scholar] [CrossRef] [PubMed]
- Gulitz, A.; Stadie, J.; Wenning, M.; Ehrmann, M.A.; Vogel, R.F. The microbial diversity of water kefir. Int. J. Food Microbiol. 2011, 151, 284–288. [Google Scholar] [CrossRef] [PubMed]
- Fiorda, F.A.; Pereira, G.V.D.M.; Thomaz-Soccol, V.; Rakshit, S.K.; Pagnoncelli, M.G.B.; Vandenberghe, L.P.D.S.; Soccol, C.R. Microbiological, biochemical, and functional aspects of sugary kefir fermentation—A review. Food Microbiol. 2017, 66, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Pidoux, M. The microbial flora of sugary kefir grain (the ginger beer plant): Biosynthesis of the grain from Lactobacillus hilgardii producing a polysaccharide gel. MIRCEN J. Appl. Microbiol. Biotechnol. 1989, 5, 223–238. [Google Scholar] [CrossRef]
- Magalhães, K.T.; Pereira, G.V.D.M.; Dias, D.R.; Schwan, R.F.; Pereira, G.V.M. Microbial communities and chemical changes during fermentation of sugary Brazilian kefir. World J. Microbiol. Biotechnol. 2010, 26, 1241–1250. [Google Scholar] [CrossRef]
- Sarikkha, P.; Nitisoravut, R.; Poljungreed, I.; Boonyarattanakalin, S. Identification of bacteria and yeast communities in a Thai sugary kefir by polymerase chain reaction-denaturing gradient gel electrophoresis (PCR-DGGE) analyses. J. Ind. Technol. 2015, 11, 25–39. [Google Scholar]
- Doulgeraki, A.I.; Paramithiotis, S.; Kagkli, D.M.; Nychas, G.-J.E. Lactic acid bacteria population dynamics during minced beef storage under aerobic or modified atmosphere packaging conditions. Food Microbiol. 2010, 27, 1028–1034. [Google Scholar] [CrossRef]
- Vogel, B.F.; Fussing, V.; Ojeniyi, B.; Gram, L.; Ahrens, P. High-Resolution Genotyping of Listeria monocytogenes by Fluorescent Amplified Fragment Length Polymorphism Analysis compared to Pulsed-Field Gel Electrophoresis, Random Amplified Polymorphic DNA Analysis, Ribotyping, and PCR–Restriction Fragment Length Polymorphism Analysis. J. Food Prot. 2004, 67, 1656–1665. [Google Scholar]
- Drosinos, E.H.; Paramithiotis, S.; Kolovos, G.; Tsikouras, I.; Metaxopoulos, I. Phenotypic and technological diversity of lactic acid bacteria and staphylococci isolated from traditionally fermented sausages in Southern Greece. Food Microbiol. 2007, 24, 260–270. [Google Scholar] [CrossRef]
- Kamzolova, S.V.; Morgunov, I.G.; Aurich, A.; Perevoznikova, O.A.; Shishkanova, N.V.; Stottmeister, U.U.; Finogenova, T.V. Lipase secretion and citric acid production in Yarrowia lipolytica yeast grown on animal and vegetable fat. Food Technol. Biotechnol. 2005, 43, 113–122. [Google Scholar]
- Paramithiotis, S.; Grivokostopoulos, N.; Skandamis, P.N. Investigating the correlation of constitutive proteins with the growth limits of Salmonella enterica isolates from feeds in response to temperature, pH, formic and lactic acid. Food Res. Int. 2013, 53, 291–296. [Google Scholar] [CrossRef]
- Hadjilouka, A.; Mantzourani, K.S.; Katsarou, A.; Cavaiuolo, M.; Ferrante, A.; Paramithiotis, S.; Mataragas, M.; Drosinos, E.H. Estimation of Listeria monocytogenes and Escherichia coli O157:H7 Prevalence and levels in naturally contaminated rocket and cucumber samples by Deterministic and Stochastic Approaches. J. Food Prot. 2015, 78, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Randazzo, W.; Corona, O.; Guarcello, R.; Francesca, N.; Germanà, M.A.; Erten, H.; Moschetti, G.; Settanni, L. Development of new non-dairy beverages from Mediterranean fruit juices fermented with water kefir microorganisms. Food Microbiol. 2016, 54, 40–51. [Google Scholar] [CrossRef]
- Witthuhn, R.C.; Schoeman, T.; Britz, T.J. Isolation and characterization of the microbial population of different South African kefir grains. Int. J. Dairy Technol. 2004, 57, 33–37. [Google Scholar] [CrossRef]
- Miguel, M.; Cardoso, P.; Magalhães-Guedes, K.; Schwan, R. Identification and assessment of kefir yeast potential for sugar/ethanol-resistance. Braz. J. Microbiol. 2013, 44, 113–118. [Google Scholar] [CrossRef][Green Version]
- Lu, M.; Wang, X.; Sun, G.; Qin, B.; Xiao, J.; Yan, S.; Pan, Y.; Wang, Y. Fine structure of tibetan kefir grains and their yeast distribution, diversity, and shift. PLoS ONE 2014, 9, e101387. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, H.; Wang, S.; Chen, T.; Huang, Y.; Chen, M. Effects of cow’s and goat’s milk as fermentation media on the microbial ecology of sugary kefir grains. Int. J. Food Microbiol. 2012, 157, 73–81. [Google Scholar] [CrossRef]
- Zanirati, D.F.; Abatemarco, M., Jr.; de Cicco Sandes, S.H.; Nicoli, J.R.; Cantini Nunes, A.; Neumann, E. Selection of lactic acid bacteria from Brazilian kefir grains for potential use as starter or probiotic cultures. Anaerobe 2015, 32, 70–76. [Google Scholar] [CrossRef]
- Gao, J.; Gu, F.; Abdella, N.H.; Ruan, H.; He, G. Investigation on culturable microflora in tibetan kefir grains from different areas of china. J. Food Sci. 2012, 77, M425–M433. [Google Scholar] [CrossRef]
- Lee, A.; Cheng, K.-C.; Liu, J.-R. Isolation and characterization of a Bacillus amyloliquefaciens strain with zearalenone removal ability and its probiotic potential. PLoS ONE 2017, 12, e0182220. [Google Scholar] [CrossRef] [PubMed]
- Hanafy, A.M.; Al-Mutairi, A.A.; Al-Reedy, R.M.; Al-Garni, S.M. Phylogenetic affiliations of Bacillus amyloliquefaciens isolates produced by a bacteriocin-like substance in goat milk. J. Taibah Univ. Sci. 2016, 10, 631–641. [Google Scholar] [CrossRef]
- Boottanun, P.; Potisap, C.; Hurdle, J.G.; Sermswan, R.W. Secondary metabolites from Bacillus amyloliquefaciens isolated from soil can kill Burkholderia pseudomallei. AMB Express 2017, 7, 16. [Google Scholar] [CrossRef] [PubMed]
- Stein, T. Bacillus subtilis antibiotics: Structures, syntheses and specific functions. Mol. Microbiol. 2005, 56, 845–857. [Google Scholar] [CrossRef]
- Kort, R.; Westerik, N.; Serrano, L.M.; Douillard, F.P.; Gottstein, W.; Mukisa, I.M.; Tuijn, C.J.; Basten, L.; Hafkamp, B.; Meijer, W.C.; et al. A novel consortium of Lactobacillus rhamnosus and Streptococcus thermophilus for increased access to functional fermented foods. Microb. Cell Factories 2015, 14, 195. [Google Scholar] [CrossRef]
- Parker, M.; Zobrist, S.; Donahue, C.; Edick, C.; Mansen, K.; Hassan Zade Nadjari, M.; Heerikhuisen, M.; Sybesma, W.; Molenaar, D.; Diallo, A.M.; et al. Naturally ermented milk from northern Senegal: Bacterial community composition and probiotic enrichment with Lactobacillus rhamnosus. Front. Microbiol. 2018, 21, 2218. [Google Scholar] [CrossRef]
- Rajoka, M.S.R.; Mehwish, H.M.; Siddiq, M.; Haobin, Z.; Zhu, J.; Yan, L.; Shao, D.; Xu, X.; Shi, J. Identification, characterization, and probiotic potential of Lactobacillus rhamnosus isolated from human milk. LWT 2017, 84, 271–280. [Google Scholar] [CrossRef]
- Ayad, E.; Nashat, S.; El-Sadek, N.; Metwaly, H.; El-Soda, M. Selection of wild lactic acid bacteria isolated from traditional Egyptian dairy products according to production and technological criteria. Food Microbiol. 2004, 21, 715–725. [Google Scholar] [CrossRef]
- Sodini, I.; Lucas, A.; Oliveira, M.; Remeuf, F.; Corrieu, G. Effect of milk base and starter culture on acidification, texture, and probiotic cell counts in fermented milk processing. J. Dairy Sci. 2002, 85, 2479–2488. [Google Scholar] [CrossRef]
- Marroki, A.; Zuñiga, M.; Kihal, M.; Martínez, G.P. Characterization of Lactobacillus from Algerian goat’s milk based on phenotypic, 16S rDNA sequencing and their technological properties. Braz. J. Microbiol. 2011, 42, 158–171. [Google Scholar] [CrossRef]
- Mora, L.; Gallego, M.; Escudero, E.; Reig, M.; Aristoy, M.-C.; Toldrá, F.; Soler, L.M. Small peptides hydrolysis in dry-cured meats. Int. J. Food Microbiol. 2015, 212, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Ebner, J.; Aşçı Arslan, A.; Fedorova, M.; Hoffmann, R.R.; Küçükçetin, A.; Pischetsrieder, M. Peptide profiling of bovine kefir reveals 236 unique peptides released from caseins during its production by starter culture or kefir grains. J. Proteom. 2015, 117, 41–57. [Google Scholar] [CrossRef] [PubMed]
- Capuani, A.; Behr, J.; Vogel, R.F. Influence of lactic acid bacteria on redox status and on proteolytic activity of buckwheat (Fagopyrum esculentum Moench) sourdoughs. Int. J. Food Microbiol. 2013, 165, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Kirilov, N.; Petkova, T.; Atanasova, J.; Danova, S.; Iliev, I.; Popov, Y.; Haertle, T.; Ivanova, I.V. Proteolytic activity in lactic acid bacteria from Iraq, Armenia and Bulgaria. Biotechnol. Biotechnol. Equip. 2009, 23, 643–646. [Google Scholar] [CrossRef]
- Dallas, D.C.; Citerne, F.; Tian, T.; Silva, V.L.M.; Kalanetra, K.M.; Frese, S.A.; Robinson, R.C.; Mills, D.A.; Barile, D. Peptidomic analysis reveals proteolytic activity of kefir microorganisms on bovine milk proteins. Food Chem. 2016, 197, 273–284. [Google Scholar] [CrossRef]
- Tulini, F.L.; Hymery, N.; Haertlé, T.; Le Blay, G.; De Martinis, E.C.P. Screening for antimicrobial and proteolytic activities of lactic acid bacteria isolated from cow, buffalo and goat milk and cheeses marketed in the southeast region of Brazil. J. Dairy Res. 2016, 83, 115–124. [Google Scholar] [CrossRef]
- El-Ghaish, S.; Dalgalarrondo, M.; Choiset, Y.; Sitohy, M.; Ivanova, I.; Haertlé, T.; Chobert, J.-M. Screening of strains of Lactococci isolated from Egyptian dairy products for their proteolytic activity. Food Chem. 2010, 120, 758–764. [Google Scholar] [CrossRef]
- Moslehishad, M.; Mirdamadi, S.; Ehsani, M.R.; Ezzatpanah, H.; Moosavi-Movahedi, A.A. The proteolytic activity of selected lactic acid bacteria in fermenting cow’s and camel’s milk and the resultant sensory characteristics of the products. Int. J. Dairy Technol. 2013, 66, 279–285. [Google Scholar] [CrossRef]
- Aljewicz, M.; Cichosz, G.; Nalepa, B.; Kowalska, M. Influence of the probiotic Lactobacillus acidophilus NCFM and Lactobacillus rhamnosus HN001 on proteolysis patterns of Edam cheese. Food Technol. Biotechnol. 2014, 52, 439–447. [Google Scholar] [CrossRef]
- Wang, H.; Yang, L.; Ping, Y.; Bai, Y.; Luo, H.; Huang, H.; Yao, B. Engineering of a Bacillus amyloliquefaciens strain with high neutral protease producing capacity and optimization of its fermentation conditions. PLoS ONE 2016, 11, 0146373. [Google Scholar] [CrossRef]
- Chaudhuri, M.; Biswas, S.; Pal, A.; Paul, A.K. Proteolytic Activity of Bacillus amyloliquefaciens UEF01 endophytic to carnivorous plant Utricularia exoleta R. Br. Biotechnol. J. Int. 2017, 17, 1–11. [Google Scholar] [CrossRef]
- Samad Abd, N.S.; Amid, A.; Jimat, D.N.; Shukor, N.A.A. Protease purification from Bacillus amyloliquefaciens B7 using Aqueous Two-Phase System (ATPS). Int. Food Res. J. 2017, 24, 292–297. [Google Scholar]
- Strongin, A.Y.; Izotova, L.S.; Abramov, Z.T.; Gorodetsky, D.I.; Ermakova, L.M.; Baratova, L.A.; Belyanova, L.P.; Stepanov, V.M. Intracellular serine protease of Bacillus subtilis: Sequence homology with extracellular subtilisins. J. Bacteriol. 1978, 133, 1401–1411. [Google Scholar] [PubMed]
- Jones, E.W. Three proteolytic systems in the yeast Saccharomyces cerevisiae. J. Boil. Chem. 1991, 266, 7963–7966. [Google Scholar]
- Atanassova, M.; Fernández-Otero, C.; Rodríguez-Alonso, P.; Fernández-No, I.; Garabal, J.; Centeno, J.A. Characterization of yeasts isolated from artisanal short-ripened cows’ cheeses produced in Galicia (NW Spain). Food Microbiol. 2016, 53, 172–181. [Google Scholar] [CrossRef]
- Klein, N.; Zourari, A.; Lortal, S. Peptidase activity of four yeast species frequently encountered in dairy products—comparison with several dairy bacteria. Int. Dairy J. 2002, 12, 853–861. [Google Scholar] [CrossRef]
- Hansen, T.K.; Jakobsen, M. Taxonomical and technological characteristics of Saccharomyces spp. associated with blue veined cheese. Int. J. Food Microbiol. 2001, 69, 59–68. [Google Scholar] [CrossRef]
- Seredyński, R.; Wolna, D.; Kędzior, M.; Gutowicz, J. Different patterns of extracellular proteolytic activity in W303a and BY4742 Saccharomyces cerevisiae strains. J. Basic Microbiol. 2017, 57, 34–40. [Google Scholar] [CrossRef]
- Beniwal, A.; Saini, P.; Kokkiligadda, Α.; Vij, S. Physiological growth and galactose utilization by dairy yeast Kluyveromyces marxianus in mixed sugars and whey during fermentation. 3 Biotech. 2017, 7, 349. [Google Scholar] [CrossRef]
- Ramírez-Zavala, B.; Mercado-Flores, Y.; Hernández-Rodríguez, C.; Villa-Tanaca, L. Purification and characterization of a lysine aminopeptidase from Kluyveromyces marxianus. FEMS Microbiol. Lett. 2004, 235, 369–375. [Google Scholar] [CrossRef]
- Deeth, H.C.; Touch, V. Methods for detecting lipase activity in milk and milk products. Aust. J. Dairy Technol. 2000, 55, 153–168. [Google Scholar]
- Chen, Q.; Kong, B.; Han, Q.; Xia, X.; Xu, L. The role of bacterial fermentation in lipolysis and lipid oxidation in Harbin dry sausages and its flavour development. LWT 2017, 77, 389–396. [Google Scholar] [CrossRef]
- Karami, M. RETRACTED: Enhancing the lipolysis of feta-type cheese made from ultrafiltered cow’s milk. LWT 2017, 80, 386–393. [Google Scholar] [CrossRef]
- Aponte, M.; Blaiotta, G. Selection of an autochthonous Saccharomyces cerevisiae strain for the vinification of “Moscato di Saracena”, a southern Italy (Calabria Region) passito wine. Food Microbiol. 2016, 54, 30–39. [Google Scholar] [CrossRef]
- Rodríguez-Gómez, F.; Arroyo-Lopez, F.N.; López-López, A.; Bautista-Gallego, J.; Garrido-Fernandez, A. Lipolytic activity of the yeast species associated with the fermentation/storage phase of ripe olive processing. Food Microbiol. 2010, 27, 604–612. [Google Scholar] [CrossRef]
- Hernández, A.; Martín, A.; Aranda, E.; Pérez Nevado, F.; Córdoba, M.G. Identification and characterization of yeasts isolated from the elaboration of seasoned green table olives. Food Microbiol. 2006, 24, 346–351. [Google Scholar] [CrossRef]
- Cardoso, V.M.; Borelli, B.M.; Lara, C.A.; Soares, M.A.; Pataro, C.; Bodevan, E.C.; Rosa, C.A. The influence of seasons and ripening time on yeast communities of a traditional Brazilian cheese. Food Res. Int. 2015, 69, 331–340. [Google Scholar] [CrossRef]
- Meng, Z.; Zhang, L.; Xin, L.; Lin, K.; Yi, H.; Han, X. Technological characterization of Lactobacillus in semihard artisanal goat cheeses from different Mediterranean areas for potential use as nonstarter lactic acid bacteria. J. Dairy Sci. 2018, 101, 2887–2896. [Google Scholar] [CrossRef]
- Collins, Y.F.; McSweeney, P.L.; Wilkinson, M.G. Lipolysis and free fatty acid catabolism in cheese: A review of current knowledge. Int. Dairy J. 2003, 13, 841–866. [Google Scholar] [CrossRef]
- Falcinelli, S.; Picchietti, S.; Rodiles, A.; Cossignani, L.; Merrifield, D.L.; Taddei, A.R.; Maradonna, F.; Olivotto, I.; Gioacchini, G.; Carnevali, O. Lactobacillus rhamnosus lowers zebrafish lipid content by changing gut microbiota and host transcription of genes involved in lipid metabolism. Sci. Rep. 2015, 5, 9336. [Google Scholar] [CrossRef]
- Cai, X.; Ma, J.; Wei, D.-Z.; Lin, J.-P.; Wei, W. Functional expression of a novel alkaline-adapted lipase of Bacillus amyloliquefaciens from stinky tofu brine and development of immobilized enzyme for biodiesel production. Antonie Leeuwenhoek 2014, 106, 1049–1060. [Google Scholar] [CrossRef]
- Saengsanga, T.; Siripornadulsil, W.; Siripornadulsil, S. Molecular and enzymatic characterization of alkaline lipase from Bacillus amyloliquefaciens E1PA isolated from lipid-rich food waste. Enzym. Microb. Technol. 2016, 82, 23–33. [Google Scholar] [CrossRef]
- Abbasiliasi, S.; Tan, J.S.; Ibrahim, T.A.T.; Bashokouh, F.; Ramakrishnan, N.R.; Mustafa, S.; Ariff, A.B. Fermentation factors influencing the production of bacteriocins by lactic acid bacteria: A review. RSC Adv. 2017, 7, 29395–29420. [Google Scholar] [CrossRef]
- Oliveira, L.D.C.; Silveira, A.M.M.; Monteiro, A.D.S.; Dos Santos, V.L.; Nicoli, J.R.; Azevedo, V.A.D.C.; Soares, S.D.C.; Dias-Souza, M.V.; Nardi, R.M.D. In silico prediction, in vitro antibacterial spectrum, and physicochemical properties of a putative bacteriocin produced by Lactobacillus rhamnosus strain L156.4. Front. Microbiol. 2017, 8, 876. [Google Scholar] [CrossRef]
- Drosinos, E.H.; Mataragas, M.; Paramithiotis, S. Antimicrobial activity of bacteriocins and their applications. In Meat Biotechnology; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2008; pp. 375–397. [Google Scholar]
| Substrate | pH0 | pHf | S0 Sugars (%, w/v) | Sf Sugars (%, w/v) | Ethanol (%, v/v) | Lactic Acid (%, w/v) | Acetic Acid (mg/L) |
|---|---|---|---|---|---|---|---|
| BSS | 6.9 ± 0.01 | 4.1 ± 0.01 | 5.03 ± 0.2 | 0.93 ±0.11 | 1.96 ± 0.11 | 162.12 ± 12.09 | nd |
| SCN | 3.9 ± 0.01 | 3.5 ± 0.02 | 13.2 ± 0.2 | 8.92 ± 0.1 | 2.14 ± 0.12 | 148.35 ± 9.46 | nd |
| NAJ | 4 ± 0.01 | 3.7 ± 0.01 | 9.2 ± 0.1 | 6.96 ± 0.1 | 0.85 ± 0.04 | 134.13 ± 8.78 | nd |
| Strain Number | Closest Relative | Identity (%) | Accession Number | |
|---|---|---|---|---|
| Yeasts | LQC 10194 | S. cerevisiae | 99 | MG017543.1 |
| LQC 10195 | S. cerevisiae | 99 | MG017587.1 | |
| LQC 10196 | S. cerevisiae | 99 | MG017572.1 | |
| LQC 10199 | S. cerevisiae | 99 | MG017572.1 | |
| LQC 10201 | S. cerevisiae | 99 | MG017587.1 | |
| LQC 10202 | S. cerevisiae | 99 | MG017588.1 | |
| LQC 10204 | S. cerevisiae | 99 | MG017572.1 | |
| LQC 10205 | S. cerevisiae | 100 | JQ964228.1 | |
| LQC 10206 | S. cerevisiae | 100 | MG017572.1 | |
| LQC 10211 | S. cerevisiae | 100 | MF406146.1 | |
| LQC 10216 | S. cerevisiae | 100 | KR069090.1 | |
| LQC 10167 | S. cerevisiae | 100 | MG017572.1 | |
| LQC 10168 | S. cerevisiae | 99 | JQ964228.1 | |
| LQC 10170 | S. cerevisiae | 100 | MF406146.1 | |
| LQC 10171 | S. cerevisiae | 100 | MF406146.1 | |
| LQC 10176 | S. cerevisiae | 99 | KJ660850.1 | |
| LQC 10177 | K. marxianus | 99 | KJ491106.1 | |
| LQC 10179 | K. marxianus | 99 | KJ491106.1 | |
| LQC 10180 | K. marxianus | 100 | KJ491106.1 | |
| LQC 10181 | S. cerevisiae | 100 | MF406146.1 | |
| LQC 10182 | K. marxianus | 100 | MH244202.1 | |
| LQC 10183 | S. cerevisiae | 100 | KJ660848.1 | |
| LQC 10184 | S. cerevisiae | 100 | MG017572.1 | |
| LQC 10185 | S. cerevisiae | 99 | MF406146.1 | |
| LQC 10187 | K. marxianus | 99 | KJ491106.1 | |
| LQC 10188 | K. marxianus | 100 | FJ896141.1 | |
| LQC 10217 | S. cerevisiae | 100 | MG017572.1 | |
| LQC 10221 | S. cerevisiae | 99 | MG017572.1 | |
| LQC 10224 | S. cerevisiae | 100 | MG017587.1 | |
| LQC 10225 | S. cerevisiae | 99 | MG017585.1 | |
| LQC 10230 | S. cerevisiae | 100 | MF406146.1 | |
| LQC 10233 | S. cerevisiae | 100 | MF406146.1 | |
| Bacteria | LQC 1986 | B. amyloliquefaciens | 99 | KY392912.1 |
| LQC 1993 | B. amyloliquefaciens | 98 | MH045777.1 | |
| LQC 2012 | B. amyloliquefaciens | 99 | KY072769.1 | |
| LQC 2014 | Lb. rhamnosus | 99 | CP020464.1 | |
| LQC 2018 | Lb. rhamnosus | 100 | HQ774719.1 | |
| LQC 2023 | Lb. rhamnosus | 99 | CP020464.1 | |
| LQC 2024 | Lb. rhamnosus | 99 | CP020464.1 | |
| LQC 2031 | Lb. rhamnosus | 99 | LC333198.1 | |
| LQC 2032 | Lb. rhamnosus | 99 | LC333198.1 | |
| LQC 2034 | Lb. rhamnosus | 99 | LC333198.1 | |
| LQC 2008 | B. amyloliquefaciens | 99 | MF953984.1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Syrokou, M.K.; Papadelli, M.; Ntaikou, I.; Paramithiotis, S.; Drosinos, E.H. Sugary Kefir: Microbial Identification and Biotechnological Properties. Beverages 2019, 5, 61. https://doi.org/10.3390/beverages5040061
Syrokou MK, Papadelli M, Ntaikou I, Paramithiotis S, Drosinos EH. Sugary Kefir: Microbial Identification and Biotechnological Properties. Beverages. 2019; 5(4):61. https://doi.org/10.3390/beverages5040061
Chicago/Turabian StyleSyrokou, Maria K., Marina Papadelli, Ioanna Ntaikou, Spiros Paramithiotis, and Eleftherios H. Drosinos. 2019. "Sugary Kefir: Microbial Identification and Biotechnological Properties" Beverages 5, no. 4: 61. https://doi.org/10.3390/beverages5040061
APA StyleSyrokou, M. K., Papadelli, M., Ntaikou, I., Paramithiotis, S., & Drosinos, E. H. (2019). Sugary Kefir: Microbial Identification and Biotechnological Properties. Beverages, 5(4), 61. https://doi.org/10.3390/beverages5040061









