Investigation of Age Gelation in UHT Milk
Abstract
:1. Introduction
2. Materials and Methods
2.1. General
2.2. Milk Parameters
2.3. Nitrogen and Mineral Analysis
2.4. Processing and Storage Conditions
2.5. Microbial Total Plate Count Analysis
2.6. Viscosity Measurements
2.7. Dynamic Light Scattering
2.8. Cryo-Transmission Electron Microscopy (Cryo-TEM)
2.9. Plasmin Activity
2.10. pH Measurements
2.11. Microbial Population Analysis Using Metagenomics
2.11.1. Processing of 16s V4 amplicon DNA Sequences
2.11.2. Analysis of 16s V4 Sequences
2.12. Top-Down Proteomics Analysis by Liquid Chromatography–mass Spectrometry (LC-MS) and Liquid Chromatography–tandem Mass Spectrometry LC-MS/MS
3. Results
3.1. Milk Parameters
3.2. Protein Genetic Variant Differences
3.3. Plasmin Activity
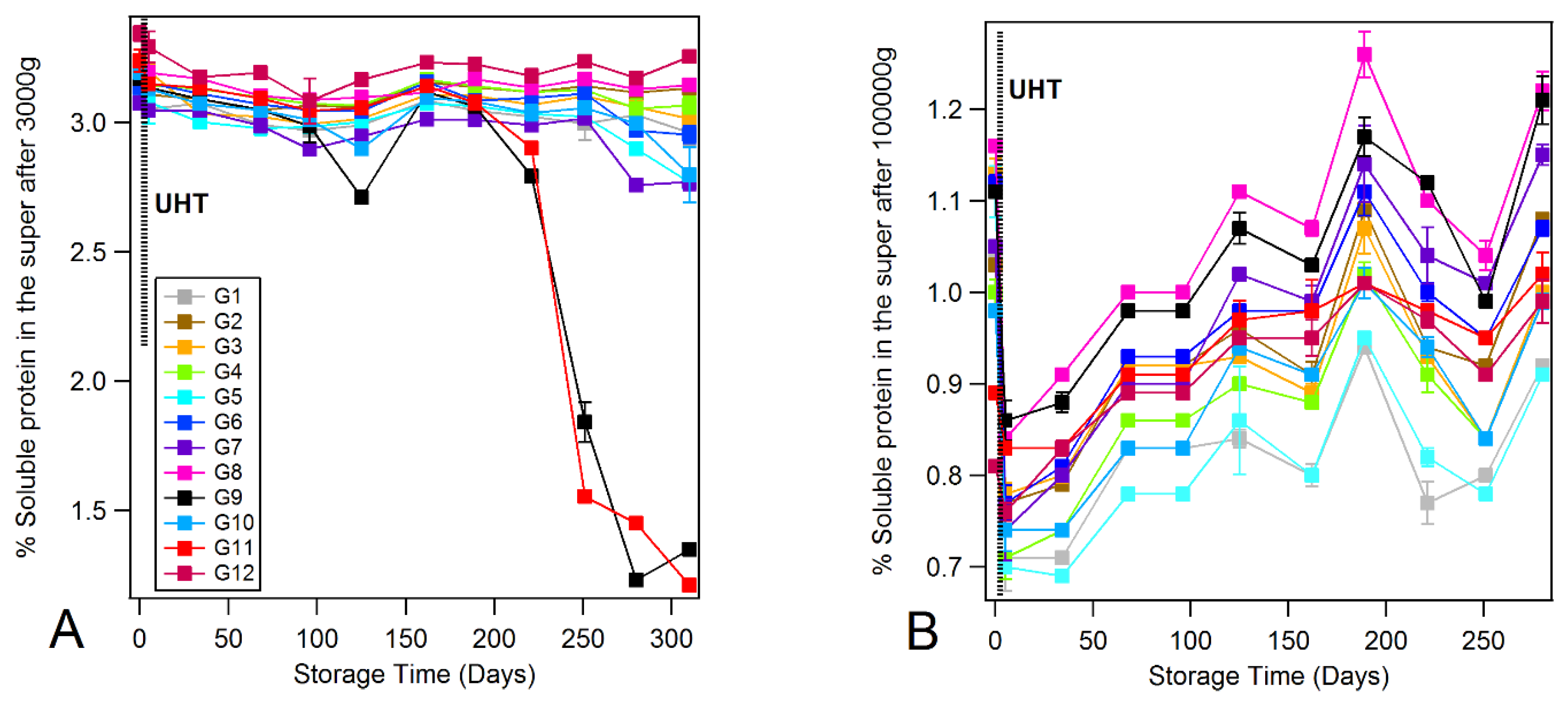
3.4. Monitoring the Protein Solubility State
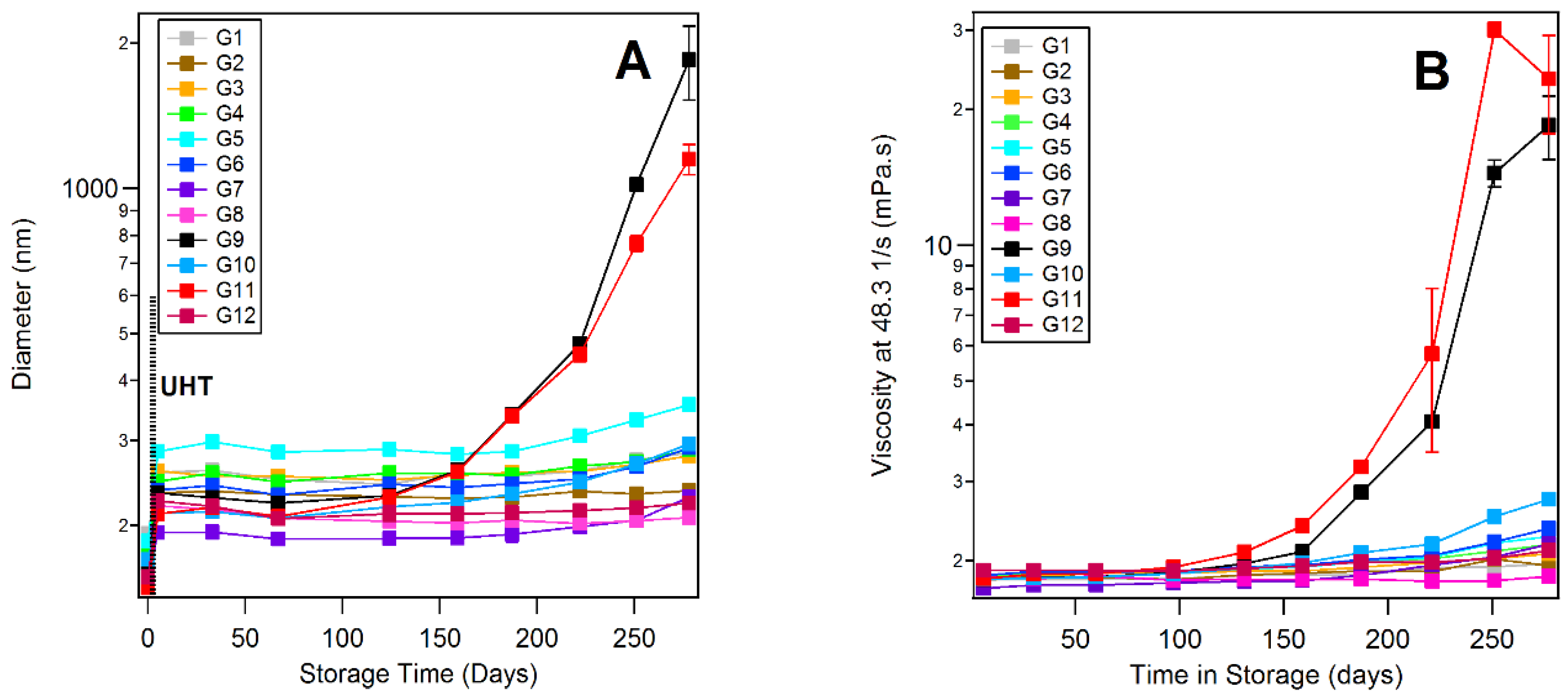
3.5. Dynamic Light Scattering and Rheology of Milk Samples
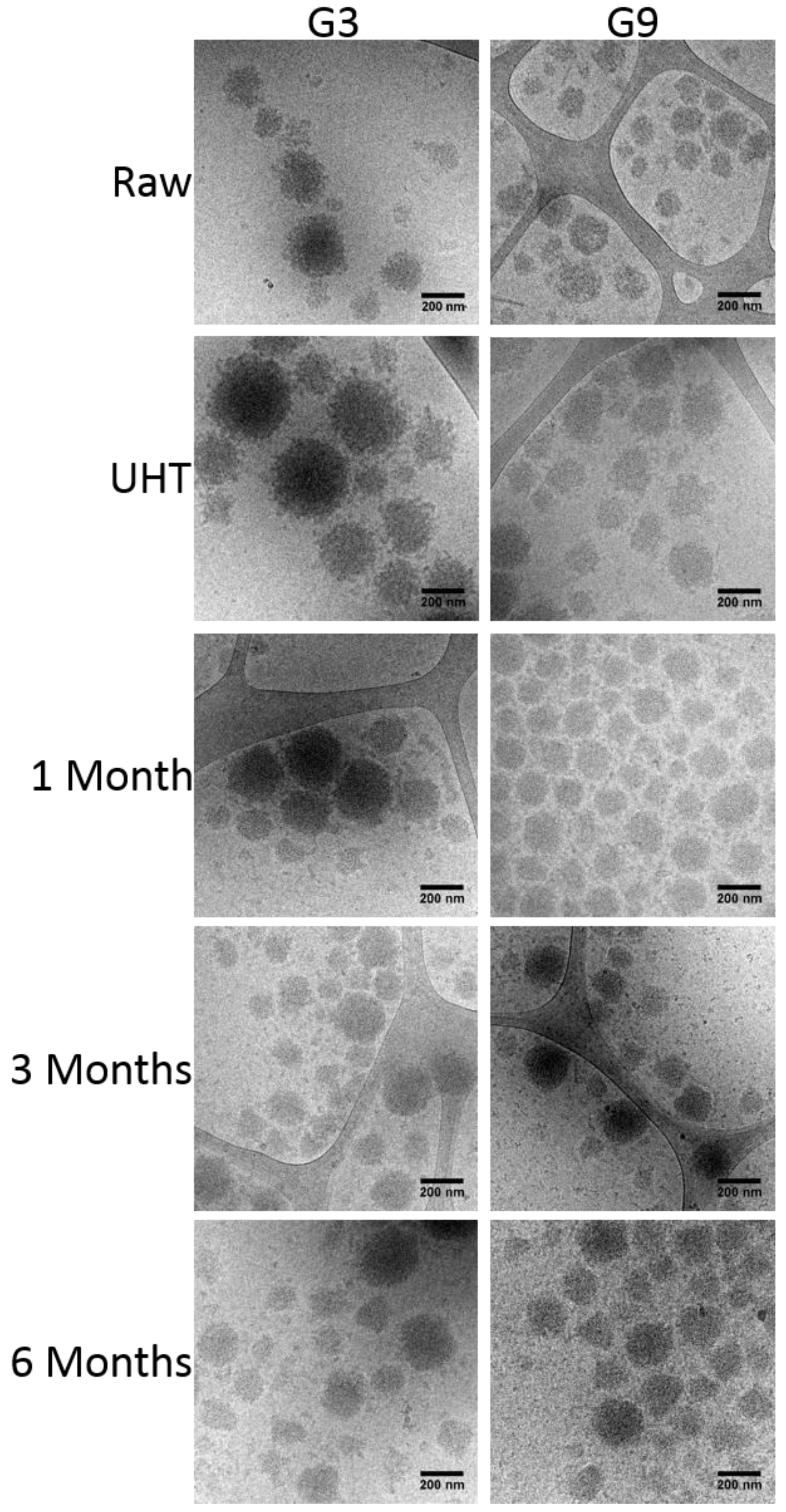
3.6. Cryo-Transmission Electron Microscopy (Cryo-TEM)
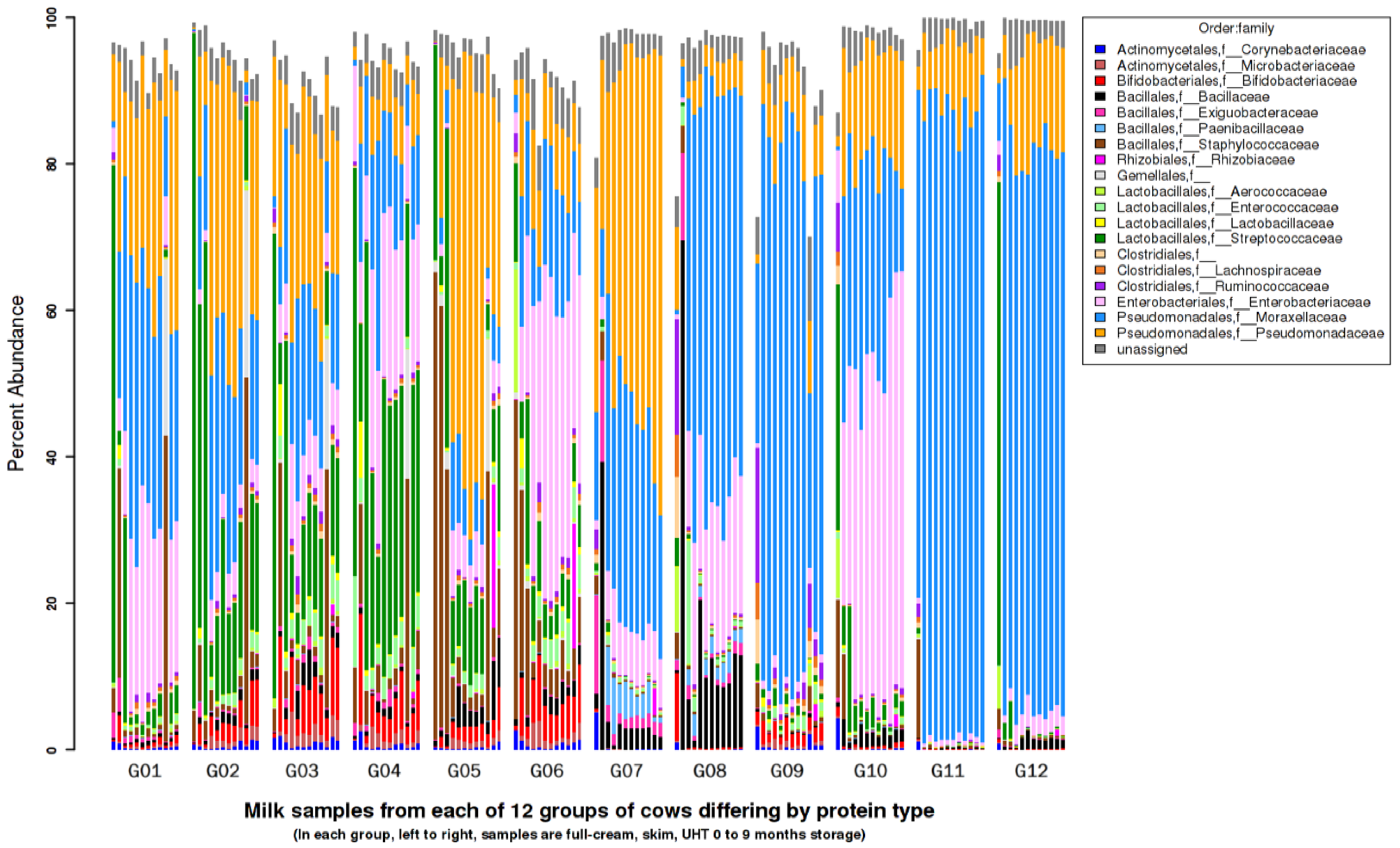
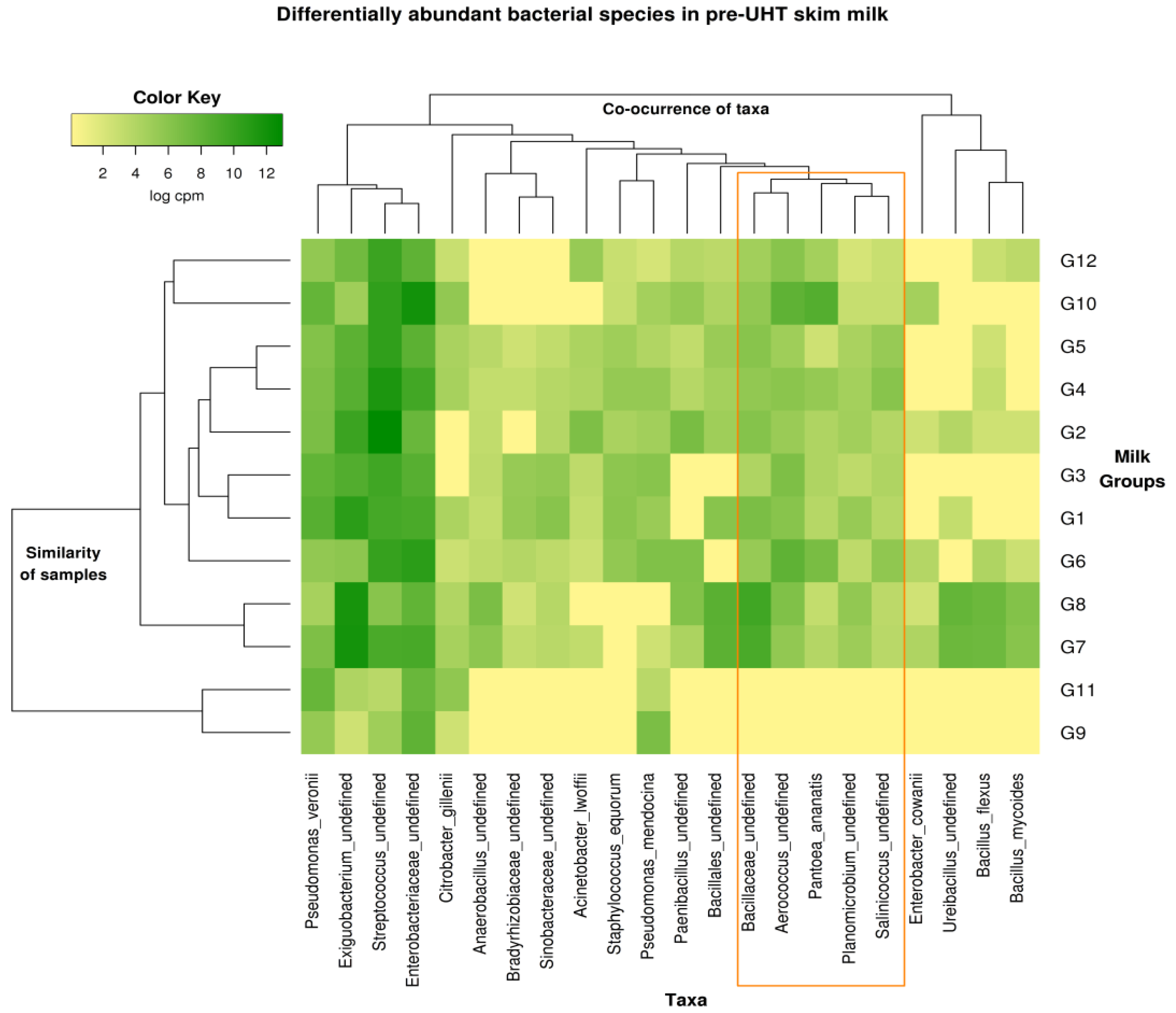
3.7. Metagenomics Microbial Population Analysis
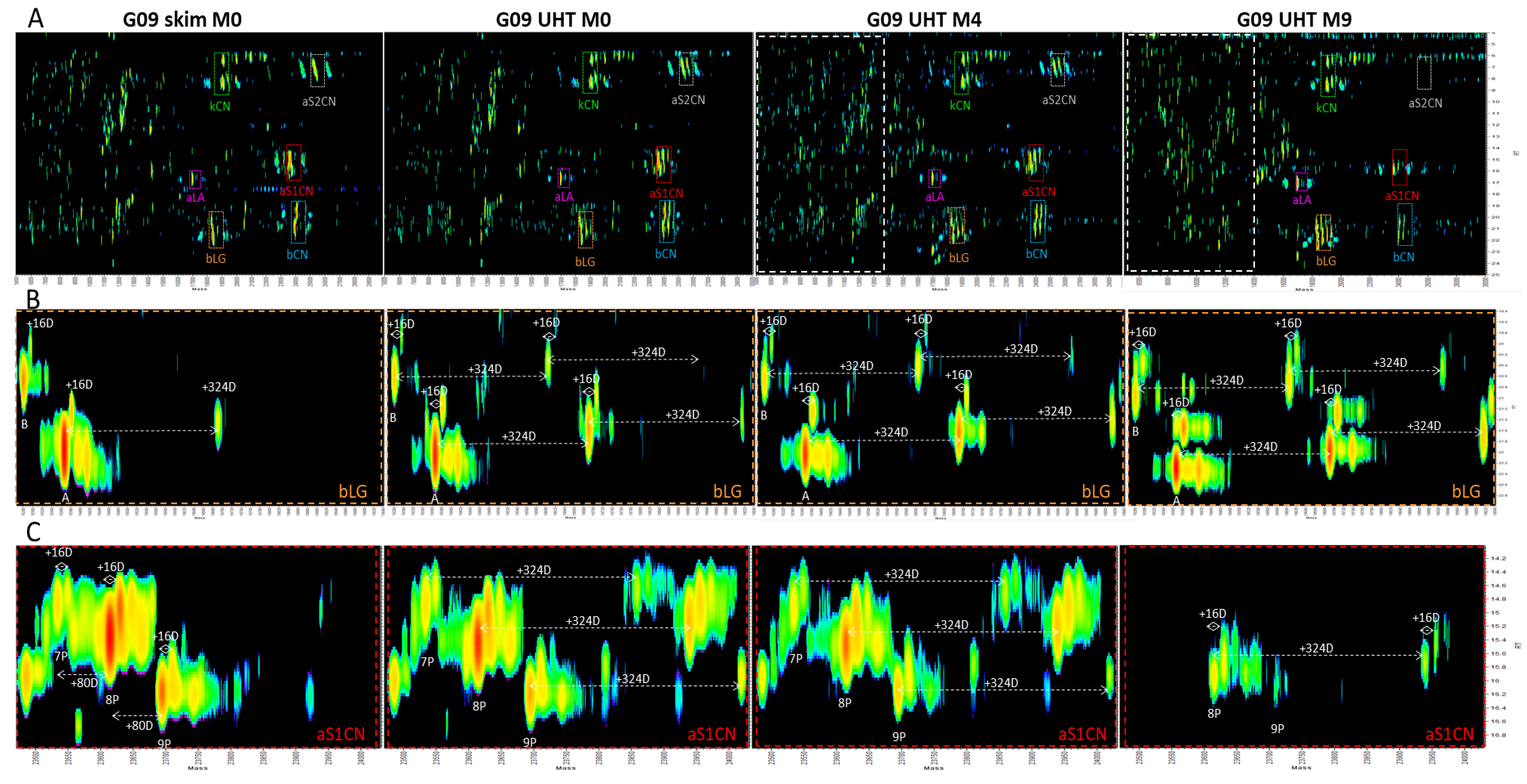
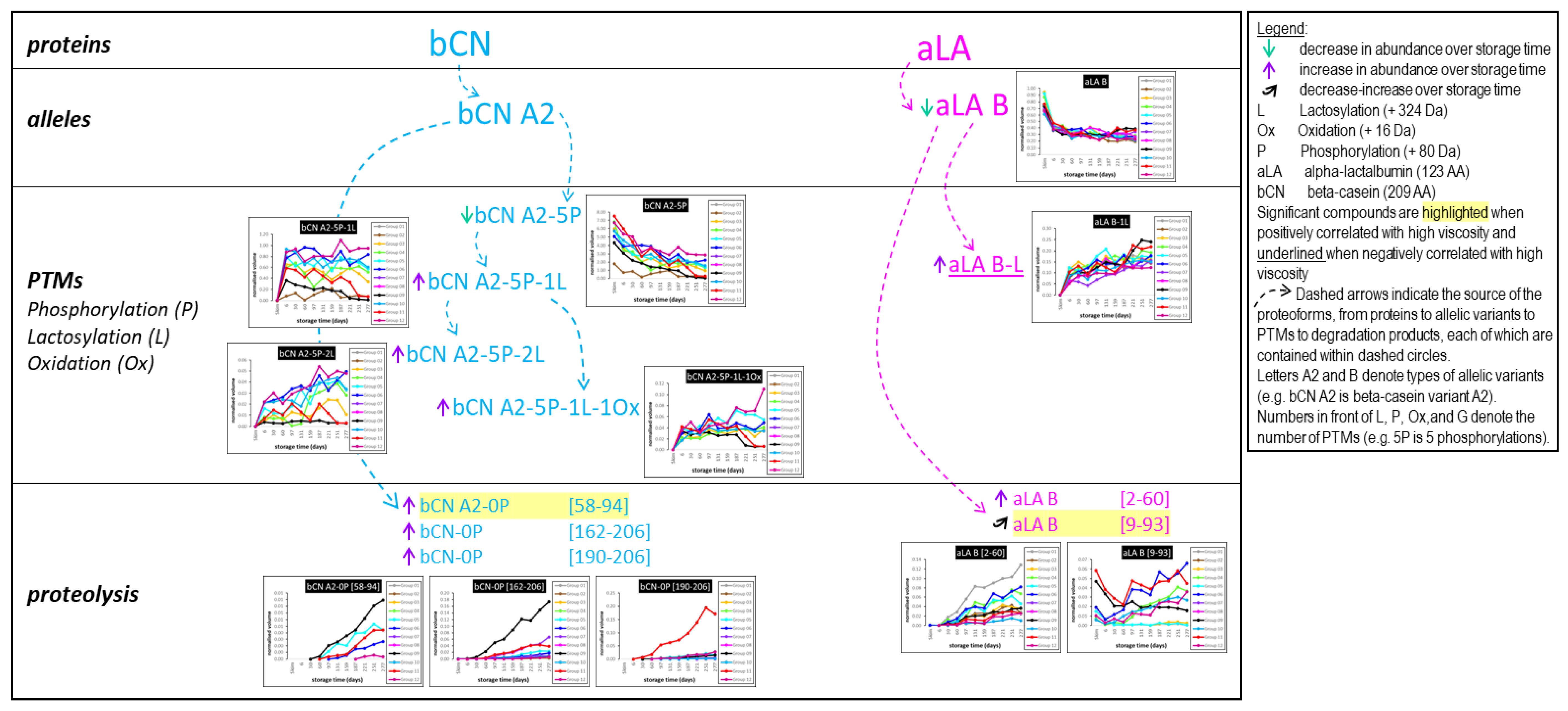
3.8. Top-Down Proteomics
- Appearance of the lactose-modified forms of the milk proteins immediately following UHT treatment.
- Decrease over time in the abundance of all the native intact milk proteins identified.
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Datta, N.; Deeth, H.C. Age Gelation of UHT Milk—A Review. Food Bioprod. Process. 2001, 79, 197–210. [Google Scholar] [CrossRef]
- Chavan, R.S.; Chavan, S.R.; Khedkar, C.D.; Jana, A.H. UHT Milk Processing and Effect of Plasmin Activity on Shelf Life: A Review. Compr. Rev. Food Sci. Food Saf. 2011, 10, 251–268. [Google Scholar] [CrossRef] [Green Version]
- Hough, G.; Garitta, L.; Sánchez, R. Determination of consumer acceptance limits to sensory defects using survival analysis. Food Qual. Prefer. 2004, 15, 729–734. [Google Scholar] [CrossRef]
- Holt, C.; Carver, J.A.; Ecroyd, H.; Thorn, D.C. Caseins and the Casein Micelle: Their Biological Functions, Structures, and Behavior in Foods. J. Dairy Sci. 2013, 96, 6127–6146. [Google Scholar] [CrossRef] [PubMed]
- Ecroyd, H.; Koudelka, T.; Thorn, D.C.; Williams, D.M.; Devlin, G.; Hoffmann, P.; Carver, J.A. Dissociation from the Oligomeric State Is the Rate-limiting Step in Fibril Formation by κ-Casein. J. Biol. Chem. 2008, 283, 9012–9022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guijarro, J.I.; Sunde, M.; Jones, J.A.; Campbell, I.D.; Dobson, C.M. Amyloid Fibril Formation by an SH3 Domain. Proc. Natl. Acad. Sci. USA 1998, 95, 4224–4228. [Google Scholar] [CrossRef] [PubMed]
- McMahon, D.J. Age-Gelation of UHT Milk: Changes That Occur During Storage, Their Effect on Shelf Life and the Mechanism by Which Age-Gelation Occurs. In Heat Treatments and Alternative Methods, IDF Symposium; International Dairy Federation: Brussels, Belgium, 1996; pp. 315–326. [Google Scholar]
- Raynes, J.K.; Day, L.; Crepin, P.; Horrocks, M.H.; Carver, J.A. Coaggregation of κ-casein and β-lactoglobulin produces morphologically distinct amyloid fibrils. Small 2017, 13, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ismail, B.; Nielsen, S.S. Invited review: Plasmin protease in milk: Current knowledge and relevance to dairy industry. J. Dairy Sci. 2010, 93, 4999–5009. [Google Scholar] [CrossRef] [PubMed]
- Manji, B.; Kakuda, Y. The Role of Protein Denaturation, Extent of Proteolysis, and Storage Temperature on the Mechanism of Age Gelation in a Model System. J. Dairy Sci. 1988, 71, 1455–1463. [Google Scholar] [CrossRef]
- Andrews, A.T. Properties of aseptically packed ultra-high-temperature milk: III. Formation of polymerized protein during storage at various temperatures. J. Dairy Res. 1975, 42, 89. [Google Scholar] [CrossRef]
- Harwalkar, V.R. Age Gelation of Sterilized Milks. In Advanced Dairy Chemistry, Volume 1: Proteins; Fox, P.E., McSweeney, P.L.H., Eds.; Elsevier Science Publisher Ltd.: Essex, UK, 1992; Volume 1, pp. 691–734. [Google Scholar]
- Malmgren, B.; Ardö, Y.; Langton, M.; Altskär, A.; Bremer, M.G.E.G.; Dejmek, P.; Paulsson, M. Changes in proteins, physical stability and structure in directly heated UHT milk during storage at different temperatures. Int. Dairy J. 2017, 71, 60–75. [Google Scholar] [CrossRef]
- Caroli, A.M.; Chessa, S.; Erhardt, G.J. Invited review: Milk protein polymorphisms in cattle: Effect on animal breeding and human nutrition. J. Dairy Sci. 2009, 92, 5335–5352. [Google Scholar] [CrossRef] [PubMed]
- Hallén, E.; Allmere, T.; Näslund, J.; Andrén, A.; Lundén, A. Effect of genetic polymorphism of milk proteins on rheology of chymosin-induced milk gels. Int. Dairy J. 2007, 17, 791–799. [Google Scholar] [CrossRef]
- Hallén, E.; Wedholm, A.; Andrén, A.; Lundén, A. Effect of β-casein, κ-casein and β-lactoglobulin genotypes on concentration of milk protein variants. J. Anim. Breed. Genet. 2008, 125, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Day, L.; Williams, R.P.W.; Otter, D.; Augustin, M.A. Casein polymorphism heterogeneity influences casein micelle size in milk of individual cows. J. Dairy Sci. 2015, 98, 3633–3644. [Google Scholar] [CrossRef] [PubMed]
- Jensen, H.B.; Pedersen, K.S.; Johansen, L.B.; Poulsen, N.A.; Bakman, M.; Chatterton, D.E.W.; Larsen, L.B. Genetic variation and posttranslational modification of bovine κ-casein: Effects on caseino-macropeptide release during renneting. J. Dairy Sci. 2015, 98, 747–758. [Google Scholar] [CrossRef] [PubMed]
- Haq, M.R.U.; Kapila, R.; Kapila, S. Release of β-casomorphin-7/5 during simulated gastrointestinal digestion of milk β-casein variants from Indian crossbred cattle (Karan Fries). Food Chem. 2015, 168, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Raynes, J.K.; Day, L.; Augustin, M.A.; Carver, J.A. Structural differences between bovine A1 and A2 β-casein alter micelle self-assembly and influence molecular chaperone activity. J. Dairy Sci. 2015, 98, 2172–2182. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.P.; Boland, M.J.; Creamer, L.K.; Anema, S.G.; Otter, D.E.; Paterson, G.R.; Lowe, R.; Motion, R.L.; Thresher, W.C. Effect of the Bovine β-Lactoglobulin Phenotype on the Properties of β-Lactoglobulin, Milk Composition, and Dairy Products. In Macromolecular Interactions in Food Technology; American Chemical Society: Washington, DC, USA, 1996; Volume 650, pp. 281–294. [Google Scholar]
- Manderson, G.A.; Hardman, M.J.; Creamer, L.K. Effect of Heat Treatment on Bovine β-Lactoglobulin A, B, and C Explored Using Thiol Availability and Fluorescence. J. Agric. Food Chem. 1999, 47, 3617–3627. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.L.; Catignani, G.L.; Foegeding, E.A.; Swaisgood, H.E. Comparison of the Gelation Properties of β-Lactoglobulin Genetic Variants A and B. J. Agric. Food Chem. 1994, 42, 1064–1067. [Google Scholar] [CrossRef]
- Mitchell, G.E.; Ewings, K.N. Quantification of bacterial proteolysis causing gelation in UHT-treated milk. N. Z. J. Dairy Sci. Technol. 1985, 20, 65–76. [Google Scholar]
- Ambrose Griffin, M.C.; Griffin, W.G. A simple turbidimetric method for the determination of the refractive index of large colloidal particles applied to casein micelles. J. Colloid Interface Sci. 1985, 104, 409–415. [Google Scholar] [CrossRef]
- Richardson, B.C.; Pearce, K.N. The determination of plasmin in dairy products. N. Z. J. Dairy Sci. Technol. 1981, 16, 209–220. [Google Scholar]
- Murphy, M.A.; Shariflou, M.R.; Moran, C. High quality genomic DNA extraction from large milk samples. J. Dairy Res. 2002, 69, 645–649. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [PubMed]
- Masella, A.P.; Bartram, A.K.; Truszkowski, J.M.; Brown, D.G.; Neufeld, J.D. PANDAseq: Paired-end assembler for illumina sequences. BMC Bioinform. 2012, 13, 31. [Google Scholar] [CrossRef] [PubMed]
- Morgulis, A.; Coulouris, G.; Raytselis, Y.; Madden, T.L.; Agarwala, R.; Schäffer, A.A. Database indexing for production MegaBLAST searches. Bioinformatics 2008, 24, 1757–1764. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package, 2.4-3 ed. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 22 November 2018).
- Vavrek, M.J. Fossil: Palaeoecological and Palaeogeographical Analysis Tools. Available online: https://palaeo-electronica.org/2011_1/238/index.html (accessed on 20 November 2018).
- Sarkar, D. Lattice Multivariate Data Visualization with R; Springer Science Business Media: New York, NY, USA, 2008. [Google Scholar]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Vincent, D.; Elkins, A.; Condina, M.R.; Ezernieks, V.; Rochfort, S. Quantitation and Identification of Intact Major Milk Proteins for High-Throughput LC-ESI-Q-TOF MS Analyses. PLoS ONE 2016, 11, e0163471. [Google Scholar] [CrossRef] [PubMed]
- Bijl, E.; de Vries, R.; van Valenberg, H.; Huppertz, T.; van Hooijdonk, T. Factors influencing casein micelle size in milk of individual cows: Genetic variants and glycosylation of κ-casein. Int. Dairy J. 2014, 34, 135–141. [Google Scholar] [CrossRef]
- Kelleher, N.L.; Lin, H.Y.; Valaskovic, G.A.; Aaserud, D.J.; Fridriksson, E.K.; McLafferty, F.W. Top Down versus Bottom Up Protein Characterization by Tandem High-Resolution Mass Spectrometry. J. Am. Chem. Soc. 1999, 121, 806–812. [Google Scholar] [CrossRef]
- Toby, T.K.; Fornelli, L.; Kelleher, N.L. Progress in Top-Down Proteomics and the Analysis of Proteoforms. Annu. Rev. Anal. Chem. 2016, 9, 499–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Brien, J. Heat-Induced Changes in Lactose: Isomerization, Degradation, Maillard Browning. Available online: http://agris.fao.org/agris-search/search.do?recordID=BE9500697 (accessed on 22 November 2018).
- Anema, S.G. Storage stability and age gelation of reconstituted ultra-high temperature skim milk. Int. Dairy J. 2017, 75, 56–67. [Google Scholar] [CrossRef]
- Schaar, J.; Hansson, B.; Pettersson, H.-E. Effects of genetic variants of K-casein and βlactoglobulin on cheesemaking. J. Dairy Res. 1985, 52, 429–437. [Google Scholar] [CrossRef]
- Rauh, V.M.; Johansen, L.B.; Ipsen, R.; Paulsson, M.; Larsen, L.B.; Hammershoj, M. Plasmin activity in UHT milk: Relationship between proteolysis, age gelation, and bitterness. J. Agric. Food Chem. 2014, 62, 6852–6860. [Google Scholar] [CrossRef] [PubMed]
- Matéos, A.; Guyard-Nicodème, M.; Baglinière, F.; Jardin, J.; Gaucheron, F.; Dary, A.; Humbert, G.; Gaillard, J.L. Proteolysis of milk proteins by AprX, an extracellular protease identified in Pseudomonas LBSA1 isolated from bulk raw milk, and implications for the stability of UHT milk. Int. Dairy J. 2015, 49, 78–88. [Google Scholar] [CrossRef]
| Group | κ-CN | β-CN | β-Lg | Age | Days in Lact | No. of Lact | 7 Day Milk | Protein% | Lactose % | Ca mg/kg | K mg/kg | Mg mg/kg | Na mg/kg | P mg/kg | pH | Casein Micelle Diameter nm | log CFU/mL | Plasmin Activity (nM AMC/min) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | AA | A1A1 | AB | 5.6 ± 1.3 | 160 ± 27 | 3 ± 1 | 23.1 ± 4.7 | 3.17 | 4.97 | 1100 | 1530 | 110 | 460 | 990 | 6.66 | 192.4 | 5.82 ± 0.16 | 0.51 ± 0.07 |
| 2 | AA | A1A1 | BB | 5.6 ± 0.9 | 162 ± 32 | 3 ± 1 | 23.6 ± 4.9 | 3.16 | 5.32 | 1080 | 1580 | 110 | 450 | 1080 | 6.61 | 177.2 | 5.98 ± 0.04 | 0.40 ± 0.11 |
| 3 | AA | A1A2 | AB | 6.6 ± 1.9 | 175 ± 19 | 4 ± 2 | 22.1 ± 2.8 | 3.14 | 5.24 | 1120 | 1700 | 110 | 520 | 1070 | 6.67 | 185.5 | 5.14 ± 0.04 | 0.85 ± 0.08 |
| 4 | AA | A1A2 | BB | 6.4 ± 1.1 | 167 ± 28 | 4 ± 1 | 22.5 ± 1.9 | 3.22 | 5.38 | 1160 | 1520 | 100 | 510 | 1070 | 6.68 | 178.8 | 5.94 ± 0.02 | 0.95 ± 0.08 |
| 5 | AA | A2A2 | AB | 6.2 ± 1.6 | 175 ± 25 | 4 ± 1 | 23.4 ± 1.2 | 3.18 | 5.27 | 1160 | 1560 | 100 | 500 | 1040 | 6.68 | 185.6 | 6.23 ± 0.12 | 0.88 ± 0.08 |
| 6 | AA | A2A2 | BB | 6 ± 1.2 | 237 ± 154 | 3 ± 1 | 20.3 ± 5.5 | 3.13 | 5.18 | 1130 | 1670 | 110 | 540 | 1040 | 6.69 | 162 | 5.05 ± 0.05 | 0.80 ± 0.05 |
| 7 | AB | A1A1 | AB | 7.5 ± 0.7 | 337 ± 245 | 5 ± 3 | 19.5 ± 4.2 | 3.08 | 4.08 | 1150 | 1150 | 90 | 430 | 900 | 6.87 | 156.3 | 4.86 ± 0.13 | 1.91 ± 0.05 |
| 8 | AB | A1A1 | BB | 7 | 171 | 5 | 25.1 | 3.21 | 4.68 | 960 | 1340 | 90 | 550 | 960 | 6.68 | 161.4 | 5.00 ± 0.13 | 1.23 ± 0.06 |
| 9 | AB | A1A2 | AB | 6.6 ± 1.1 | 177 ± 27 | 4 ± 1 | 23.2 ± 3.2 | 3.17 | 5.11 | 1150 | 1430 | 100 | 560 | 1040 | 6.71 | 165.1 | 4.11 ± 0.05 | 2.39 ± 0.07 |
| 10 | AB | A1A2 | BB | 4.2 ± 0.4 | 172 ± 25 | 2 ± 0 | 20.0 ± 1.8 | 3.20 | 5.17 | 1220 | 1590 | 100 | 450 | 1100 | 6.72 | 170.2 | 5.81 ± 0.09 | 0.32 ± 0.08 |
| 11 | AB | A2A2 | AB | 5.8 ± 2.2 | 179 ± 17 | 3 ± 2 | 23.7 ± 4.5 | 3.24 | 5.20 | 1230 | 1510 | 100 | 490 | 1070 | 6.7 | 148.6 | 6.34 ± 0.03 | 0.77 ± 0.09 |
| 12 | AB | A2A2 | BB | 4.4 ± 0.5 | 239 ± 155 | 2 ± 0 | 20.3 ± 3.4 | 3.35 | 4.94 | 1030 | 1280 | 90 | 390 | 910 | 6.7 | 156.3 | 6.07 ± 0.05 | 0.16 ± 0.05 |
| Proteoform | Mass | p-Values | Positive Correlation | Proteoform | Mass | p-Values | Negative Correlation |
|---|---|---|---|---|---|---|---|
| bLG [127–153] * | 3125.7 | 5.2 × 10−34 | 0.79 | ppc5.1 A1-2P [1–105] | 11,970.3 | 5.2 × 10−34 | −0.71 |
| bCN-0P [190–206] * | 1928.1 | 5.2 × 10−34 | 0.73 | aS1CN B-8P-1L-1Ox | 23,940.4 | 5.2 × 10−34 | −0.69 |
| bCN-0P-1L [125–138] | 1898.0 | 4.6 × 10−15 | 0.67 | aS1CN B-8P-1L | 23,924.4 | 8.3 × 10−13 | −0.62 |
| bCN-0P-1Ox [37–59] | 2727.4 | 5.0 × 10−14 | 0.65 | aS1CN B-8P-1L-2Ox | 23,956.5 | 1.2 × 10−10 | −0.57 |
| bCN-0P [133–161] * | 3364.9 | 1.2 × 10−13 | 0.64 | aS1CN B-8P-1Ox | 23,616.3 | 1.0 × 10−09 | −0.55 |
| bCN-0P-1Ox [139–162] | 2810.4 | 2.3 × 10−13 | 0.63 | aS1CN B-8P-2Ox | 23,632.3 | 2.6 × 10−09 | −0.54 |
| aS1CN B-0P [180–199] * | 2215.1 | 3.7 × 10−13 | 0.63 | aS1CN B-0P [158–199] * | 4650.2 | 4.6 × 10−06 | −0.43 |
| bCN A2-0P [58–94] * | 3992.2 | 5.4 × 10−12 | 0.60 | aS1CN B-9P-1L | 24,004.4 | 1.4 × 10−05 | −0.42 |
| bCN-0P-1Ox [162–206] | 5014.8 | 1.4 × 10−11 | 0.59 | aS1CN B-8P | 23,600.4 | 3.8 × 10−05 | −0.40 |
| bCN-0P [127–138] * | 1359.8 | 2.5 × 10−11 | 0.59 | aS1CN B-8P [39–135] | 11,800.2 | 5.1 × 10−05 | −0.39 |
| bCN-0P-1Ox [107–154] | 5604.0 | 4.2 × 10−11 | 0.58 | aS1CNB-8P-2Ox [39–135] | 11,832.3 | 2.0 × 10−04 | −0.37 |
| bCN-0P [192–206] * | 1667.9 | 5.5 × 10−11 | 0.58 | bCN A2-5P-1Ox | 23,984.1 | 2.5 × 10−04 | −0.36 |
| bCN-0P [139–161] * | 2695.4 | 1.4 × 10−10 | 0.57 | aS1CN B-0P [125–199] | 8633.3 | 6.6 × 10−04 | −0.34 |
| bCN-0P-1Ox [96–138] | 4918.5 | 1.4 × 10−10 | 0.57 | aS1CN B-0P [1–34] * | 3981.2 | 7.3 × 10−04 | −0.34 |
| bCN-0P [139–162] * | 2794.4 | 5.8 × 10−10 | 0.55 | bCN-0P [170–209] * | 4481.5 | 9.2 × 10−04 | −0.33 |
| bCN-0P [107–154] | 5588.0 | 4.8 × 10−09 | 0.53 | g3CN-0P [108–209] | 11,551.1 | 9.8 × 10−04 | −0.33 |
| bCN-0P [125–138] * | 1573.9 | 5.1 × 10−09 | 0.53 | aS1CNB-8P-1Ox [39–135] | 11,816.3 | 1.2 × 10−03 | −0.33 |
| bCN-0P [162–206] * | 4998.8 | 5.4 × 10−08 | 0.50 | aS1CN B-9P-1Ox | 23,696.1 | 3.3 × 10−03 | −0.30 |
| bCN-0P [37–59] * | 2711.4 | 3.2 × 10−07 | 0.48 | bLG B | 18,269.5 | 5.5 × 10−03 | −0.29 |
| bCN-0P [190–209] * | 2253.3 | 3.4 × 10−07 | 0.48 | aS1CN B-7P [22–110] | 10,871.7 | 6.0 × 10−03 | −0.28 |
| aLA B-1L | 14,501.0 | 1.4 × 10−06 | 0.45 | aS1CN B-0P [1–36] | 4238.3 | 7.0 × 10−03 | −0.28 |
| bCN-0P [96–138] * | 4902.5 | 1.7 × 10−06 | 0.45 | aS1CN B-8P-2L | 24,248.6 | 7.0 × 10−03 | −0.28 |
| bCN-0P-1L [106–138] | 4157.1 | 3.0 × 10−06 | 0.44 | aS1CN B-9P | 23,680.3 | 9.4 × 10−03 | −0.27 |
| bCN A2-0P-1Ox [58–105] | 5176.8 | 4.2 × 10−06 | 0.44 | bCN-0P [177–209] * | 3720.0 | 1.0 × 10−02 | −0.27 |
| bCN-0P-1Ox [108–138] | 3583.8 | 6.5 × 10−06 | 0.43 | aS1CN B-0P [1–33] * | 3853.1 | 1.2 × 10−02 | −0.26 |
| bCN-0P [108–138] * | 3567.8 | 1.1 × 10−05 | 0.42 | aS1CN B-8P [22–109] | 10,822.8 | 2.0 × 10−02 | −0.24 |
| aS1CN B-5P-1L [9–67] | 7350.4 | 5.6 × 10−05 | 0.39 | bCN A1-5P-1L–1Ox | 24,347.3 | 2.0 × 10−02 | −0.24 |
| aS1CN B-2P-1L [24–63] | 4985.6 | 2.5 × 10−04 | 0.36 | bCN A1-5P-1L | 24,332.3 | 2.1 × 10−02 | −0.24 |
| bCN-0P-1Ox [162–209] | 5340.0 | 3.7 × 10−04 | 0.35 | aS1CN B-7P [32–89] | 7051.0 | 2.1 × 10−02 | −0.24 |
| bCN-0P [192–209] * | 1993.2 | 4.1 × 10−04 | 0.35 | kCN A-1P-1G(947.3D) | 19,971.9 | 2.3 × 10−02 | −0.24 |
| aS2CN A-0P [71–117] | 5725.1 | 6.6 × 10−04 | 0.34 | g3CN-0P-1Ox [108–209] | 11,567.1 | 2.4 × 10−02 | −0.24 |
| bCN-0P [106–138] * | 3833.0 | 1.4 × 10−03 | 0.32 | bLG B-1L | 18,593.7 | 2.4 × 10−02 | −0.24 |
| aLA B [9–93] | 9707.9 | 1.4 × 10−03 | 0.32 | bCN A1-5P-1L-2Ox | 24,364.3 | 2.4 × 10−02 | −0.24 |
| bCN-0P [139–164] * | 2994.5 | 2.1 × 10−03 | 0.31 | aS1CNB-8P-3Ox [39–135] | 11,848.3 | 3.0 × 10−02 | −0.23 |
| bCN-0P [191–209] * | 2106.2 | 2.2 × 10−03 | 0.31 | bCN A1-5P | 24,007.3 | 3.7 × 10−02 | −0.22 |
| bCN-0P [108–124] * | 2011.9 | 2.7 × 10−03 | 0.31 | kCN A-1P | 19,025.6 | 3.8 × 10−02 | −0.22 |
| aS1CN B-0P [3–37] * | 4084.3 | 3.3 × 10−03 | 0.30 | bCN-0P [98–203] | 12,004.7 | 4.0 × 10−02 | −0.22 |
| kCN A-0P [125–150] * | 2589.3 | 3.3 × 10−03 | 0.30 | aS1CN B-0P [1–38] * | 4451.5 | 4.4 × 10−02 | −0.21 |
| bCN A2-0P [58–105] * | 5160.8 | 6.0 × 10−03 | 0.28 | bCN A1-5P-1Ox | 24,023.3 | 4.5 × 10−02 | −0.21 |
| bCN-0P-1L [162–209] | 5648.1 | 6.5 × 10−03 | 0.28 | kCN A-1P-1L | 19,349.7 | 4.6 × 10−02 | −0.21 |
| bLG A-2L | 19,003.8 | 7.0 × 10−03 | 0.28 | bCN-0P-1Ox [98–203] | 12,020.7 | 4.7 × 10−02 | −0.21 |
| aLA B-1Allys [9–93] | 9706.7 | 7.4 × 10−03 | 0.28 | ||||
| bCN-0P [198–209] * | 1263.8 | 7.4 × 10−03 | 0.28 | ||||
| bLG A-3Ox-1L | 18,727.6 | 8.9 × 10−03 | 0.27 | ||||
| aS1CN B-2P-1Ox [1–56] | 6592.3 | 1.1 × 10−02 | 0.26 | ||||
| bCN-0P [71–169] | 11,100.9 | 1.2 × 10−02 | 0.26 | ||||
| bCN A2-0P [58–106] | 5298.9 | 1.2 × 10−02 | 0.26 | ||||
| bLG A-1Ox | 18,371.5 | 1.5 × 10−02 | 0.25 | ||||
| aS1CN B-5P [9–67] | 7026.3 | 1.6 × 10−02 | 0.25 | ||||
| aS1CN B-2P [24–63] | 4661.5 | 2.1 × 10−02 | 0.24 | ||||
| aS1CNB-0P-1Ox [176–199] | 2633.3 | 4.2 × 10−02 | 0.22 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raynes, J.K.; Vincent, D.; Zawadzki, J.L.; Savin, K.; Mertens, D.; Logan, A.; Williams, R.P.W. Investigation of Age Gelation in UHT Milk. Beverages 2018, 4, 95. https://doi.org/10.3390/beverages4040095
Raynes JK, Vincent D, Zawadzki JL, Savin K, Mertens D, Logan A, Williams RPW. Investigation of Age Gelation in UHT Milk. Beverages. 2018; 4(4):95. https://doi.org/10.3390/beverages4040095
Chicago/Turabian StyleRaynes, Jared K., Delphine Vincent, Jody L. Zawadzki, Keith Savin, Dominik Mertens, Amy Logan, and Roderick P.W. Williams. 2018. "Investigation of Age Gelation in UHT Milk" Beverages 4, no. 4: 95. https://doi.org/10.3390/beverages4040095
APA StyleRaynes, J. K., Vincent, D., Zawadzki, J. L., Savin, K., Mertens, D., Logan, A., & Williams, R. P. W. (2018). Investigation of Age Gelation in UHT Milk. Beverages, 4(4), 95. https://doi.org/10.3390/beverages4040095







