1. Introduction
The flavour and aroma of any beer is, in large part, determined by the yeast strain employed together with the wort composition. In addition, yeast properties such as flocculation, fermentation ability (including the uptake of wort sugars, amino acids, small peptides, and ammonium ions), osmotic pressure, ethanol tolerance, and oxygen requirements have a critical impact on fermentation performance. Proprietary strains, belonging to individual breweries, are usually jealously guarded and conserved. However, this is not always the situation. In Germany, for example, most of the beer is produced with only four individual yeast strains and approximately 65% of it is produced with a single strain sourced from the Weihenstephan Brewing School, which is located in the Munich Technical University.
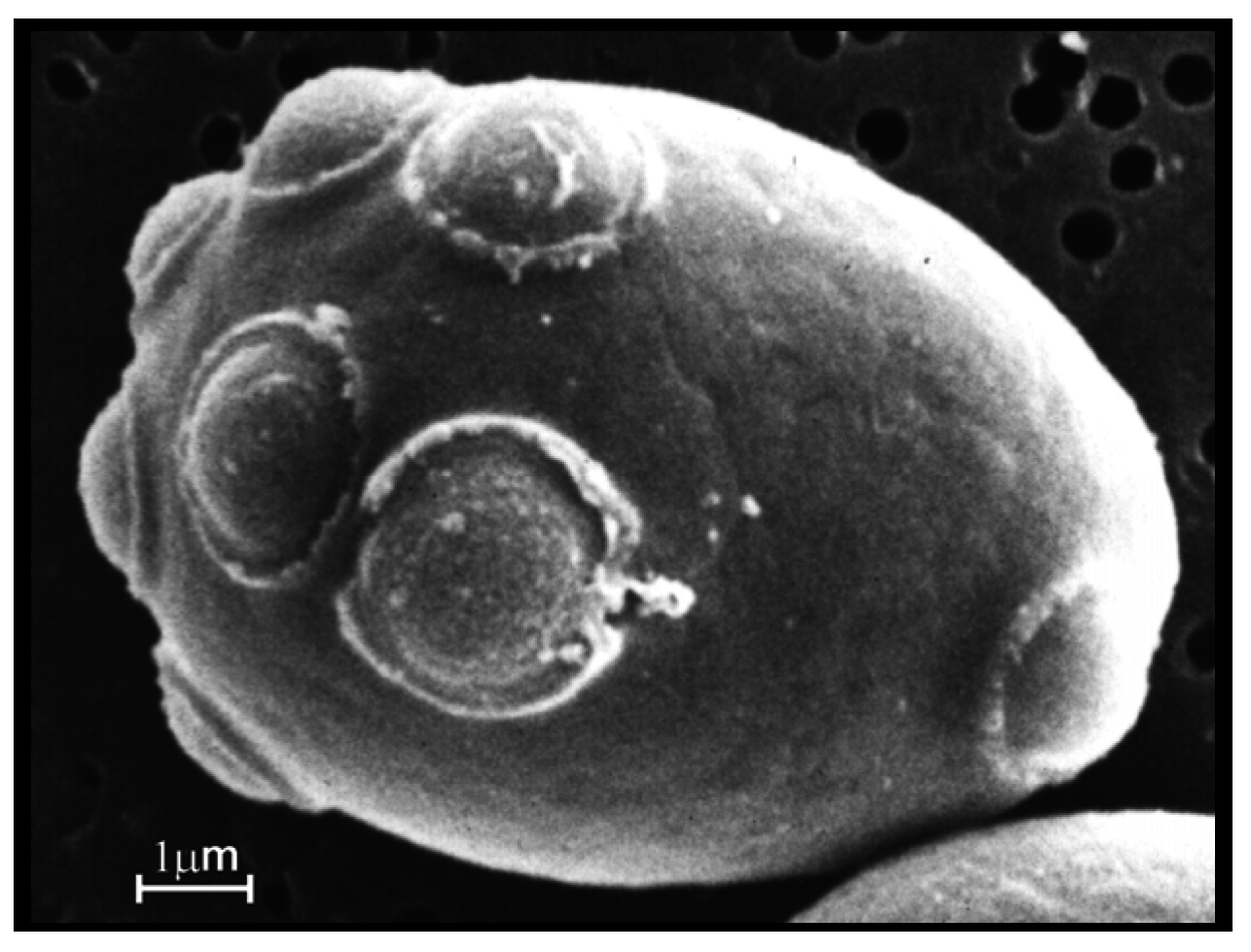
No satisfactory definition of yeast exists that encompasses commonly encountered properties such as alcoholic fermentation and growth by budding. The latter is not unusual in yeast and nearly all brewer’s yeast strains multiply by budding (
Figure 1). Brewer’s yeast cultures predominantly come from the genus
Saccharomyces—a minority of non-
Saccharomyces yeast cultures employed in brewing will be discussed in
Section 8. Yeast is cultured in an acidic aqueous sugary solution called
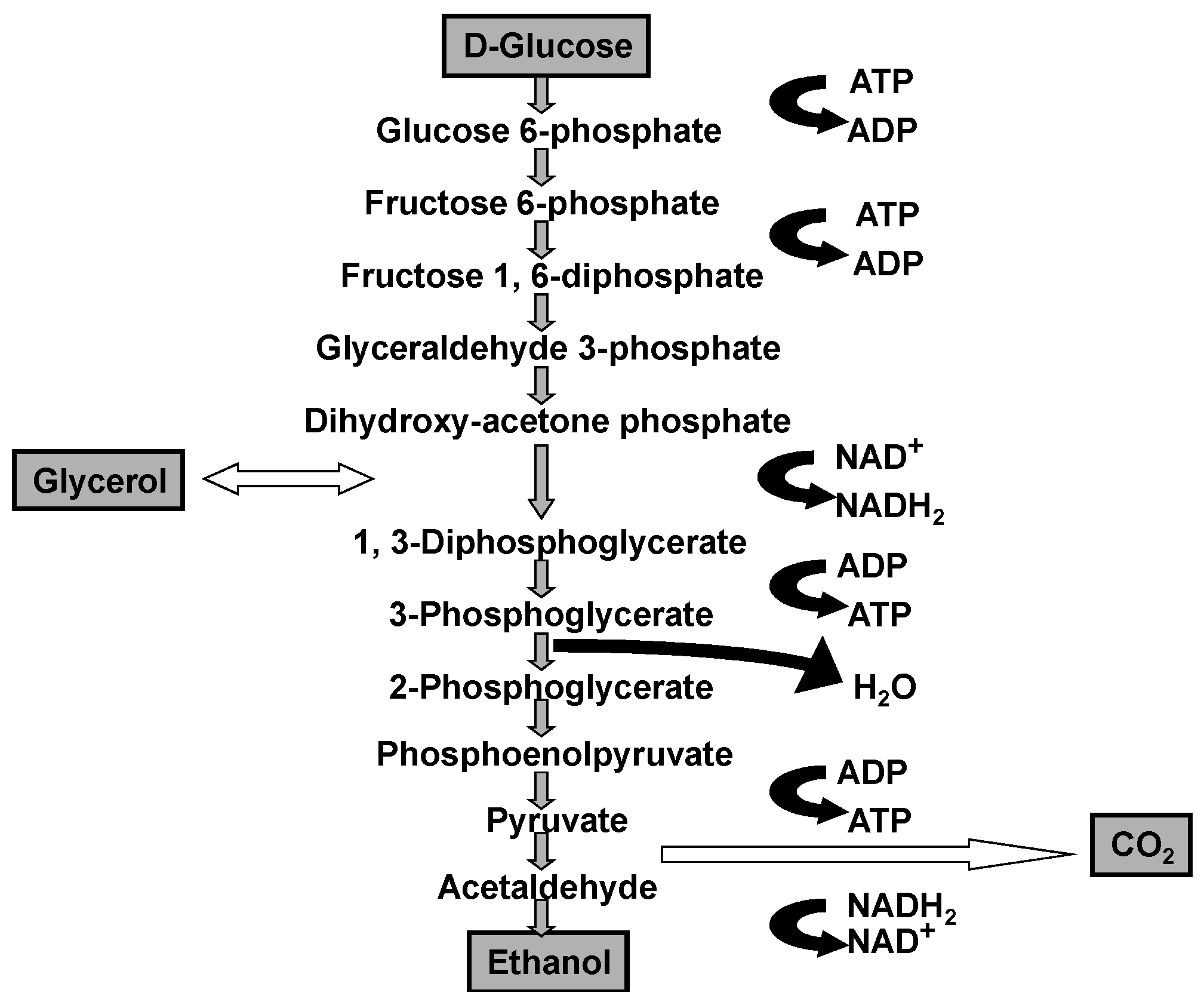
wort prepared from barley malt and other cereals such as corn (maize), wheat, rice, sorghum, and also cane and beet sugar. The cells absorb dissolved sugars, simple nitrogenous matter (amino acids, ammonium ions, and small peptides), vitamins, and ions through their plasma membrane. Subsequently, they employ a series of reactions known as metabolic pathways (glycolysis, biosynthesis of cellular constituents, etc.) and use these nutrient materials for growth and fermentation. It is important to emphasise that the primary products of glycolysis are: ethanol, glycerol, and carbon dioxide (
Figure 2).
Saccharomyces cerevisiae (ale yeast) has the ability to take up a wide range of sugars, for example, glucose, fructose, mannose, galactose, sucrose, maltose, maltotriose, and raffinose (in part). In addition, as will be described later, a sub-species of
S. cerevisiae,
Saccharomyces diastaticus, is able to utilise dextrins (partially hydrolysed starch). Also,
Saccharomyces pastorianus (lager yeast) strains are able to utilise the disaccharide melibiose (glucose—galactose) in addition to the sugar spectra taken up by
S. cerevisiae. This melibiose utilisation property can be used in diagnostic tests to distinguish between ale and lager yeast strains. The enzymatic hydrolysis of starch, as would occur during mashing (
Figure 3), leads to a fermentable medium (wort) that consists of a number of simple sugars—glucose, fructose, sucrose, maltose, and maltotriose together with unfermentable dextrins. The predominant sugars in most brewer’s worts are: glucose, maltose, and maltotriose (
Table 1) [
1].
2. The Malting and Brewing Processes and Wort Composition
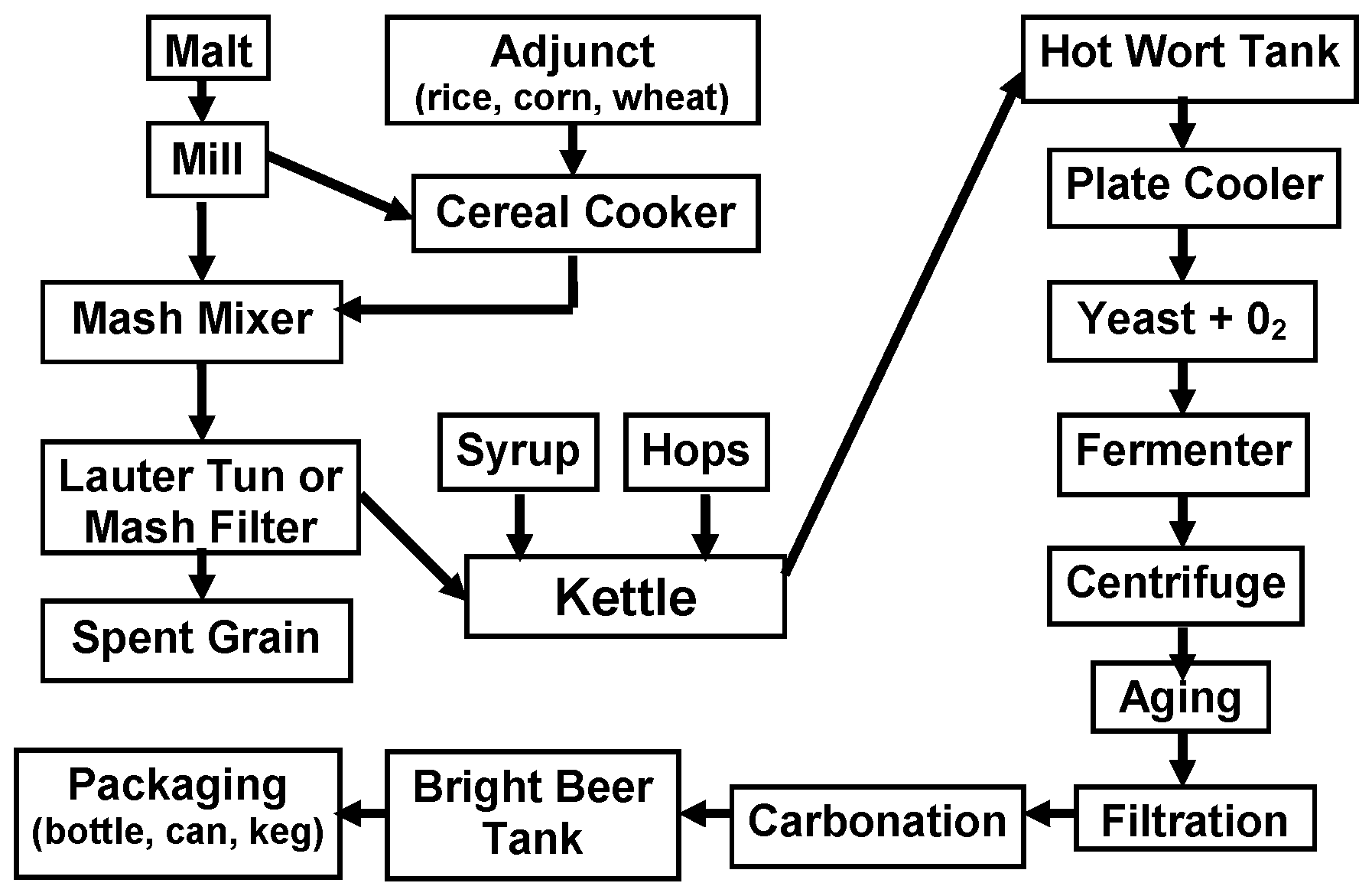
The beer production process is outlined in
Figure 3. Malting, mashing, and fermentation/aging are essentially enzymatic processes. The major brewing raw material is malt usually (not always) from barley and it contains extract components (starch, proteins, etc.) and enzymes (amylases, proteases, glucanases, etc.). However, malt is an expensive raw material and unmalted cereals (adjuncts) [
2] are extensively used in most brewing countries.
The exception to this use of adjuncts is Germany, where the Purity Law (also called the Reinheitsgebot), was adopted in Bavaria in 1516. As Germany unified, Bavaria campaigned for adoption of this law nationally. The only ingredients that could be used in Germany for the production of beer were water, barley, and hops. The principal purpose of the law was to prevent price competition with bakers for wheat and rye and to ensure the availability of bread [
3]. When yeast was described by Antonie van Leewenhoek in 1680 and its role in fermentation was described, it was added to modern versions of the Reinheitsgebot. It is worth noting that Norway and Greece also commonly subscribe to this law [
2].
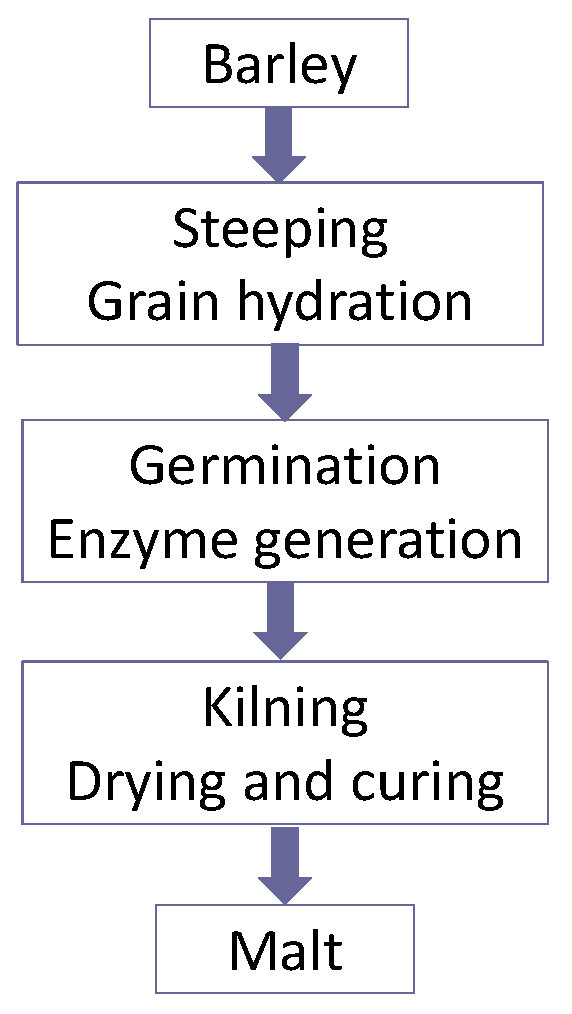
The objectives of malting are to develop a spectrum of enzymes in cereals such as barley and wheat. These enzymes are able to hydrolyse the cereal constituents in order to produce a fermentable extract called wort (
Figure 4). As already discussed, wort is a medium that will support yeast growth and fermentation with beer as the end product. It is important that beer is drinkable (beer is not usually supped, it is drunk!) and it exhibits a number of stability properties, i.e., flavour, physical, foam, and biological characteristics [
4]. Malt contributes a large number of materials to wort [
5]. The principal wort components (not the only ones) are free amino nitrogen (FAN), fermentable sugars, and unfermentable dextrins.
Compared to other media used in the production of fermentation alcohol (both industrial and potable), wort is by far the most intricate. Therefore, when yeast is pitched (inoculated) into wort it is introduced into a complex environment because it consists of simple sugars, dextrins, amino acids, peptides, proteins, vitamins, ions, nucleic acids, and other constituents too numerous to mention. One of the major advances in brewing science during the past 40 years has been the elucidation of the mechanisms by which yeast cells utilise in an orderly manner, the plethora of wort nutrients [
1,
6].
FAN is the sum of the individual wort amino acids, ammonium ions, and small peptides (di- and tripeptides). FAN is an important general measure of yeast nutrients which constitute the yeast assimilable nitrogen during brewery fermentations [
6]. Wort also contains the sugars sucrose, fructose, glucose, maltose, and maltotriose, together with dextrin material. A typical percentage sugar spectrum of brewer’s wort is shown in
Table 1.
3. Wort Fermentation
The objectives of wort fermentation are to consistently metabolise wort constituents into ethanol, carbon dioxide, and other fermentation products in order to produce beer with satisfactory quality and stability. Another objective is to produce yeast crops that can be confidently re-pitched into subsequent brews [
7]. It is noteworthy that brewing is the only major fermentation process that recycles its yeast culture from one fermentation to another. Fermentation processes such as most potable and industrial distilled alcohol production, viticulture, saké brewing, and cider production only use their yeast culture once. As will be discussed later, the management of brewer’s yeast culture between fermentations is a critical part of the brewing process. It is important to jealously protect the quality of cropped yeast because it will be used to pitch a subsequent wort fermentation and will, therefore, have a profound effect on the quality of the beer resulting from it (see
Section 7).
Over the years, considerable effort has been devoted to studies on the biochemistry and genetic/molecular biology of brewer’s yeast (together with industrial strains in general). The objectives of these studies have been two-fold:
To learn more about the biochemical and genetic makeup of brewing yeast strains and
To improve the overall performance of such strains, with particular emphasis on broader substrate utilisation capabilities, increased ethanol production, and improved tolerance to environmental stress conditions such as temperature, elevated osmotic pressure, and ethanol, and finally to understand the mechanism(s) of flocculation.
There are several differences in the production of ale and lager beers, one of the main ones being the characteristics of the ale and lager yeast strains employed and the fermentation temperatures. Considerable research by many breweries and institutions on this topic has been conducted [
8,
9,
10] and the typical differences between the ale and lager yeast strains have been established (
Table 2). With the advent of molecular biology-based methodologies, gene sequencing of ale and lager brewing strains has shown that
S. pastorianus is an interspecies hybrid between
S. cerevisiae and
S. eubayanus, with homologous relationships to one another and also to
Saccharomyces bayanus, a yeast species used in wine fermentation and identified as a wild yeast in brewing fermentations.
Gallone et al. [
8] have proposed that current industrial yeasts (including brewing and distilling) can be divided into five sublinkages that are genetically and phenotypically separated from wild strains and originate from only a few ancestors through complex patterns of domestic and local divergence. Large-scale phenotyping and genome analysis further show strong industry-specific selection for stress tolerance, sugar utilization, and flavor production—details to follow.
In 2011, a paper entitled “Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast” was published [
11]. This study confirmed that
S. pastorianus is a domesticated yeast species created by the fusion of
S. cerevisiae with a previously unknown species that has now been designated
Saccharomyces eubayanus because of its close relationship to
S. bayanus. They proposed
S. eubayanus exists exclusively in the forests of Patagonia! Since this publication,
S. eubayanus has also been isolated and identified in Tibet [
12] and in the USA [
13]. The initial paper from Argentina contains a draft genome sequence of
S. eubayanus; it is 99.5% identical to the non-
S. cerevisiae DNA fragment of the
S. pastorianus genome sequence. This suggests specific changes in wort sugar and sulphate metabolism, compared to ale strains, that are critical for determining lager beer characteristics. The complete DNA sequence of
S. eubayanus has recently been published [
14].
4. Uptake and Metabolism of Wort Sugars
It is very important that wort sugars and the FAN complex are efficiently taken up by a brewer’s yeast culture. In normal situations, brewing yeast strains (both ale and lager strains) are capable of utilising sucrose, glucose, fructose, maltose, and maltotriose in this approximate sequence (or priority) (
Figure 5), although some degree of overlap does occur. The majority of brewing yeast strains leave the maltotetraose (G4) and other dextrins unfermented. However,
S. diastaticus is able to utilise dextrin material, albeit inefficiently. Wort dextrin utilisation is possible by this yeast sub-species due to the extracellular secretion of glucoamylase. However, this enzyme produced by
S. diastaticus, is incapable of hydrolysing the α-1,6 bonds of dextrins whereas it is able to hydrolyse the α-1,4 bonds [
15,
16]—details to follow.
The initial step in the utilisation of any wort sugar is either its passage intact across the yeast plasma membrane, or its hydrolysis outside the cell membrane, followed by entry into the cells of some or all of the hydrolysis products (
Figure 6). Maltose and maltotriose (
Figure 7) are examples of sugars that pass intact across the cell membrane, whereas sucrose and dextrins are hydrolysed by extracellular enzymes (invertase for sucrose and glucoamylase (amyloglucosidase) for dextrins) and the resultant simple sugars taken into the cell. An important metabolic difference between the uptake of monosaccharides such as glucose and fructose and disaccharides such as maltose and maltotriose uptake is that energy (ATP conversion to ADP) is required for maltose and maltotriose uptake (active transport) whereas glucose and fructose are taken up passively with no energy requirement [
17]. As maltose and maltotriose (
Table 1) are the major sugars in brewer’s wort, the ability of a brewing yeast strain to use these two sugars depends on the correct genetic complement. Brewer’s yeast possesses independent uptake mechanisms (maltose and maltotriose permeases) to transport these two sugars across the cell membrane into the cell. Once inside the cell, both sugars are hydrolysed to glucose units by the α-glucosidase system (
Figure 6). The transport, hydrolysis, and maltose fermentation systems are particularly important in brewing, distilling, and baking since maltose is the major sugar component of brewing wort, spirit mash, and wheat dough. Maltose fermentation by brewing, distilling, and baking yeasts requires at least one of five unlinked
MAL loci, each consisting of three genes encoding:
the structural gene for α-glucosidase (maltase) (MALS)
maltose permease (MALT) and
an activator whose product co-ordinately regulates the expression of the α-glucosidase and permease genes.
The expression of
MALS and
MALT is regulated by maltose induction and repression by glucose. When glucose concentrations are high (>10 mg/L), the
MAL genes are repressed, and only when 40%–50% of the glucose has been taken up from the wort will maltose and maltotriose uptake commence (
Figure 5).
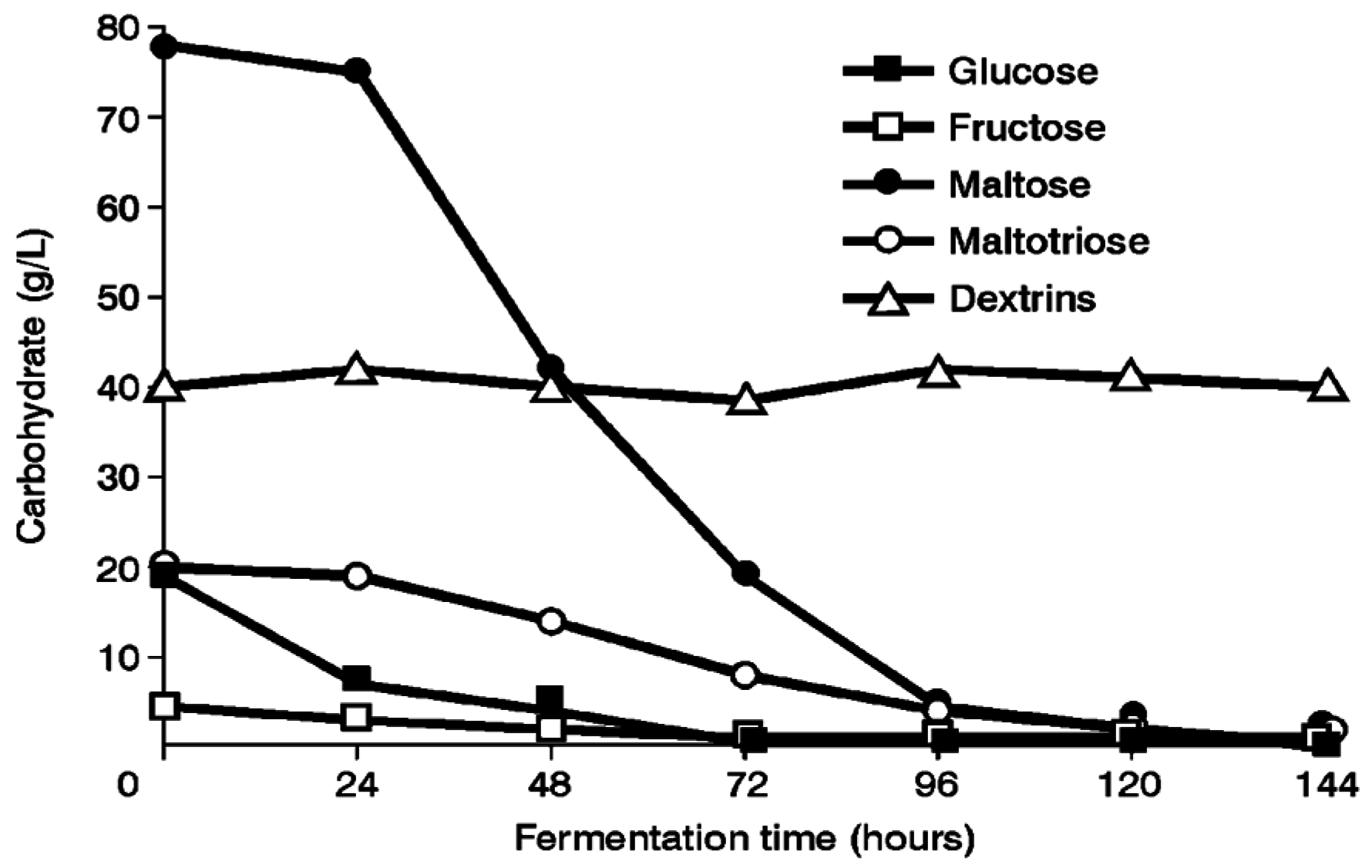
Several ale and lager yeast strains have been used to explore maltose and maltotriose uptake mechanisms in wort. For example, a 16°Plato all-malt wort was fermented in a 30 liter static vessel at 15 °C (
Figure 8). Under these conditions, lager strains utilised maltotriose more efficiently than ale strains, whereas maltose utilisation efficiency was not dependent on the type of brewing strain [
18,
19]. This supports the proposal that maltotriose and maltose possess independent, but closely linked, uptake (permease) systems [
18,
19]. In addition, this consistent difference between ale and lager strains supports the observation that ale strains appear to have greater difficulty than lager strains to completely ferment wort, particularly high gravity wort (>16°Plato) [
20].
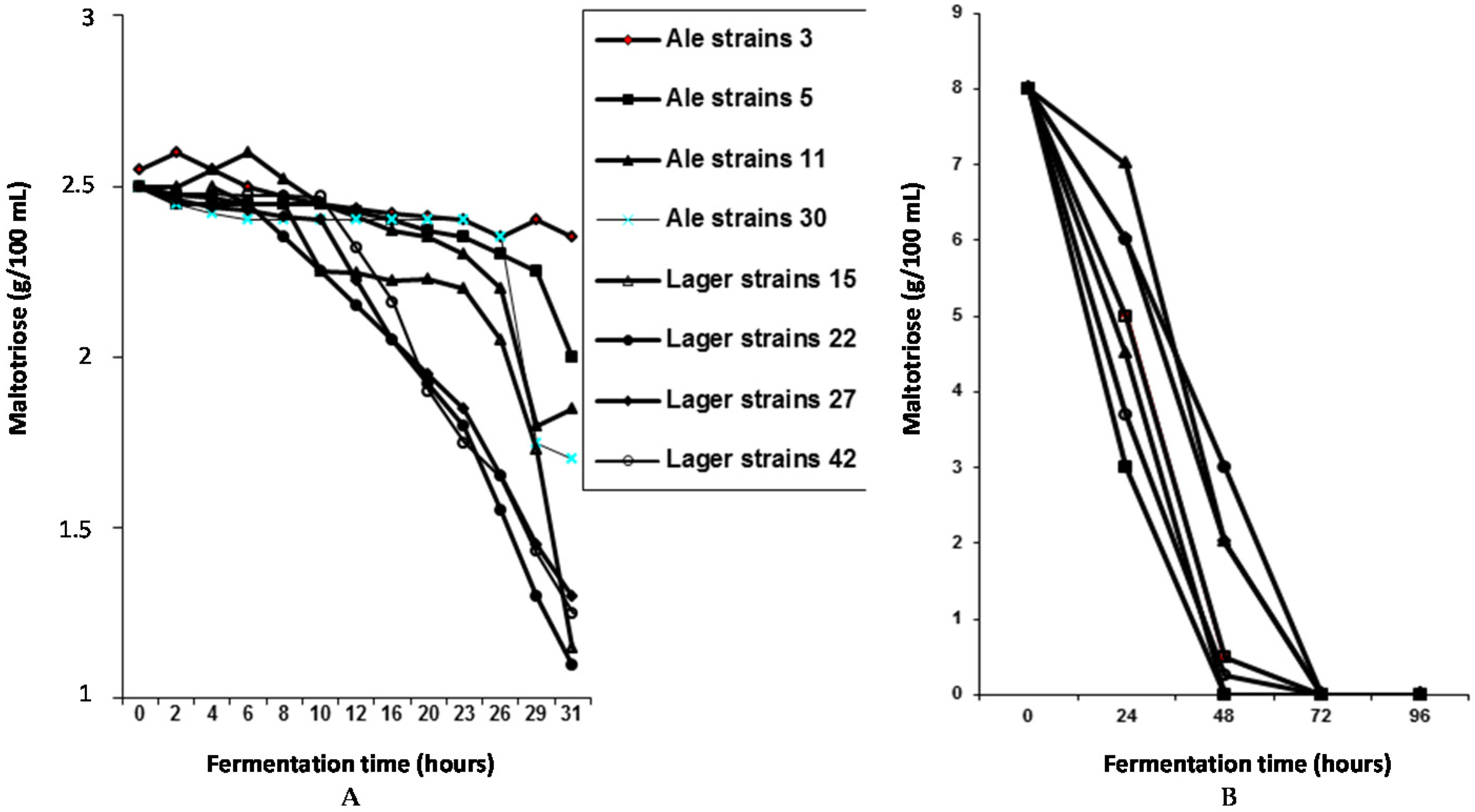
In order to investigate the
MAL-gene cassettes further, a strain containing two
MAL2 and two
MAL4 gene copies was constructed using hybridisation techniques [
21]. The wort (16°Plato) fermentation rate was compared to a strain that only contained one copy of
MAL2. As expected, the overall fermentation rate with the strain containing multiple
MAL genes was considerably faster than the strain containing the single
MAL2 copy (
Figure 9). The principal reason for this faster fermentation rate was due to an increased rate of maltose uptake and subsequent metabolism compared to the yeast strain containing a single
MAL2 copy (
Figure 9).
5. Carbohydrate Metabolism
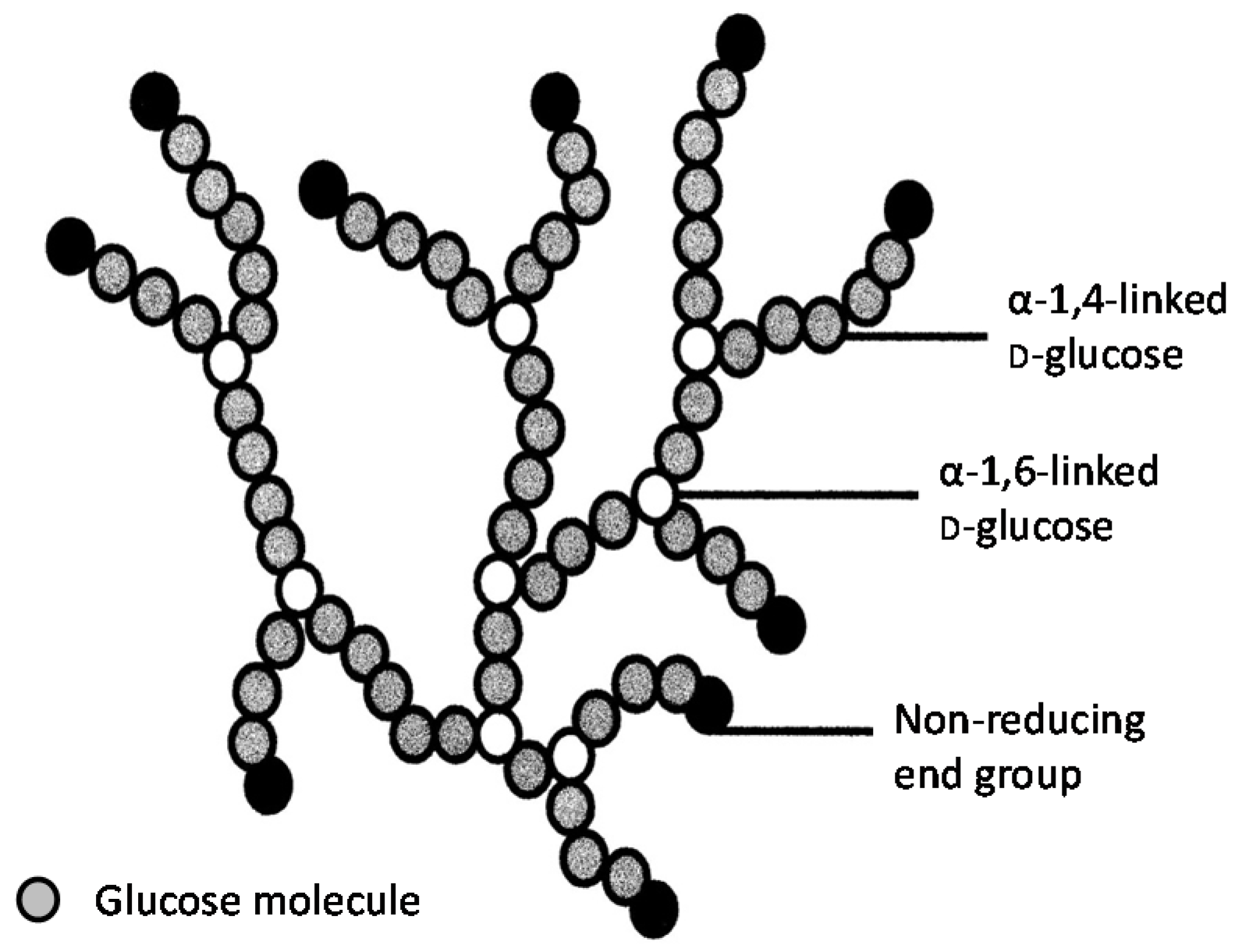
It has already been described that
S. diastaticus is a subspecies of
S. cerevisiae. It differs from the parental species by producing the extracellular enzyme glucoamylase (α-1,4-glucan glucohydrolase) [
22]. This enzyme removes successive glucose units from the non-reducing ends of dextrins (partially hydrolysed starch). Using hybridisation techniques, three non-allelic genes have been identified and characterised from strains of
S. diastaticus [
23]. Two
DEX genes (
DEX1 and
DEX2) have been identified that codes for the production of
S. diastaticus glucoamylase, with glucose being the sole hydrolysis product. A third dextrinase gene (
DEX3) was found to be allelic to
STA3, a gene reported to control starch fermentation in
Saccharomyces sp. [
24,
25].
Using hybridisation techniques [
21], a number of diploid strains have been developed, each being either heterozygous or homozygous for the individual
DEX genes. Wort fermentations with the
DEX containing cultures showed that in addition to the wort sugars (glucose, fructose, sucrose, maltose, and maltotriose), the dextrins (G4–G15) were partially fermented. There was residual dextrin in the fermented worts because the glucoamylase from
S. diastaticus strains was unable hydrolyse the α(1-6) branch points, only α(1-4) linkages.
Beer produced with
DEX-containing yeast strains has been studied. Glucose concentration in the pasteurized beer increased during storage for three months at room temperature—this indicates that this glucoamylase is not heat sensitive [
24]. Also, overall beer flavour produced by these
DEX-containing yeast strains was unacceptable because it contained 4-vinylguaiacol (clove-like), estery, sulphury, and musty aromas [
25]. Nevertheless, the brewing industry has continued to be interested in the glucoamylases produced by
S. diastaticus strains. Consequently, recombinant DNA (rDNA), plasmids, and transformation techniques have been employed to clone
DEX genes into brewing strains [
26]. In order to enhance glucoamylase activity, the glucoamylase gene from
Aspergillus niger was also cloned into a brewing yeast along with the
STA2 gene. The
A. niger glucoamylase has the advantage of possessing debranching activity, unlike the yeast enzyme, so that it can hydrolyse more of the wort dextrins [
26]. This enzyme is also sensitive to pasteurisation conditions.
The cloned amylolytic strains have received approval by the U.K. Agriculture and Health Ministries, who concluded in 1995 that there were no food safety reasons why this yeast should not be used [
27]. The reduced dextrin beer produced with this cloned yeast was called Nutfield Lyte (
Figure 10) and was the first beer in the world to be produced from this genetically modified yeast to receive approval. After considerable deliberation, it was decided that specific labelling was unnecessary. However, a back label to Nutfield Lyte bottles explained the process by which it had been produced and the advantages of doing so. Reaction to this beer from the general public has been neutral or positive. Media reports were largely factual and supportive. There was some criticism from the environmental lobby. This criticism has not diminished over the years and the production of modified foodstuffs (including alcoholic beverages) in Europe has been, and still is, severely restricted [
28].
6. Uptake and Metabolism of Free Amino Nitrogen (FAN)
FAN is a general measure of a yeast culture’s assimilable nitrogen efficiency. It is a good index for yeast growth and consequently fermentation efficiency and sugar uptake [
29]. Wort FAN is essential for the formation of new yeast amino acids, the synthesis of new structural and enzymatic proteins, cell proliferation, and cell viability and vitality. Lastly, FAN levels have a direct influence on beer flavour compounds (for example, higher alcohols, carbonyls, and esters).
There are differences between lager and ale yeast strains with respect to wort assimilable nitrogen uptake characteristics [
30]. Nevertheless, with all brewing strains the amount of wort FAN content required by yeast under normal brewery fermentation conditions is directly proportional to yeast growth and also affects certain aspects of beer maturation. There has been considerable problems regarding the minimal wort FAN required to achieve satisfactory yeast growth and fermentation performance in conventional gravity (10–12°Plato) wort, considered to be 130 mg FAN/L. For rapid attenuation of high gravity wort (>16°Plato), increased levels of FAN are required [
31]. However, optimum wort FAN levels differ from fermentation to fermentation and from yeast strain to yeast strain. Furthermore, the optimum FAN values are dependent on different wort sugar levels and type [
6].
Studies on the uptake of wort nitrogen by brewer’s yeast strains began over 50 years ago. During the 1960s, Margaret Jones and John Pierce, working in the Research Department of the Guinness Brewery in Park Royal, London, conducted notable studies on nitrogen metabolism during brewing, mashing, and fermentation [
32]. They reported that the absorption and utilisation of exogenous nitrogenous wort compounds and their synthesis intracellularly are controlled by three main factors:
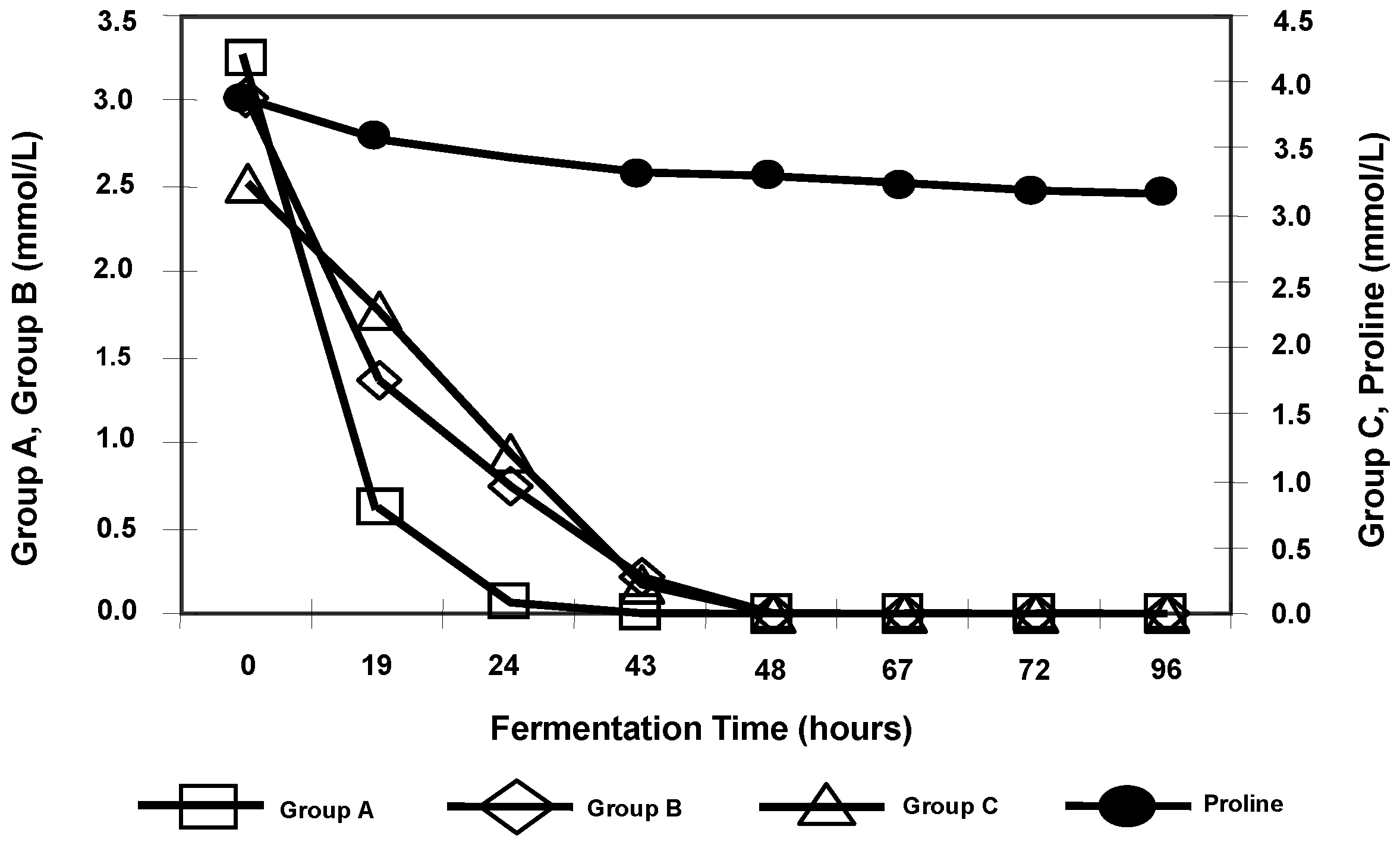
A unique classification of amino acids according to their rates of consumption during brewing wort fermentation has been established. There are four uptake groups of amino acids. Three groups of wort amino acids are taken up at different stages of fermentation and the fourth group consists of only one amino acid, proline (the largest concentration amino acid in wort), which is not taken up during brewing fermentations (
Figure 11). This is because of the anaerobic conditions that prevail late in the fermentation (oxygen is necessary for proline uptake). When this classification was developed, the methodology employed liquid chromatography for measuring individual amino acids and was iconic. Similar measurements today employ automated computerized high-performance liquid chromatography (HPLC), and it is difficult to envisage and appreciate the challenges that were faced and overcome 50 years ago! Indeed, advances in analytical methods have been a primary reason for our increasing knowledge of wort fermentation control.
The Jones and Pierce amino acid classification [
32,
33] continues to be the basis of our current understanding of the relative importance of individual wort amino acids during fermentation. This information has assisted manipulation of wort nitrogen levels by the addition of supplements such as yeast extract or specific amino acids, particularly during high gravity brewing. However, this assimilation pattern is often specific to the conditions employed. The nutritional preferences of a particular yeast strain are paramount. The absorption and utilisation of wort amino acids has recently been re-examined [
34] and the overall Jones and Pierce classification was confirmed using contemporary analytical techniques. However, the order of methionine uptake has been transferred from Group B to Group A (
Table 3).
It has also been reported [
35] that approximately 30% of incorporated nitrogen compounds come from sources other than amino acids. Although the utilisation of small peptides by brewing yeasts was confirmed over 50 years ago, an understanding of their role in yeast nitrogen requirements is still limited. Small peptides can be used as nutritional sources of amino acids as both carbon or nitrogen sources and precursors of cell wall peptides during yeast growth, although, growth is much slower when they are the sole nitrogen source [
36]. Polypeptides may also used as a substrate because yeasts can generate proteolytic enzymes extracellularly to provide additional assimilable nitrogen to the cells—see below and reference [
37].
Most brewing strains transport peptides with no more than three amino acid residues but this limit is strain dependent [
35]. Nevertheless, small wort peptides are an important source of assimilable nitrogen and 20%–40% of wort oligopeptides are used during fermentation. Similar to single amino acids, peptides probably contribute to beer character and flavour. Wort peptides are taken up in a specific order dependent on their amino acid composition. The determination of small (two and three amino acids) peptides has been an impediment to this area of research. However, a method to measure these small oligopeptides has now been developed [
35]. In brief, the sample is deproteinised, the supernatant ultrafiltered through a membrane with a 500 Da exclusion limit, the filtrate is acid and alkaline hydrolysed, and the hydrolysate is subjected to amino acid analysis by HPLC.
It has already been briefly discussed that yeast secretes proteinase into fermenting wort (mainly proteinases). Therefore, the hydrolysis of medium-sized peptides into smaller peptides occurs, which can be taken up by yeast cultures, and this activity continues throughout fermentation [
35]. This fact highlights a difference between the uptake of sugars and FAN during brewing. The spectrum of wort sugars at the beginning of fermentation is fixed because after wort boiling, all malt amylases have been inactivated. Whereas, because of proteinase secretion by yeast, the spectrum of nitrogen compounds is dynamic. This is not the case during malt and grain whisky fermentations where the wort is not boiled. Consequently, the wort still contains amylase activity (along with proteinase activity) and microbial contaminants. Consequently, the spectrum of wort carbohydrate is flexible [
38]. Additionally, stress effects on yeast can increase proteinase secretion and this occurs to a greater extent during the fermentation of high gravity worts [
37,
38].
Regarding wort fermentation by brewing yeast, the metabolism of wort sugars and FAN during fermentation has already been discussed but a plethora of other metabolic reactions also occur. As there are many reactions, they will not be discussed in detail here, but are covered in greater detail in reference [
39]. As a consequence, only three reactions will be considered:
7. Yeast Cropping—Flocculation and/or Centrifugation
Flocculation is one of the major factors when considering important characteristics of yeast strains during brewing fermentations [
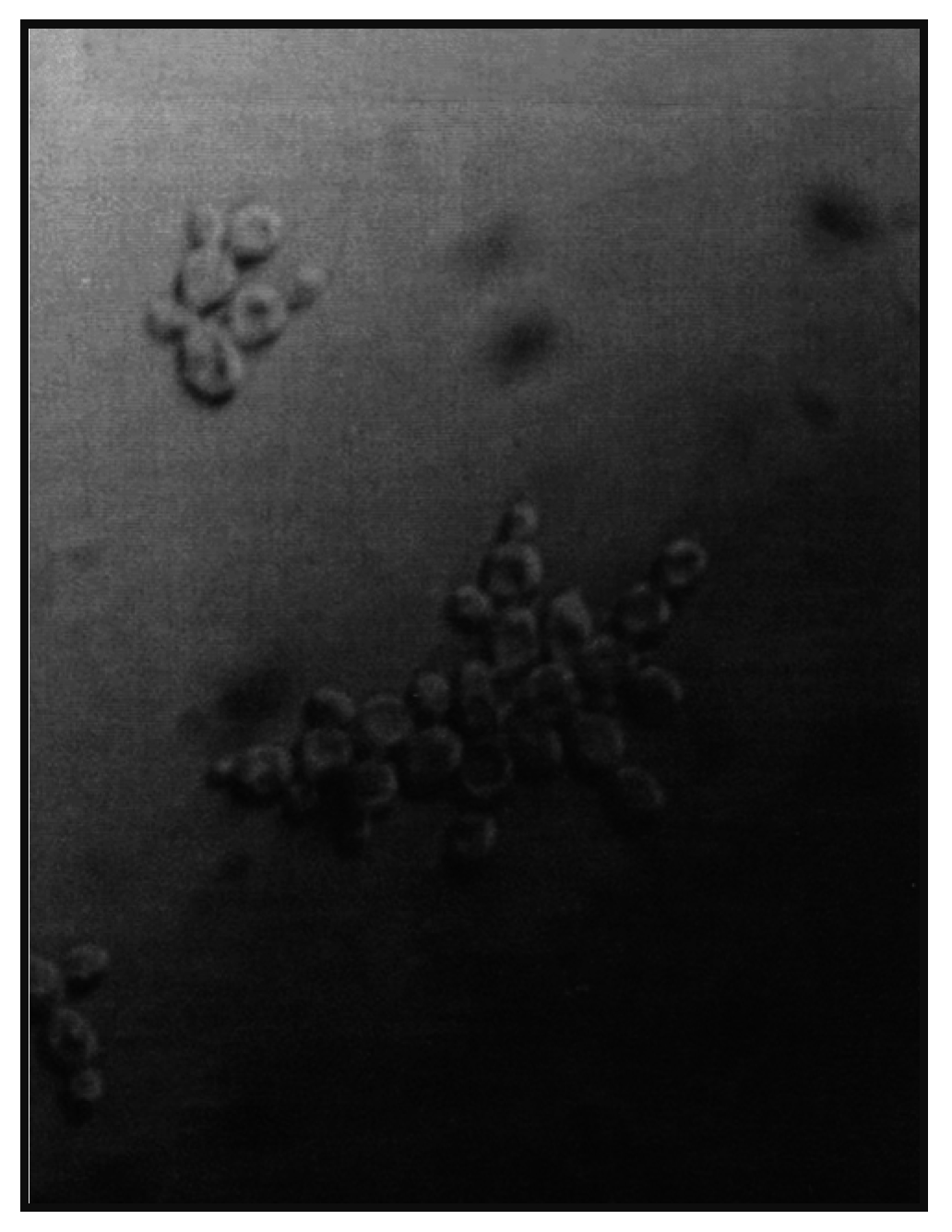
7]. Most brewing yeast strains are classified as either flocculent or non-flocculent (powdery) (
Figure 14) [
41,
42,
43]. One definition of flocculation that has been used for many years is: “the phenomenon wherein yeast cells adhere in clumps and either sediment from the medium in which they are suspended or rise to the medium’s surface” [
43]. This definition excludes forms of “clumpy growth” and “chain formation” [
44] which will not be discussed further here (
Figure 15).
It is important to crop a yeast culture at the end of fermentation either using a culture’s flocculation characteristics (
Figure 16) or with a centrifuge (
Figure 17). Flocculation is a cell surface phenomenon and the cell wall structure of a culture is critical (
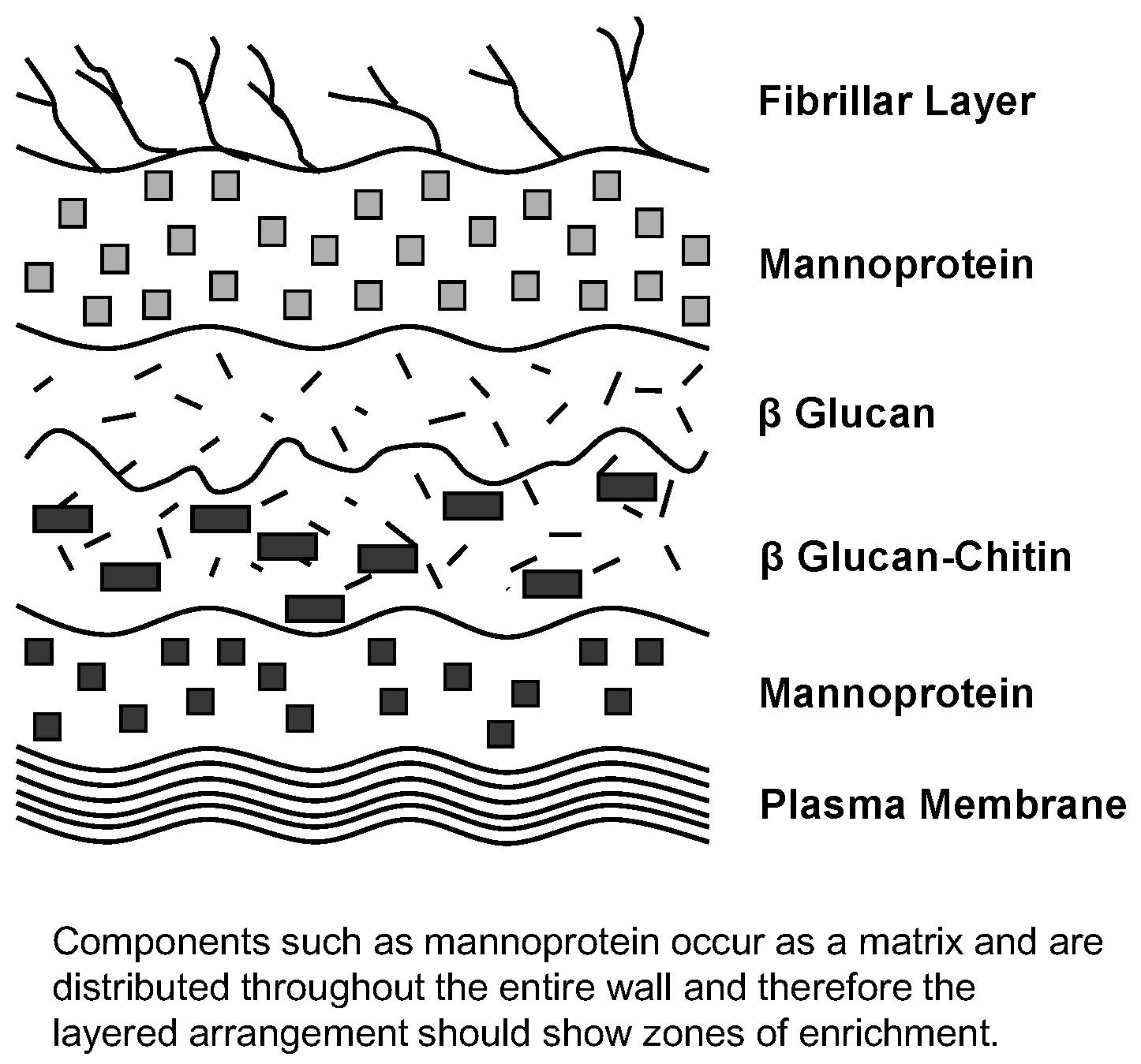
Figure 18). Components such as mannoprotein are found distributed throughout the entire yeast wall and therefore the wall’s layered arrangement exhibits zones of enrichment. It is a matrix, not a sandwich! Nevertheless, the mannoprotein is an integral structure that determines a yeast culture’s flocculation characteristics [
7].
Flocculation usually occurs in the absence of cell division (not always) during late logarithmic and early stationary growth phases and only under rather circumscribed environmental conditions involving specific yeast cell surface structures (mainly protein and carbohydrate (mannan) components) and also an interaction of calcium ions [
45]. Only a microamount of calcium ions is necessary for the flocculation of brewer’s yeast strains. Cells can be deflocculated as a result of the addition of sugars such as mannose and also treatment with chelating agents such as ethylenediamine tetraacetic acid (EDTA) that complex with Ca
++ ions. Cells will usually reflocculate rapidly upon the addition of Ca
++ ions [
45].
Although yeast separation often occurs by sedimentation, it may also be by flotation because of cell aggregation entrapping bubbles of CO
2 as is the case of hydrophobic top-cropping ale brewing yeast strains. Co-flocculation [
46] (also called mutual agglutination [
47], mutual aggregation, and mutual flocculation) is an aggregation process between two yeast strains (yeast-bacteria co-flocculation has also been reported [
48,
49]). When two ale strains (one being non-flocculent and the other weakly flocculent) are mixed together in the presence of Ca
++, flocs form and the culture rapidly settles out of suspension or rises to the surface during static wort fermentation. Co-flocculation has not been reported in lager yeast strains [
50].
Genetic studies on yeast flocculation began over 60 years ago. Thorne [
51] and Gilliland [
52] independently confirmed that this phenomenon was an inherited characteristic with the flocculation phenotype being dominant over non-flocculence. The first flocculation gene (
FLO gene) to be studied in detail was
FLO1. Using traditional gene mapping techniques (mating, sporulation, micromanipulation, tetrad analysis, etc.) [
21], it was shown that
FLO1 is located on chromosome I, 33cM from the centromere on the right-hand side of the chromosome [
53].
Novel genetic techniques have been developed, the principal of which is the sequencing of the
Saccharomyces genome [
54]. A sequenced laboratory yeast strain contains five
FLO genes, four located near the chromosome telomeres
FLO1, FLO5, FLO9, and
FLO10 and one neither at the centromere nor at the telomeres,
FLO11 [
55]. At least nine genes—
FLO1, FLO5, FLO9, FLO10, FLO11, FLONL, FLONS, and
LgFLO in
S. cerevisiae and
S. pastorianus—encode flocculin proteins. The flocculin encoded by
FLO11 differs from other
FLO genes in that it is involved in filamentous growth, adhesion to solid surfaces, and floc formation rather flocculation per se [
56]. Another
FLO gene,
FLO8, encodes for a transposable factor regulating the expression of other
FLO genes [
57]. It has already been stated that many yeast strains usually possess several
FLO genes in their genome [
56].
FLO genes are very long (up to 4 Kbp) because of the number of tandem repeated DNA sequences of about 100 nucleotides that are repeated 10–20 times in each gene [
56]. Tandem repeated DNA sequences of the
FLO genes are highly dynamic components of the genome—they change more rapidly than other parts of the genome. They enable rearrangements both between and within flocculin genes. The longer the
FLO protein, the stronger the strain’s flocculation ability [
56].
FLO1 has been identified as the longest
FLO gene, with the most repeats and confers the strongest flocculation phenotype to a yeast strain [
57].
The sedimentation performance of a brewer’s yeast strain often changes during repeated re-pitching in a brewery [
39,
58]. This could be due to irreversible or reversible genetic change. Alternatively, it could be due to long lasting physiological changes—perhaps modifications in yeast handling and fermentation environments (for example, the use of high gravity worts). When a genetic change, conferring a non-flocculent phenotype, occurs in a yeast culture, it will gradually become a mixture of flocculent and non-flocculent cells [
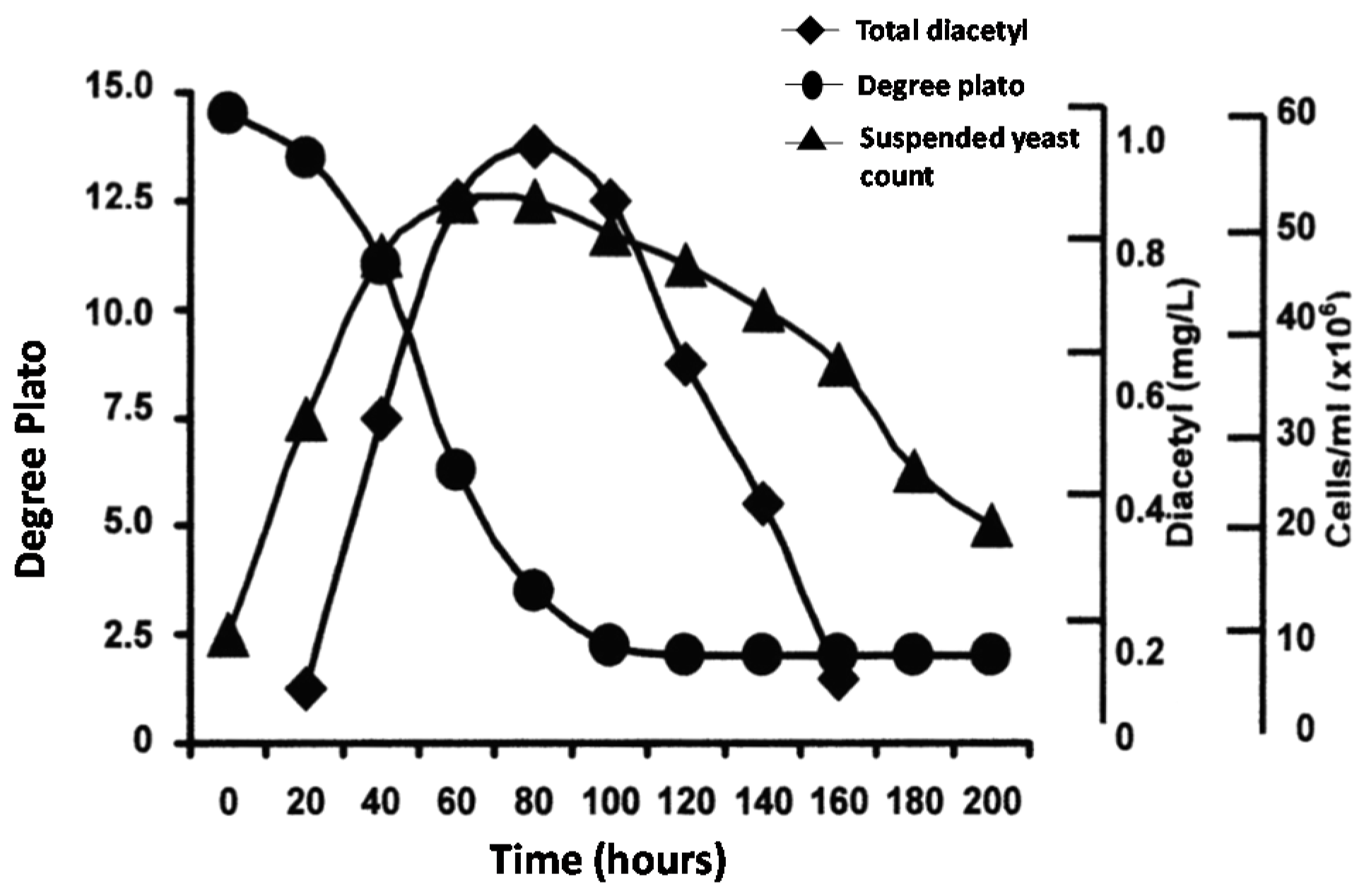
39]. Often within a lager yeast population, exhibiting moderate flocculent characteristics, a more flocculent variant culture can be isolated. An example of this development was when a Canadian brewing company began brewing its beer, under contract, in breweries in the United Kingdom. Most of the contracted breweries employed vertical fermenters. However, at that time, the Canadian breweries only possessed horizontal tanks (as both fermentation and maturation vessels). This difference in tank geometry influenced the yeast culture’s sedimentation characters. In vertical fermenters, this yeast culture was too non-flocculent (powdery), with too much yeast remaining in suspension at the end of fermentation (centrifuges were not available at the time). It was thought possible that the culture contained a spectrum of isolates that exhibited various flocculation intensities. Consequently, one of the variants from this strain, with more intensive flocculation characteristics, was successfully isolated and employed in the vertical fermenters. The result was less yeast in suspension at the end of fermentation. However, care had to be taken to ensure that the flocculent variant used was not too flocculent because under-fermented wort and residual unwanted beer flavours (for example, diacetyl and other VDKs—
Figure 16) could have been the result [
39].
The alternative way to crop yeast for reuse and to clarify primary fermented wort is with the use of a disk stack centrifuge (
Figure 17). However, when a yeast culture is passed through a centrifuge it can experience mechanical and hydrodynamic shear stresses unless the centrifuge is correctly operated under the appropriate conditions. This shear stress can cause a decrease in cell viability/vitality, cell wall damage, injury to the mitochondrial DNA leading to formation of respiratory deficient mutants, increased extracellular proteinase A levels, hazier beer, and reduced foam stability [
59].