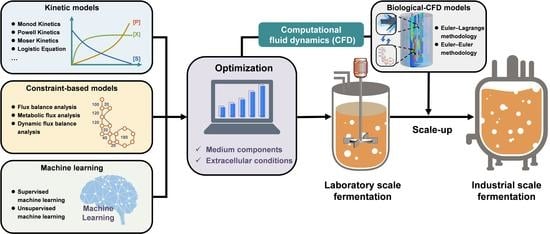
Optimization and Scale-Up of Fermentation Processes Driven by Models
Abstract
:1. Introduction
2. Methods and Applications of Mechanistic Modeling
2.1. Application of Kinetic Modeling to Fermentation Processes
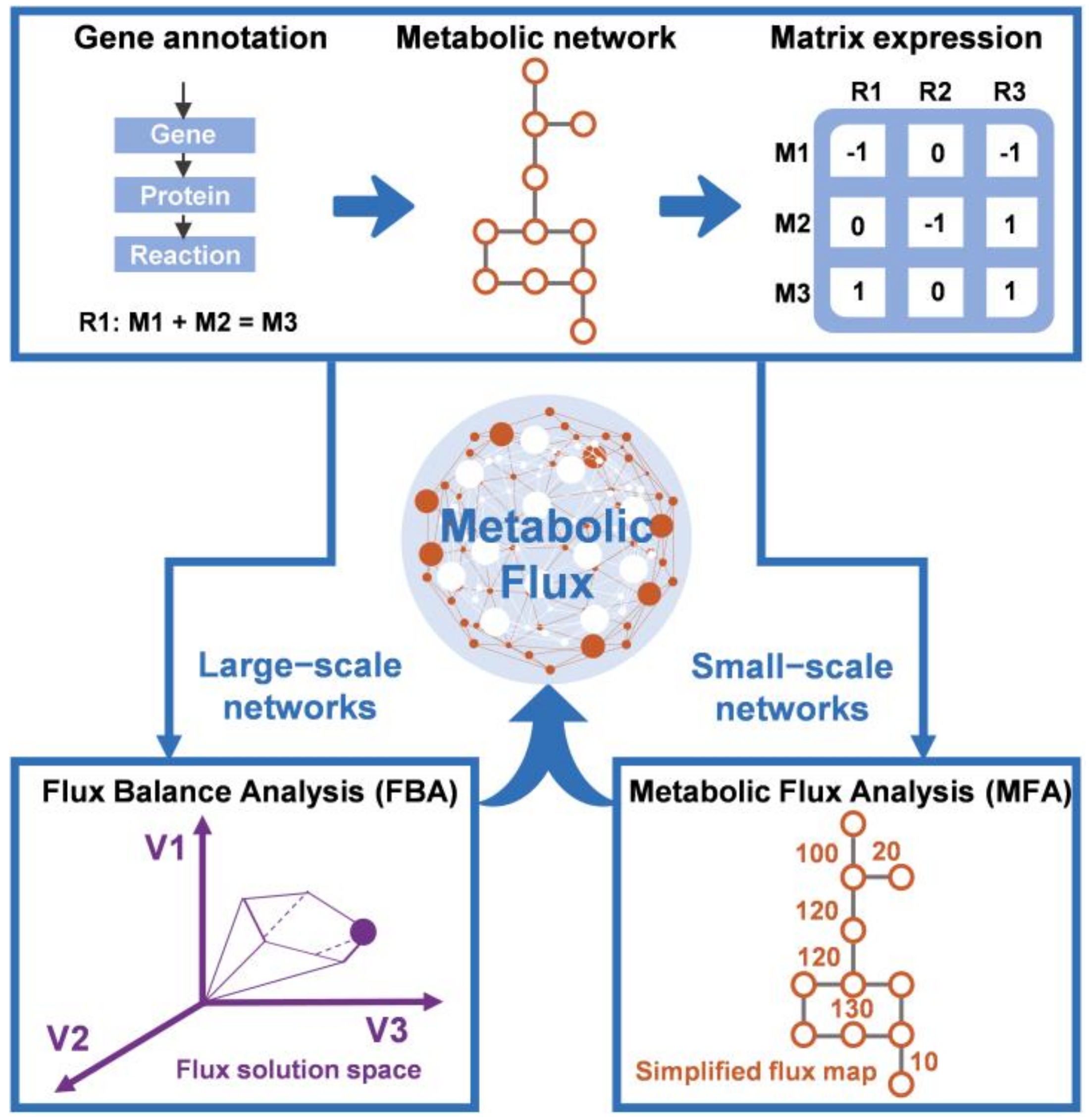
2.2. Application of Constraint-Based Modeling to Fermentation Processes
2.2.1. Flux Balance Analysis
2.2.2. Metabolic Flux Analysis
2.2.3. Dynamic Flux Balance Analysis
| Parameter | Approach | Case | Refs. |
|---|---|---|---|
| Theoretical maximum | FBA | The relationship between various products and biomass in the process of butyric acid fermentation was described, and the theoretical yield of several fermentation products of butyric acid bacteria was predicted accurately. | [66] |
| Theoretical maximum | FBA | Quantitative prediction of maximum cell growth rate and cell density of wild-type E. coli W3110 were clarified. | [67] |
| Culture medium | FBA | Amino acids and carbon sources that have a significant influence on the yield were identified, and the yield of siderophore compounds in recombinant E. coli was improved by medium optimization. | [68] |
| Culture medium | FBA | The effects of glucose, glycerol, and the mixture of glucose and glycerol on the distribution of carbon flux in the simultaneous production of ethanol and butanol by Clostridium sporogenes NCIM 2918 were studied. | [69] |
| Culture medium | FBA | The effects of amino acid composition in a culture medium on the catabolism of Chinese hamster ovary (CHO) cells were analyzed to optimize culture medium formulation and increase antibody production. | [70] |
| pH | MFA | By analyzing the effect of pH on the intracellular metabolic network of β-lactamase producing Bacillus licheniformis, a pH manipulation strategy was proposed to improve the yield of β -lactamase. | [77] |
| Ultrasound | MFA | The effect of ultrasound promoting biological hydrogen production from glycerol fermentation was understood to a significant extent, and an optimal strategy of enhancing glycerol uptake and blocking the butyric acid pathway under the guidance of the MFA model was proposed. | [78] |
| Temperature | MFA | By quantifying the flux during l-lactic acid production from glucose, a temperature control strategy was proposed to maximize the productivity of L-lactic acid. | [79] |
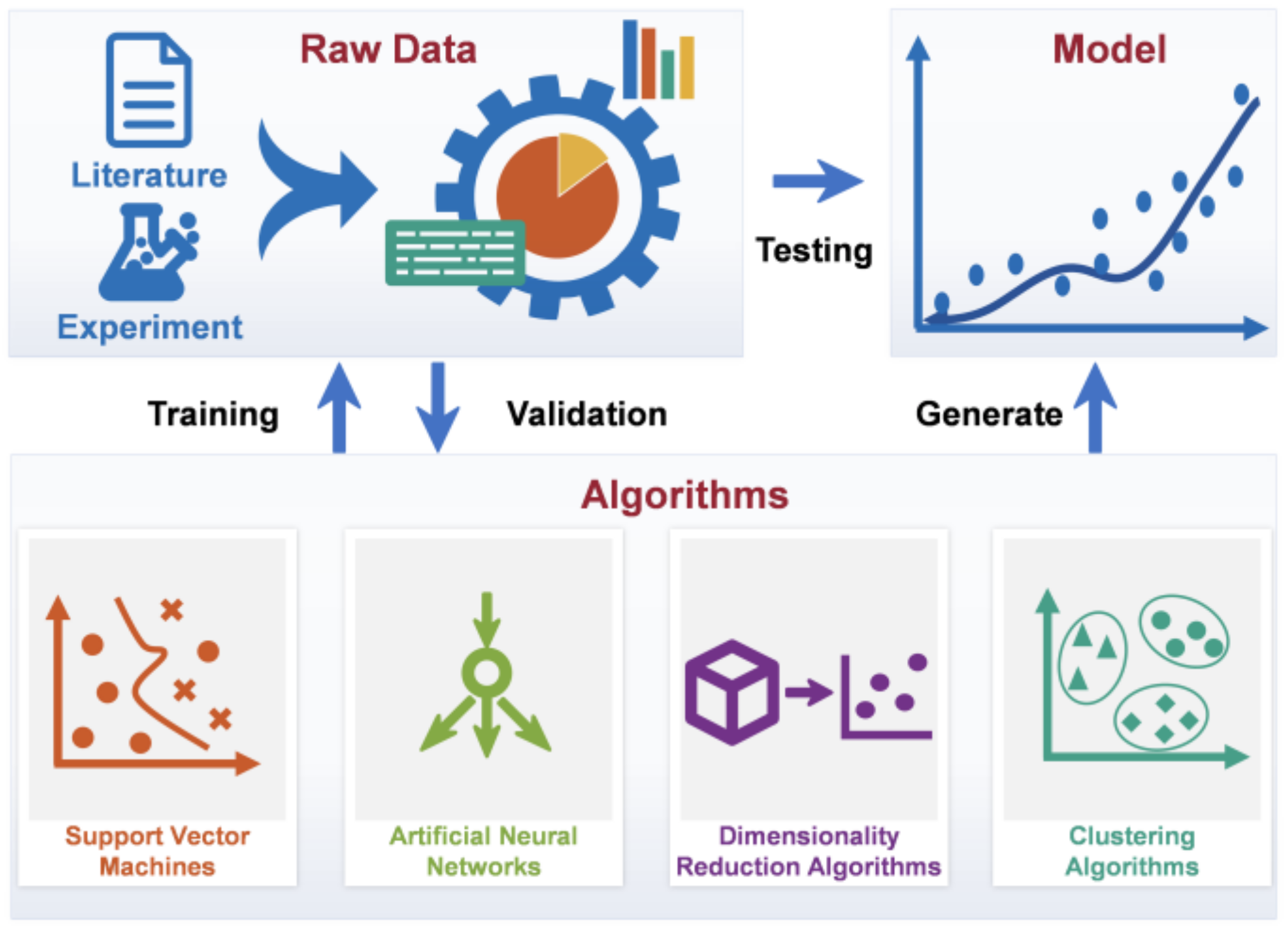
3. Methods and Applications of Data-Driven Modeling
3.1. Supervised Machine Learning
3.2. Unsupervised Machine Learning
4. Hybrid Models and Modeling Methods
- (a)
- CBM models were constructed using ML by merging and analyzing omics data from different sources, whereas CBM was trained and reassembled by obtaining genomic data under specific conditions [123]. This method is suitable for situations wherein mechanistic models are not accurate enough. Vijayakumar et al. used ML to analyze RNA sequencing data extracted under 23 different growth conditions, which was then combined with flux data obtained by using FBA to elucidate the mechanisms underlying cyanobacterial responses to fluctuations in light intensity and salinity [124]. The growth rates of yeasts, such as S. cerevisiae, have also been predicted using this technique. For example, Culley et al. obtained reliable results for the growth rate prediction of S. cerevisiae by combining CBM-derived flux omics data with transcriptomics using ANNs [116].
- (b)
- Metabolic flux data obtained from CBM was trained by the ML method to gain more biological insights into the required system [125]. With this method, potential phenomena that cannot be mechanistically described can be analyzed. For example, Sridhara et al. used ML to analyze the metabolic flux data generated by CBM, and they realized that the retroversion of the culture medium components, which occurred during bacterial growth, could not be achieved using CBM alone [126].
- (c)
- ML can be used to analyze multi-omics data so as to provide data preprocessing services for CBM model construction. In 2016, Wu et al. used the ML method to analyze and integrate heterotrophic bacterial metabolic data from about 100 papers, finally constructing MFlux, a Web-based platform that can analyze metabolic fluxes [127].
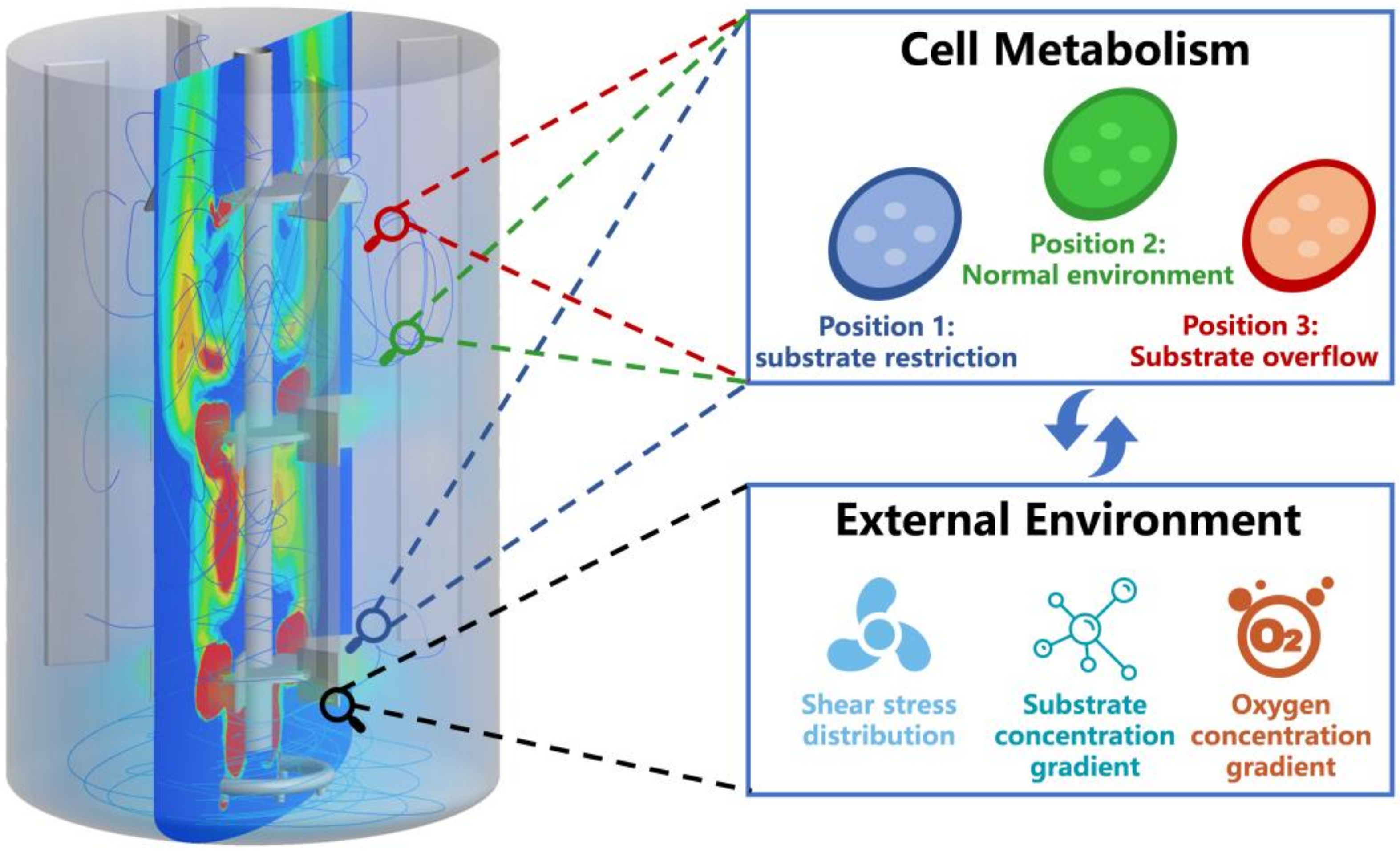
5. Coupling Biological Models with Computational Fluid Dynamics Models Enabling Rational Fermentation Scale-Up
| Approach | Application | Refs. |
|---|---|---|
| ELM | The transcriptional changes of Clostridium ljungdahlii cells subjected to CO restriction in a 125 m3 bubble column bioreactor was predicted, which guided the scaling-up of production. | [140] |
| ELM | The decrease in penicillin production when using P. chrysogenum due to glucose gradient in a 54 m3 stirred tank reactor was predicted. | [57] |
| ELM | The formation of population heterogeneity in E. coli in a 54 m3 bioreactor was predicted. | [141] |
| ELM | The difference in microalgae biomass in different photoreactors caused by different light distributions was predicted. | [143] |
| EEM | The reason for the decrease in the gluconic acid yield during the production of gluconic acid by Aspergillus Niger was revealed, which was due to the decrease in oxygen mass transfer due to the increase in medium viscosity during fermentation. | [145] |
| EEM | The performance degradation of the industrial bioreactor under poor mixing conditions was explained by comparing the flow field environment of the laboratory bioreactor (70 L) with that of the industrial (70 m3) bioreactor. | [146] |
| EEM | The effects of the size of the bioreactor and the operating conditions on DHA fermentation were predicted, and DHA fermentation was scaled up from 5 L to 35 m3. | [49] |
| EEM | The biological production process was fine-tuned by coupling CFD and biokinetics, and the scale required to turn ferulic acid into vanillin (scaling it up from shaker to bioreactor) was realized, with a conversion rate up to 94%. | [147] |
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- European Technology and Innovation Platform: Bioenergy for Industry. Available online: https://etipbioenergy.eu/industry/production-facilities (accessed on 30 July 2022).
- The Engineering Chemical Technology Centre: Biodegradable Plastic and World Biopolymers Market 2019–2020. Available online: https://ect-center.com/blog/biopolymers-market-2019 (accessed on 30 July 2022).
- Du, J.; Shao, Z.; Zhao, H. Engineering Microbial Factories for Synthesis of Value-Added Products. J. Ind. Microbiol. Biotechnol. 2011, 38, 873–890. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.; Teo, W.; Chen, B.; Leong, S.; Chang, M. Microbial Tolerance Engineering toward Biochemical Production: From Lignocellulose to Products. Curr. Opin. Biotechnol. 2014, 29, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Magocha, T.; Zabed, H.; Yang, M.; Yun, J.; Zhang, H.; Qi, X. Improvement of Industrially Important Microbial Strains by Genome Shuffling: Current Status and Future Prospects. Bioresour. Technol. 2018, 257, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.; Hirz, M.; Pichler, H.; Schwab, H. Protein Expression in Pichia Pastoris: Recent Achievements and Perspectives for Heterologous Protein Production. Appl. Microbiol. Biotechnol. 2014, 98, 5301–5317. [Google Scholar] [CrossRef]
- Sawant, A.M.; Vamkudoth, K.R. Biosynthetic Process and Strain Improvement Approaches for Industrial Penicillin Production. Biotechnol. Lett. 2022, 44, 179–192. [Google Scholar] [CrossRef]
- Favaro, L.; Jansen, T.; van Zyl, W.H. Exploring Industrial and Natural Saccharomyces Cerevisiae Strains for the Bio-Based Economy from Biomass: The Case of Bioethanol. Crit. Rev. Biotechnol. 2019, 39, 800–816. [Google Scholar] [CrossRef]
- Jansen, M.L.A.; Bracher, J.M.; Papapetridis, I.; Verhoeven, M.D.; de Bruijn, H.; de Waal, P.P.; van Maris, A.J.A.; Klaassen, P.; Pronk, J.T. Saccharomyces Cerevisiae Strains for Second-Generation Ethanol Production: From Academic Exploration to Industrial Implementation. FEMS Yeast Res. 2017, 17, fox044. [Google Scholar] [CrossRef]
- Nielsen, J.; Keasling, J.D. Engineering Cellular Metabolism. Cell 2016, 164, 1185–1197. [Google Scholar] [CrossRef]
- Wu, T.; Li, S.; Ye, L.; Zhao, D.; Fan, F.; Li, Q.; Zhang, B.; Bi, C.; Zhang, X. Engineering an Artificial Membrane Vesicle Trafficking System (AMVTS) for the Excretion of β-Carotene in Escherichia coli. ACS Synth. Biol. 2019, 8, 1037–1046. [Google Scholar] [CrossRef]
- Baeshen, N.A.; Baeshen, M.N.; Sheikh, A.; Bora, R.S.; Ahmed, M.M.M.; Ramadan, H.A.I.; Saini, K.S.; Redwan, E.M. Cell Factories for Insulin Production. Microb. Cell Factories 2014, 13, 141. [Google Scholar] [CrossRef] [Green Version]
- Jiang, G.Z.; Yao, M.D.; Wang, Y.; Zhou, L.; Song, T.Q.; Liu, H.; Xiao, W.H.; Yuan, Y.J. Manipulation of GES and ERG20 for Geraniol Overproduction in Saccharomyces cerevisiae. Metab. Eng. 2017, 41, 57–66. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Y.; Liu, L.; Wang, M.; Li, J.; Du, G.; Chen, J. Modular Pathway Engineering of Key Carbon-precursor Supply-pathways for Improved N-acetylneuraminic Acid Production in Bacillus subtilis. Biotechnol. Bioeng. 2018, 115, 2217–2231. [Google Scholar] [CrossRef]
- Westbrook, A.W.; Ren, X.; Oh, J.; Moo-Young, M.; Chou, C.P. Metabolic Engineering to Enhance Heterologous Production of Hyaluronic Acid in Bacillus subtilis. Metab. Eng. 2018, 47, 401–413. [Google Scholar] [CrossRef]
- De, T.; Sikder, J.; Narayanan, C.M. Biodiesel Synthesis Using Immobilised Lipase Enzyme in Semi-fluidised Bed Bioreactors-Bioreactor Design and Performance Analysis. Environ. Prog. Sustain. Energy 2017, 36, 1537–1545. [Google Scholar] [CrossRef]
- Brindhadevi, K.; Shanmuganathan, R.; Pugazhendhi, A.; Gunasekar, P.; Manigandan, S. Biohydrogen Production using Horizontal and Vertical Continuous Stirred Tank Reactor—A Numerical Optimization. Int. J. Hydrog. Energy 2021, 46, 11305–11312. [Google Scholar] [CrossRef]
- Chu, R.Y.; Li, S.X.; Zhu, L.D.; Yin, Z.H.; Hu, D.; Liu, C.C.; Mo, F. A review on Co-cultivation of Microalgae with Filamentous Fungi: Efficient Harvesting, Wastewater Treatment and Biofuel Production. Renew. Sust. Energy Rev. 2021, 139, 110689. [Google Scholar] [CrossRef]
- Pistikopoulos, E.N.; Barbosa-Povoa, A.; Lee, J.H.; Misener, R.; Mitsos, A.; Reklaitis, G.V.; Venkatasubramanian, V.; You, F.; Gani, R. Process Systems Engineering—The Generation Next? Comput. Chem. Eng. 2021, 147, 107252. [Google Scholar] [CrossRef]
- Shih, W.; Chai, S. Data-Driven vs. Hypothesis-Driven Research: Making sense of big data. Acad. Manag. J. 2016, 2016, 14843. [Google Scholar] [CrossRef]
- Straathof, A.J.J.; Wahl, S.A.; Benjamin, K.R.; Takors, R.; Wierckx, N.; Noorman, H.J. Grand Research Challenges for Sustainable Industrial Biotechnology. Trends Biotechnol. 2019, 37, 1042–1050. [Google Scholar] [CrossRef]
- Xia, J.Y.; Wang, G.; Lin, J.H.; Wang, Y.H.; Chu, J.; Zhuang, Y.P.; Zhang, S.L. Advances and Practices of Bioprocess Scale-up. In Bioreactor Engineering Research and Industrial Applications II; Bao, J., Ye, Q., Zhong, J.J., Eds.; Advances in Biochemical Engineering-Biotechnology; Springer: Berlin/Heidelberg, Germany, 2016; Volume 152, pp. 137–151. [Google Scholar]
- Neubauer, P.; Cruz, N.; Glauche, F.; Junne, S.; Knepper, A.; Raven, M. Consistent development of bioprocesses from microliter cultures to the industrial scale. Eng. Life Sci. 2013, 13, 224–238. [Google Scholar] [CrossRef]
- Solle, D.; Hitzmann, B.; Herwig, C.; Pereira Remelhe, M.; Ulonska, S.; Wuerth, L.; Prata, A.; Steckenreiter, T. Between the Poles of Data-Driven and Mechanistic Modeling for Process Operation. Chem. Ing. Tech. 2017, 89, 542–561. [Google Scholar] [CrossRef]
- Flevaris, K.; Chatzidoukas, C. Facilitating the industrial transition to microbial and microalgal factories through mechanistic modelling within the Industry 4.0 paradigm. Curr. Opin. Chem. Eng. 2021, 33, 100713. [Google Scholar] [CrossRef]
- Yasemi, M.; Jolicoeur, M. Modelling Cell Metabolism: A Review on Constraint-Based Steady-State and Kinetic Approaches. Processes 2021, 9, 322. [Google Scholar] [CrossRef]
- Khaleghi, M.K.; Savizi, I.S.P.; Lewis, N.E.; Shojaosadati, S.A. Synergisms of machine learning and constraint-based modeling of metabolism for analysis and optimization of fermentation parameters. Biotechnol. J. 2021, 16, 2100212. [Google Scholar] [CrossRef]
- Helmy, M.; Smith, D.; Selvarajoo, K. Systems biology approaches integrated with artificial intelligence for optimized metabolic engineering. Metab. Eng. Commun. 2020, 11, e00149. [Google Scholar] [CrossRef]
- Kim, Y.; Kim, G.B.; Lee, S.Y. Machine learning applications in genome-scale metabolic modeling. Curr. Opin. Struct. Biol. 2021, 25, 42–49. [Google Scholar] [CrossRef]
- Antonakoudis, A.; Barbosa, R.; Kotidis, P.; Kontoravdi, C. The era of big data: Genome-scale modelling meets machine learning. Comput. Struct. Biotechnol. J. 2020, 18, 3287–3300. [Google Scholar] [CrossRef]
- Nadal-Rey, G.; McClure, D.D.; Kavanagh, J.M.; Cornelissen, S.; Fletcher, D.F.; Gernaey, K.V. Understanding gradients in industrial bioreactors. Biotechnol. Adv. 2021, 46, 107660. [Google Scholar] [CrossRef]
- Wang, G.; Haringa, C.; Tang, W.; Noorman, H.; Chu, J.; Zhuang, Y.; Zhang, S. Coupled metabolic-hydrodynamic modeling enabling rational scale-up of industrial bioprocesses. Biotechnol. Bioeng. 2020, 117, 844–867. [Google Scholar] [CrossRef]
- Joyce, A.R.; Palsson, B. The model organism as a system: Integrating ‘omics’ data sets. Nat. Rev. Mol. Cell Biol. 2006, 7, 198–210. [Google Scholar] [CrossRef]
- Kim, O.D.; Rocha, M.; Maia, P. A review of dynamic modeling approaches and their application in computational strain optimization for metabolic engineering. Front. Microbiol. 2018, 9, 1690. [Google Scholar] [CrossRef] [PubMed]
- Theodoropoulos, C.; Sun, C. 2.45—Bioreactor Models and Modeling Approaches. In Comprehensive Biotechnology, 3rd ed.; Moo-Young, M., Ed.; Pergamon: Oxford, UK, 2019; pp. 663–680. [Google Scholar]
- Zhang, A.H.; Huang, S.Y.; Zhuang, X.Y.; Wang, K.; Yao, C.Y.; Fang, B.S. A novel kinetic model to describe 1,3-propanediol production fermentation by Clostridium butyricum. AIChE J. 2019, 65, e16587. [Google Scholar] [CrossRef]
- Garnier, A.; Gaillet, B. Analytical solution of Luedeking–Piret equation for a batch fermentation obeying Monod growth kinetics. Biotechnol. Bioeng. 2015, 112, 2468–2474. [Google Scholar] [CrossRef] [PubMed]
- Bhowmik, S.K.; Alqahtani, R.T. Mathematical analysis of bioethanol production through continuous reactor with a settling unit. Comput. Chem. Eng. 2018, 111, 241–251. [Google Scholar] [CrossRef]
- Zentou, H.; Zainal Abidin, Z.; Yunus, R.; Awang Biak, D.R.; Zouanti, M.; Hassani, A. Modelling of Molasses Fermentation for Bioethanol Production: A Comparative Investigation of Monod and Andrews Models Accuracy Assessment. Biomolecules 2019, 9, 308. [Google Scholar] [CrossRef]
- Moodley, P.; Gueguim Kana, E.B. Bioethanol production from sugarcane leaf waste: Effect of various optimized pretreatments and fermentation conditions on process kinetics. Biotechnol. Rep. 2019, 22, e00329. [Google Scholar] [CrossRef]
- Ranjbar, S.; Hejazi, P. Modeling and validating Pseudomonas aeruginosa kinetic parameters based on simultaneous effect of bed temperature and moisture content using lignocellulosic substrate in packed-bed bioreactor. Food Bioprod. Process. 2019, 117, 51–63. [Google Scholar] [CrossRef]
- Ray, M.; Kumar, V.; Banerjee, C. Kinetic modelling, production optimization, functional characterization and phyto-toxicity evaluation of biosurfactant derived from crude oil biodegrading Pseudomonas sp. IITISM 19. J. Environ. Chem. Eng. 2022, 10, 107190. [Google Scholar] [CrossRef]
- Beagan, N.; O’Connor, K.E.; Del Val, I.J. Model-based operational optimisation of a microbial bioprocess converting terephthalic acid to biomass. Biochem. Eng. J. 2020, 158, 107576. [Google Scholar] [CrossRef]
- Finkler, A.T.J.; de Lima Luz, L.F.; Krieger, N.; Mitchell, D.A.; Jorge, L.M. A model-based strategy for scaling-up traditional packed-bed bioreactors for solid-state fermentation based on measurement of O2 uptake rates. Biochem. Eng. J. 2021, 166, 107854. [Google Scholar] [CrossRef]
- Feng, F.; Li, Y.; Latimer, B.; Zhang, C.; Nair, S.S.; Hu, Z. Prediction of maximum algal productivity in membrane bioreactors with a light-dependent growth model. Sci. Total Environ. 2021, 753, 141922. [Google Scholar] [CrossRef]
- Yu, L.; Li, T.T.; Ma, J.W.; Zhao, Q.B.; Wensel, P.; Lian, J.N.; Chen, S.L. A kinetic model of heterotrophic and mixotrophic cultivation of the potential biofuel organism microalgae Chlorella sorokiniana. Algal Res. 2022, 64, 102701. [Google Scholar] [CrossRef]
- Montenegro-Herrera, C.A.; Portillo, F.V.L.; Hernandez-Chavez, G.T.; Martinez, A. Single-cell protein production potential with the extremophilic red microalgae Galdieria sulphuraria: Growth and biochemical characterization. J. Appl. Phycol. 2022, 34, 1341–1352. [Google Scholar] [CrossRef]
- Najafpour, G.; Younesi, H. Ethanol fermentation in an immobilized cell reactor using Saccharomyces cerevisiae. Bioresour. Technol. 2004, 92, 251–260. [Google Scholar] [CrossRef]
- Du, Y.H.; Tong, L.L.; Wang, Y.; Liu, M.Z.; Yuan, L.; Mu, X.Y.; He, S.J.; Wei, S.X.; Zhang, Y.D.; Chen, Z.L.; et al. Development of a kinetics-integrated CFD model for the industrial scale-up of DHA fermentation using Schizochytrium sp. AIChE J. 2022, 68, e17750. [Google Scholar] [CrossRef]
- Bezerra, R.M.F.; Dias, A.A. Enzymatic kinetic of cellulose hydrolysis—Inhibition by ethanol and cellobiose. Appl. Biochem. Biotechnol. 2005, 126, 49–59. [Google Scholar] [CrossRef]
- Dong, Y.M.; Yan, X.F.; Lu, F.; Guo, M.J.; Zhuang, Y.P. Development and Optimization of an Unstructured Kinetic Model for Sodium Gluconate Fermentation Process. Comput. Mater. Contin. 2015, 48, 43–55. [Google Scholar] [CrossRef]
- Costa, F.; Quintelas, C.; Tavares, T. An approach to the metabolic degradation of diethylketone (DEK) by Streptococcus equisimilis: Effect of DEK on the growth, biodegradation kinetics and efficiency. Ecol. Eng. 2014, 70, 183–188. [Google Scholar] [CrossRef]
- Ghoshal, G.; Banerjee, U.C.; Shivhare, U.S. Xylanase Production by Penicillium citrinum in Laboratory-scale Stirred Tank Reactor. Chem. Biochem. Eng. Q. 2014, 28, 399–408. [Google Scholar] [CrossRef]
- Buendia-Kandia, F.; Rondags, E.; Framboisier, X.; Mauviel, G.; Dufour, A.; Guedon, E. Diauxic growth of Clostridium acetobutylicum ATCC 824 when grown on mixtures of glucose and cellobiose. AMB Express 2018, 8, 85. [Google Scholar] [CrossRef]
- Sun, H.Y.; Zhang, X.Y.; Wang, D.L.; Lin, Z.F. Insights into the role of energy source in hormesis through diauxic growth of bacteria in mixed cultivation systems. Chemosphere 2020, 261, 127669. [Google Scholar] [CrossRef]
- Wang, D.; Lai, Y.C.; Karam, A.L.; de Los Reyes, F.L., 3rd; Ducoste, J.J. Dynamic modeling of microalgae growth and lipid production under transient light and nitrogen conditions. Environ. Sci. Technol. 2019, 53, 11560–11568. [Google Scholar] [CrossRef]
- Haringa, C.; Tang, W.; Wang, G.; Deshmukh, A.T.; van Winden, W.A.; Chu, J.; van Gulik, W.M.; Heijnen, J.J.; Mudde, R.F.; Noorman, H.J. Computational fluid dynamics simulation of an industrial P. chrysogenum fermentation with a coupled 9-pool metabolic model: Towards rational scale-down and design optimization. Chem. Eng. Sci. 2018, 175, 12–24. [Google Scholar] [CrossRef]
- Cvijovic, M.; Bordel, S.; Nielsen, J. Mathematical models of cell factories: Moving towards the core of industrial biotechnology. Microb. Biotechnol. 2011, 4, 572–584. [Google Scholar] [CrossRef]
- Stephanopoulos, G. Metabolic fluxes and metabolic engineering. Metab. Eng. 1999, 1, 1–11. [Google Scholar] [CrossRef]
- Bordbar, A.; Monk, J.M.; King, Z.A.; Palsson, B.O. Constraint-based models predict metabolic and associated cellular functions. Nat. Rev. Genet. 2014, 15, 107–120. [Google Scholar] [CrossRef]
- Ramon, C.; Gollub, M.G.; Stelling, J. Integrating -omics data into genome-scale metabolic network models: Principles and challenges. Essays Biochem. 2018, 62, 563–574. [Google Scholar] [CrossRef]
- Llaneras, F.; Pico, J. Stoichiometric modelling of cell metabolism. J. Biosci. Bioeng. 2008, 105, 1–11. [Google Scholar] [CrossRef]
- Orth, J.D.; Thiele, I.; Palsson, B.O. What is flux balance analysis? Nat. Biotechnol. 2010, 28, 245–248. [Google Scholar] [CrossRef]
- Varma, A.; Palsson, B.O. Metabolic flux balancing: Basic concepts, scientific and practical use. Nat. Biotechnol. 1994, 12, 994–998. [Google Scholar] [CrossRef]
- Schuetz, R.; Kuepfer, L.; Sauer, U. Systematic evaluation of objective functions for predicting intracellular fluxes in Escherichia coli. Mol. Syst. Biol. 2007, 3, 119. [Google Scholar] [CrossRef] [PubMed]
- Papoutsakis, E.T. Equations and calculations for fermentations of butyric acid bacteria. Biotechnol. Bioeng. 1984, 26, 174–187. [Google Scholar] [CrossRef] [PubMed]
- Varma, A.; Palsson, B.O. Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110. Appl. Environ. Microbiol. 1994, 60, 3724–3731. [Google Scholar] [CrossRef] [PubMed]
- Swayambhu, G.; Moscatello, N.; Atilla-Gokcumen, G.E.; Pfeifer, B.A. Flux balance analysis for media optimization and genetic targets to improve heterologous siderophore production. iScience 2020, 23, 101016. [Google Scholar] [CrossRef]
- Kaushal, M.; Chary, K.V.N.; Ahlawat, S.; Palabhanvi, B.; Goswami, G.; Das, D. Understanding regulation in substrate dependent modulation of growth and production of alcohols in Clostridium sporogenes NCIM 2918 through metabolic network reconstruction and flux balance analysis. Bioresour. Technol. 2018, 249, 767–776. [Google Scholar] [CrossRef]
- Huang, Z.; Xu, J.; Yongky, A.; Morris, C.S.; Polanco, A.L.; Reily, M.; Borys, M.C.; Li, Z.J.; Yoon, S. CHO cell productivity improvement by genome-scale modeling and pathway analysis: Application to feed supplements. Biochem. Eng. J. 2020, 160, 107638. [Google Scholar] [CrossRef]
- Long, C.P.; Gonzalez, J.E.; Sandoval, N.R.; Antoniewicz, M.R. Characterization of physiological responses to 22 gene knockouts in Escherichia coli central carbon metabolism. Metab. Eng. 2016, 37, 102–113. [Google Scholar] [CrossRef]
- Long, C.P.; Antoniewicz, M.R. Metabolic flux responses to deletion of 20 core enzymes reveal flexibility and limits of E. coli metabolism. Metab. Eng. 2019, 55, 249–257. [Google Scholar] [CrossRef]
- Shlomi, T.; Berkman, O.; Ruppin, E. Regulatory on/off minimization of metabolic flux changes after genetic perturbations. Proc. Natl. Acad. Sci. USA 2005, 102, 7695–7700. [Google Scholar] [CrossRef]
- Segrè, D.; Vitkup, D.; Church, G.M. Analysis of optimality in natural and perturbed metabolic networks. Proc. Natl. Acad. Sci. USA 2002, 99, 15112–15117. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Reed, J.L. RELATCH: Relative optimality in metabolic networks explains robust metabolic and regulatory responses to perturbations. Genome Biol. 2012, 13, R78. [Google Scholar] [CrossRef]
- Antoniewicz, M.R. A guide to metabolic flux analysis in metabolic engineering: Methods, tools and applications. Metab. Eng. 2021, 63, 2–12. [Google Scholar] [CrossRef]
- Calik, P.; Ileri, N. PH influences intracellular reaction network of beta-lactamase producing Bacillus licheniformis. Chem. Eng. Sci. 2007, 62, 5206–5211. [Google Scholar] [CrossRef]
- Sarma, S.; Anand, A.; Dubey, V.K.; Moholkar, V.S. Metabolic flux network analysis of hydrogen production from crude glycerol by Clostridium pasteurianum. Bioresour. Technol. 2017, 242, 169–177. [Google Scholar] [CrossRef]
- Xiong, X.J.; Li, Y.F. Study on temperature regulation of lactic acid bacteria using metabolic flux analysis. In Proceedings of the International Conference on Energy, Environment and Sustainable Development (ICEESD 2011), Shanghai, China, 21–23 October 2011; pp. 127–132. [Google Scholar]
- Li, J.H.; Yang, Y.M.; Chu, J.; Huang, M.Z.; Li, L.A.; Zhang, X.C.; Wang, Y.H.; Zhuang, Y.P.; Zhang, S.L. Quantitative metabolic flux analysis revealed uneconomical utilization of ATP and NADPH in Acremonium chrysogenum fed with soybean oil. Bioprocess Biosyst. Eng. 2010, 33, 1119–1129. [Google Scholar] [CrossRef]
- Yang, S.-T.; Liu, X.; Zhang, Y. Chapter 4—Metabolic Engineering—Applications, Methods, and Challenges. In Bioprocessing for Value-Added Products from Renewable Resources; Yang, S.-T., Ed.; Elsevier: Amsterdam, The Netherlands, 2007; pp. 73–118. [Google Scholar]
- Vallino, J.J.; Stephanopoulos, G. Metabolic flux distributions in Corynebacterium glutamicum during growth and lysine overproduction. Biotechnol. Bioeng. 1993, 41, 633–646. [Google Scholar] [CrossRef]
- Takaç, S.; Çalık, G.; Mavituna, F.; Dervakos, G. Metabolic flux distribution for the optimized production of l-glutamate. Enzyme Microb. Technol. 1998, 23, 286–300. [Google Scholar] [CrossRef]
- Jørgensen, H.; Nielsen, J.; Villadsen, J.; Møllgaard, H. Metabolic flux distributions in Penicillium chrysogenum during fed-batch cultivations. Biotechnol. Bioeng. 1995, 46, 117–131. [Google Scholar] [CrossRef]
- Shimizu, H.; Shimizu, N.; Shioya, S. Roles of glucose and acetate as carbon sources in l-histidine production with Brevibacterium flavum FERM1564 revealed by metabolic flux analysis. Biotechnol. Bioprocess Eng. 2002, 7, 171. [Google Scholar] [CrossRef]
- Shirai, T.; Nakato, A.; Izutani, N.; Nagahisa, K.; Shioya, S.; Kimura, E.; Kawarabayasi, Y.; Yamagishi, A.; Gojobori, T.; Shimizu, H. Comparative study of flux redistribution of metabolic pathway in glutamate production by two coryneform bacteria. Metab. Eng. 2005, 7, 59–69. [Google Scholar] [CrossRef]
- Mahadevan, R.; Schilling, C.H. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab. Eng. 2003, 5, 264–276. [Google Scholar] [CrossRef]
- Heirendt, L.; Arreckx, S.; Pfau, T.; Mendoza, S.N.; Richelle, A.; Heinken, A.; Haraldsdóttir, H.S.; Wachowiak, J.; Keating, S.M.; Vlasov, V.; et al. Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v.3.0. Nat. Protoc. 2019, 14, 639–702. [Google Scholar] [CrossRef]
- Mahadevan, R.; Edwards, J.S.; Doyle, F.J. Dynamic Flux Balance Analysis of Diauxic Growth in Escherichia coli. Biophys. J. 2002, 83, 1331–1340. [Google Scholar] [CrossRef]
- Kuriya, Y.; Araki, M. Dynamic Flux Balance Analysis to Evaluate the Strain Production Performance on Shikimic Acid Production in Escherichia coli. Metabolites 2020, 10, 198. [Google Scholar] [CrossRef]
- Hanly, T.J.; Henson, M.A. Dynamic model-based analysis of furfural and HMF detoxification by pure and mixed batch cultures of S. cerevisiae and S. stipitis. Biotechnol. Bioeng. 2014, 111, 272–284. [Google Scholar] [CrossRef]
- Valverde, J.R.; Gullón, S.; García-Herrero, C.A.; Campoy, I.; Mellado, R.P. Dynamic metabolic modelling of overproduced protein secretion in Streptomyces lividans using adaptive DFBA. BMC Microbiol. 2019, 19, 233. [Google Scholar] [CrossRef]
- Henson, M.A.; Hanly, T.J. Dynamic flux balance analysis for synthetic microbial communities. IET Syst. Biol. 2014, 8, 214–229. [Google Scholar] [CrossRef]
- Chen, L.; Liu, F. Recursive parameter identification for fermentation processes with the multiple model technique. Appl. Math. Model. 2012, 36, 2275–2285. [Google Scholar] [CrossRef]
- Li, X.; Zhang, S.; Xu, Z.; Feng, E. Parameter Identification Model with the Control Term in Batch Anaerobic Fermentation. In Proceedings of the 2nd International Conference on Advanced Design and Manufacturing Engineering (ADME 2012), Taiyuan, China, 16–18 August 2012; pp. 1535–1540. [Google Scholar]
- Greener, J.G.; Kandathil, S.M.; Moffat, L.; Jones, D.T. A guide to machine learning for biologists. Nat. Rev. Mol. Cell Biol. 2022, 23, 40–55. [Google Scholar] [CrossRef]
- Xu, R.Z.; Cao, J.S.; Wu, Y.; Wang, S.N.; Luo, J.Y.; Chen, X.; Fang, F. An integrated approach based on virtual data augmentation and deep neural networks modeling for VFA production prediction in anaerobic fermentation process. Water. Res. 2020, 184, 116103. [Google Scholar] [CrossRef]
- de Vazelhes, W.; Carey, C.J.; Tang, Y.; Vauquier, N.; Bellet, A. Metric-learn: Metric Learning Algorithms in Python. J. Mach. Learn. Res. 2020, 21, 138-1. [Google Scholar]
- Kuhn, M. Building Predictive Models in R Using the caret Package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Blaom, A.D.; Király, F.J.; Lienart, T.; Simillides, Y.; Arenas, D.; Vollmer, S.J.J.A. MLJ: A Julia package for composable Machine Learning. Comput. Sci. 2020, 5, 2704–2711. [Google Scholar] [CrossRef]
- Noble, W.S. What is a support vector machine? Nat. Biotechnol. 2006, 24, 1565–1567. [Google Scholar] [CrossRef]
- Dong, C.; Chen, J. Optimization of process parameters for anaerobic fermentation of corn stalk based on least squares support vector machine. Bioresour. Technol. 2019, 271, 174–181. [Google Scholar] [CrossRef]
- Zhang, L.; Chao, B.; Zhang, X. Modeling and optimization of microbial lipid fermentation from cellulosic ethanol wastewater by Rhodotorula glutinis based on the support vector machine. Bioresour. Technol. 2020, 301, 122781. [Google Scholar] [CrossRef]
- Uslan, V.; Seker, H. Binding affinity prediction of S. cerevisiae 14-3-3 and GYF peptide-recognition domains using support vector regression. In Proceedings of the 38th Annual International Conference of the IEEE-Engineering-in-Medicine-and-Biology-Society (EMBC), Orlando, FL, USA, 16–20 August 2016; pp. 3445–3448. [Google Scholar]
- Lewis, D.P.; Jebara, T.; Noble, W.S. Support vector machine learning from heterogeneous data: An empirical analysis using protein sequence and structure. Bioinformatics 2006, 22, 2753–2760. [Google Scholar] [CrossRef]
- Shah, A.R.; Oehmen, C.S.; Webb-Robertson, B.J. SVM-HUSTLE—An iterative semi-supervised machine learning approach for pairwise protein remote homology detection. Bioinformatics 2008, 24, 783–790. [Google Scholar] [CrossRef]
- Shahid, N.; Rappon, T.; Berta, W. Applications of artificial neural networks in health care organizational decision-making: A scoping review. PLoS ONE 2019, 14, e0212356. [Google Scholar] [CrossRef]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.M.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. In Proceedings of the 33rd Conference on Neural Information Processing Systems (NeurIPS), Vancouver, BC, Canada, 8–14 December 2019. [Google Scholar]
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; et al. TensorFlow: A system for large-scale machine learning. In Proceedings of the 12th USENIX conference on Operating Systems Design and Implementation, Savannah, GA, USA, 2–4 November 2016; pp. 265–283. [Google Scholar]
- Darsey, J.A.; Griffn, W.O.; Joginipelli, S.; Melapu, V.K. Architecture and biological applications of artifcial neural networks: A tuberculosis perspective. Methods Mol. Biol. 2015, 1260, 269–283. [Google Scholar] [CrossRef]
- Melcher, M.; Scharl, T.; Spangl, B.; Luchner, M.; Cserjan, M.; Bayer, K.; Leisch, F.; Striedner, G. The potential of random forest and neural networks for biomass and recombinant protein modeling in Escherichia coli fed-batch fermentations. Biotechnol. J. 2015, 10, 1770–1782. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, G.; Sage, V.; Xu, J.; Sun, G.; He, J.; Sun, Y. Optimization of dark fermentation for biohydrogen production using a hybrid artificial neural network (ANN) and response surface methodology (RSM) approach. Environ. Prog. Sustain. Energy 2021, 40, e13485. [Google Scholar] [CrossRef]
- Tavasoli, T.; Arjmand, S.; Ranaei Siadat, S.O.; Shojaosadati, S.A.; Sahebghadam Lotfi, A. A robust feeding control strategy adjusted and optimized by a neural network for enhancing of alpha 1-antitrypsin production in Pichia pastoris. Biochem. Eng. J. 2019, 144, 18–27. [Google Scholar] [CrossRef]
- Schinn, S.M.; Morrison, C.; Wei, W.; Zhang, L.; Lewis, N.E. A genome-scale metabolic network model and machine learning predict amino acid concentrations in Chinese Hamster Ovary cell cultures. Biotechnol. Bioeng. 2021, 118, 2118–2123. [Google Scholar] [CrossRef]
- Oyetunde, T.; Liu, D.; Martin, H.G.; Tang, Y.J. Machine learning framework for assessment of microbial factory performance. PLoS ONE 2019, 14, e0210558. [Google Scholar] [CrossRef]
- Culley, C.; Vijayakumar, S.; Zampieri, G.; Angione, C. A mechanism-aware and multiomic machine-learning pipeline characterizes yeast cell growth. Proc. Natl. Acad. Sci. USA 2020, 117, 18869–18879. [Google Scholar] [CrossRef]
- Luo, Y.; Kurian, V.; Ogunnaike, B.A. Bioprocess systems analysis, modeling, estimation, and control. Curr. Opin. Chem. Eng. 2021, 33, 100705. [Google Scholar] [CrossRef]
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. R. Soc. A 2016, 374, 20150202. [Google Scholar] [CrossRef]
- Arian, R.; Hariri, A.; Mehridehnavi, A.; Fassihi, A.; Ghasemi, F. Protein kinase inhibitors’ classification using K-Nearest neighbor algorithm. Comput. Biol. Chem. 2020, 86, 107269. [Google Scholar] [CrossRef]
- Thompson, M.L.; Kramer, M.A. Modeling chemical processes using prior knowledge and neural networks. AIChE J. 1994, 40, 1328–1340. [Google Scholar] [CrossRef]
- Xu, C.M.; Jackson, S.A. Machine learning and complex biological data. Genome Biol. 2019, 20, 76. [Google Scholar] [CrossRef]
- von Stosch, M.; Oliveira, R.; Peres, J.; Feyo de Azevedo, S. Hybrid semi-parametric modeling in process systems engineering: Past, present and future. Comput. Chem. Eng. 2014, 60, 86–101. [Google Scholar] [CrossRef]
- Reel, P.S.; Reel, S.; Pearson, E.; Trucco, E.; Jefferson, E. Using machine learning approaches for multi-omics data analysis: A review. Biotechnol. Adv. 2021, 49, 107739. [Google Scholar] [CrossRef]
- Vijayakumar, S.; Rahman, P.K.S.M.; Angione, C. A Hybrid Flux Balance Analysis and Machine Learning Pipeline Elucidates Metabolic Adaptation in Cyanobacteria. iScience 2020, 23, 101818. [Google Scholar] [CrossRef]
- Vernon, I.; Liu, J.; Goldstein, M.; Rowe, J.; Topping, J.; Lindsey, K. Bayesian uncertainty analysis for complex systems biology models: Emulation, global parameter searches and evaluation of gene functions. BMC Syst. Biol. 2018, 12, 1. [Google Scholar] [CrossRef]
- Sridhara, V.; Meyer, A.G.; Rai, P.; Barrick, J.E.; Ravikumar, P.; Segrè, D.; Wilke, C.O. Predicting growth conditions from internal metabolic fluxes in an in-silico model of E. coli. PLoS ONE 2014, 9, e114608. [Google Scholar] [CrossRef]
- Wu, S.G.; Wang, Y.; Jiang, W.; Oyetunde, T.; Yao, R.; Zhang, X.; Shimizu, K.; Tang, Y.J.; Bao, F.S. Rapid Prediction of Bacterial Heterotrophic Fluxomics Using Machine Learning and Constraint Programming. PLoS Comput. Biol. 2016, 12, e1004838. [Google Scholar] [CrossRef]
- Noor, E.; Cherkaoui, S.; Sauer, U. Biological insights through omics data integration. Curr. Opin. Syst. Biol. 2019, 15, 39–47. [Google Scholar] [CrossRef]
- Hewitt, C.J.; Nienow, A.W. The scale-up of microbial batch and fed-batch fermentation processes. Adv. Appl. Microbiol. 2007, 62, 105–135. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, Z.-J.; Xia, J.-Y.; Haringa, C.; Liu, Y.-P.; Chu, J.; Zhuang, Y.-P.; Zhang, S.-L. Application of Euler-Lagrange CFD for quantitative evaluating the effect of shear force on Carthamus tinctorius L. cell in a stirred tank bioreactor. Biochem. Eng. J. 2016, 114, 212–220. [Google Scholar] [CrossRef]
- Lara, A.R.; Galindo, E.; Ramírez, O.T.; Palomares, L.A. Living with heterogeneities in bioreactors: Understanding the effects of environmental gradients on cells. Mol. Biotechnol. 2006, 34, 355–381. [Google Scholar] [CrossRef]
- Heins, A.L.; Weuster-Botz, D. Population heterogeneity in microbial bioprocesses: Origin, analysis, mechanisms, and future perspectives. Bioprocess Biosyst. Eng. 2018, 41, 889–916. [Google Scholar] [CrossRef]
- Wang, G.; Zhao, J.; Haringa, C.; Tang, W.; Xia, J.; Chu, J.; Zhuang, Y.; Zhang, S.; Deshmukh, A.T.; van Gulik, W.; et al. Comparative performance of different scale-down simulators of substrate gradients in Penicillium chrysogenum cultures: The need of a biological systems response analysis. Microb. Biotechnol. 2018, 11, 486–497. [Google Scholar] [CrossRef]
- Bolic, A.; Larsson, H.; Hugelier, S.; Lantz, A.E.; Kruhne, U.; Gernaey, K.V. A flexible well-mixed milliliter-scale reactor with high oxygen transfer rate for microbial cultivations. Chem. Eng. J. 2016, 303, 655–666. [Google Scholar] [CrossRef]
- Mishra, S.; Kumar, V.; Sarkar, J.; Rathore, A.S. CFD based mass transfer modeling of a single use bioreactor for production of monoclonal antibody biotherapeutics. Chem. Eng. J. 2021, 412, 128592. [Google Scholar] [CrossRef]
- Villiger, T.K.; Neunstoecklin, B.; Karst, D.J.; Lucas, E.; Stettler, M.; Broly, H.; Morbidelli, M.; Soos, M. Experimental and CFD physical characterization of animal cell bioreactors: From micro- to production scale. Biochem. Eng. J. 2018, 131, 84–94. [Google Scholar] [CrossRef]
- Wang, G.; Haringa, C.; Noorman, H.; Chu, J.; Zhuang, Y. Developing a Computational Framework to Advance Bioprocess Scale-Up. Trends Biotechnol. 2020, 38, 846–856. [Google Scholar] [CrossRef]
- Lapin, A.; Muller, D.; Reuss, M. Dynamic behavior of microbial populations in stirred bioreactors simulated with Euler-Lagrange methods: Traveling along the lifelines of single cells. Ind. Eng. Chem. Res. 2004, 43, 4647–4656. [Google Scholar] [CrossRef]
- Haringa, C.; Mudde, R.F.; Noorman, H.J. From industrial fermenter to CFD-guided downscaling: What have we learned? Biochem. Eng. J. 2018, 140, 57–71. [Google Scholar] [CrossRef]
- Siebler, F.; Lapin, A.; Hermann, M.; Takors, R. The impact of CO gradients on C. ljungdahlii in a 125 m3 bubble column: Mass transfer, circulation time and lifeline analysis. Chem. Eng. Sci. 2019, 207, 410–423. [Google Scholar] [CrossRef]
- Kuschel, M.; Siebler, F.; Takors, R. Lagrangian Trajectories to Predict the Formation of Population Heterogeneity in Large-Scale Bioreactors. Bioengineering 2017, 4, 27. [Google Scholar] [CrossRef]
- Gao, X.; Kong, B.; Vigil, R.D. Simulation of algal photobioreactors: Recent developments and challenges. Biotechnol. Lett. 2018, 40, 1311–1327. [Google Scholar] [CrossRef]
- Gernigon, V.; Chekroun, M.A.; Cockx, A.; Guiraud, P.; Morchain, J. How Mixing and Light Heterogeneity Impact the Overall Growth Rate in Photobioreactors. Chem. Eng. Technol. 2019, 42, 1663–1669. [Google Scholar] [CrossRef]
- Guha, D.; Ramachandran, P.A.; Dudukovic, M.P.; Derksen, J.J. Evaluation of large eddy simulation and Euler-Euler CFD models for solids flow dynamics in a stirred tank reactor. AIChE J. 2008, 54, 766–778. [Google Scholar] [CrossRef]
- Elqotbi, M.; Vlaev, S.D.; Montastruc, L.; Nikov, I. CFD modelling of two-phase stirred bioreaction systems by segregated solution of the Euler–Euler model. Comput. Chem. Eng. 2013, 48, 113–120. [Google Scholar] [CrossRef]
- Morchain, J.; Gabelle, J.-C.; Cockx, A. A coupled population balance model and CFD approach for the simulation of mixing issues in lab-scale and industrial bioreactors. AIChE J. 2014, 60, 27–40. [Google Scholar] [CrossRef]
- Yeoh, J.W.; Jayaraman, S.S.O.; Tan, S.G.-D.; Jayaraman, P.; Holowko, M.B.; Zhang, J.; Kang, C.-W.; Leo, H.L.; Poh, C.L. A model-driven approach towards rational microbial bioprocess optimization. Biotechnol. Bioeng. 2021, 118, 305–318. [Google Scholar] [CrossRef]
- Tajsoleiman, T.; Spann, R.; Bach, C.; Gernaey, K.V.; Huusom, J.K.; Krühne, U. A CFD based automatic method for compartment model development. Comput. Chem. Eng. 2019, 123, 236–245. [Google Scholar] [CrossRef]
- Venkatasubramanian, V. The promise of artificial intelligence in chemical engineering: Is it here, finally? AIChE J. 2019, 65, 466–478. [Google Scholar] [CrossRef]
- del Rio-Chanona, E.A.; Wagner, J.L.; Ali, H.; Fiorelli, F.; Zhang, D.; Hellgardt, K. Deep learning-based surrogate modeling and optimization for microalgal biofuel production and photobioreactor design. AIChE J. 2019, 65, 915–923. [Google Scholar] [CrossRef]
- Zhang, D.; Del Rio-Chanona, E.A.; Petsagkourakis, P.; Wagner, J. Hybrid physics-based and data-driven modeling for bioprocess online simulation and optimization. Biotechnol. Bioeng. 2019, 116, 2919–2930. [Google Scholar] [CrossRef] [PubMed]

| Name | Expression | Function | Refs. |
|---|---|---|---|
| Monod Kinetics | To describe microbial growth based on the consumption of one substrate. | [48] | |
| Double Monod Kinetics | To describe microbial growth based on the consumption of multiple substrates. | [49] | |
| Enzyme inhibition Kinetics | To describe microbial growth in the presence of competitive substrate inhibition. | [50] | |
| Contois Kinetics | To describe microbial growth in a high-density culture. | [51] | |
| Powell Kinetics | To describe microbial growth while considering the basal metabolic consumption of cells (e.g., metabolite turnover). | [52] | |
| Moser Kinetics | To describe microbial growth in situations where cells have multiple pathways to utilize substrates. | [53] | |
| Logistic Equation | To describe microbial growth without any biological explanation other than the assumption that there is a maximum cell growth concentration. | [41,42] | |
| Haldane–Andrew Model | To describe microbial growth while considering that some substrates are toxic to cells and can inhibit cell growth at high concentrations. | [39,52] | |
| Diauxic Growth | To describe microbial growth while considering that there are two carbon sources, S1 and S2, during cell growth and that the cell preferentially uses S1. | [54,55] | |
| Luedeking–Piret Equation | To describe the production rate of product P in the case where product synthesis is related to the growth rate and cell density of microbial cells. | [54,55] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, Y.-H.; Wang, M.-Y.; Yang, L.-H.; Tong, L.-L.; Guo, D.-S.; Ji, X.-J. Optimization and Scale-Up of Fermentation Processes Driven by Models. Bioengineering 2022, 9, 473. https://doi.org/10.3390/bioengineering9090473
Du Y-H, Wang M-Y, Yang L-H, Tong L-L, Guo D-S, Ji X-J. Optimization and Scale-Up of Fermentation Processes Driven by Models. Bioengineering. 2022; 9(9):473. https://doi.org/10.3390/bioengineering9090473
Chicago/Turabian StyleDu, Yuan-Hang, Min-Yu Wang, Lin-Hui Yang, Ling-Ling Tong, Dong-Sheng Guo, and Xiao-Jun Ji. 2022. "Optimization and Scale-Up of Fermentation Processes Driven by Models" Bioengineering 9, no. 9: 473. https://doi.org/10.3390/bioengineering9090473
APA StyleDu, Y.-H., Wang, M.-Y., Yang, L.-H., Tong, L.-L., Guo, D.-S., & Ji, X.-J. (2022). Optimization and Scale-Up of Fermentation Processes Driven by Models. Bioengineering, 9(9), 473. https://doi.org/10.3390/bioengineering9090473