Designer Exosomes: Smart Nano-Communication Tools for Translational Medicine
Abstract
1. Introduction
Prelims of Exosome Biogenesis and Cargo Loading Patterns
2. Scenario Oof Exosomes in Translational Medicine
2.1. Relationship between Cancer and Exosomes
2.2. Neurogenerative Diseases and Exosomes
2.3. Cardiovascular Complications and Exosomes
2.4. Skeletal Muscles, Bones, and Exosomes
2.5. Inflammatory Disorders and Exosomes
3. Empowering of Exosome by Engineering
4. Summary and Future Road Map
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Properzi, F.; Logozzi, M.; Fais, S. Exosomes: The future of biomarkers in medicine. Biomark. Med. 2013, 7, 769–778. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Kang, M.H.; Jeyaraj, M.; Qasim, M.; Kim, J.H. Review of the Isolation, Characterization, Biological Function, and Multifarious Therapeutic Approaches of Exosomes. Cells 2019, 8, 307. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 3–22. [Google Scholar] [CrossRef]
- Ayala-Mar, S.; Donoso-Quezada, J.; Gallo-Villanueva, R.C.; Perez-Gonzalez, V.H.; González-Valdez, J. Recent advances and challenges in the recovery and purification of cellular exosomes. Electrophoresis 2019, 40, 3036–3049. [Google Scholar] [CrossRef] [PubMed]
- Kanwar, S.S.; Dunlay, C.J.; Simeone, D.M.; Nagrath, S. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab. Chip 2014, 14, 1891–1900. [Google Scholar] [CrossRef]
- Koritzinsky, E.H.; Street, J.M.; Star, R.A.; Yuen, P.S. Quantification of Exosomes. J. Cell Physiol. 2017, 232, 1587–1590. [Google Scholar] [CrossRef] [PubMed]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef] [PubMed]
- Van Niel, G.; Porto-Carreiro, I.; Simoes, S.; Raposo, G. Exosomes: A common pathway for a specialized function. J. Biochem. 2006, 140, 13–21. [Google Scholar] [CrossRef]
- Alenquer, M.; Amorim, M.J. Exosome Biogenesis, Regulation, and Function in Viral Infection. Viruses 2015, 7, 5066–5083. [Google Scholar] [CrossRef]
- Hassanpour, M.; Rezaie, J.; Nouri, M.; Panahi, Y. The role of extracellular vesicles in COVID-19 virus infection. Infect. Genet. Evol 2020, 85, 104422. [Google Scholar] [CrossRef]
- Patil, M.; Singh, S.; Henderson, J.; Krishnamurthy, P. Mechanisms of COVID-19-induced cardiovascular disease: Is sepsis or exosome the missing link? J. Cell Physiol. 2021, 236, 3366–3382. [Google Scholar] [CrossRef] [PubMed]
- Schorey, J.S.; Cheng, Y.; Singh, P.P.; Smith, V.L. Exosomes and other extracellular vesicles in host-pathogen interactions. EMBO Rep. 2015, 16, 24–43. [Google Scholar] [CrossRef] [PubMed]
- Raimondo, S.; Saieva, L.; Corrado, C.; Fontana, S.; Flugy, A.; Rizzo, A.; De Leo, G.; Alessandro, R. Chronic myeloid leukemia-derived exosomes promote tumor growth through an autocrine mechanism. Cell Commun. Signal. 2015, 13, 1–2. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- Tengda, L.; Shuping, L.; Mingli, G.; Jie, G.; Yun, L.; Weiwei, Z.; Anmei, D. Serum exosomal microRNAs as potent circulating biomarkers for melanoma. Melanoma Res. 2018, 28, 295–303. [Google Scholar] [CrossRef]
- Kumar, S.A.; Harishkumar, M. Polymeric nanoparticles for vaccine delivery. In Integrating Biologically-Inspired Nanotechnology into Medical Practice, 3rd ed.; BK, N., Amin, N., Amin, B.A., Eds.; IGI-Global: Hershey, PA, USA, 2017; pp. 32–49. [Google Scholar]
- Hendrix, A.; Maynard, D.; Pauwels, P.; Braems, G.; Denys, H.; Van den Broecke, R.; Lambert, J.; Van Belle, S.; Cocquyt, V.; Gespach, C.; et al. Effect of the secretory small GTPase Rab27B on breast cancer growth, invasion, and metastasis. J. Natl. Cancer Inst. 2010, 102, 866–880. [Google Scholar] [CrossRef]
- Harris, D.A.; Patel, S.H.; Gucek, M.; Hendrix, A.; Westbroek, W.; Taraska, J.W. Exosomes released from breast cancer carcinomas stimulate cell movement. PLoS ONE 2015, 10, e0117495. [Google Scholar] [CrossRef]
- Safaei, R.; Larson, B.J.; Cheng, T.C.; Gibson, M.A.; Otani, S.; Naerdemann, W.; Howell, S.B. Abnormal lysosomal trafficking and enhanced exosomal export of cisplatin in drug-resistant human ovarian carcinoma cells. Mol. Cancer Ther. 2005, 4, 1595–1604. [Google Scholar] [CrossRef] [PubMed]
- Eyo, U.B.; Wu, L.J. Bidirectional microglia-neuron communication in the healthy brain. Neural Plast. 2013, 2013, 456857. [Google Scholar] [CrossRef]
- Videira, R.F.; Da Costa Martins, P.A. Non-coding RNAs in Cardiac Intercellular Communication. Front. Physiol. 2020, 11, 738. [Google Scholar] [CrossRef]
- Weilner, S.; Schraml, E.; Wieser, M.; Messner, P.; Schneider, K.; Wassermann, K.; Micutkova, L.; Fortschegger, K.; Maier, A.B.; Westendorp, R.; et al. Secreted microvesicular miR-31 inhibits osteogenic differentiation of mesenchymal stem cells. Aging Cell 2016, 15, 744–754. [Google Scholar] [CrossRef]
- Behera, J.; Tyagi, N. Exosomes: Mediators of bone diseases, protection, and therapeutics potential. Oncoscience 2018, 5, 181–195. [Google Scholar] [CrossRef] [PubMed]
- Karimi, Z.; Seyedjafari, E.; Khojasteh, A.; Hashemi, S.M.; Kazemi, B.; Mohammadi-Yeganeh, S. MicroRNA-218 competes with differentiation media in the induction of osteogenic differentiation of mesenchymal stem cell by regulating β-catenin inhibitors. Mol. Biol. Rep. 2020, 47, 8451–8463. [Google Scholar] [CrossRef]
- Jones, H.R.; Robb, C.T.; Perretti, M.; Rossi, A.G. The role of neutrophils in inflammation resolution. Semin. Immunol. 2016, 28, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Smyth, T.; Petrova, K.; Payton, N.M.; Persaud, I.; Redzic, J.S.; Graner, M.W.; Smith-Jones, P.; Anchordoquy, T.J. Surface functionalization of exosomes using click chemistry. Bioconjugate Chem. 2014, 25, 1777–1784. [Google Scholar] [CrossRef] [PubMed]
- Basma, H.; Johanson, A.N.; Dhar, K.; Anderson, D.; Qiu, F.; Rennard, S.; Lowes, B.D. TGF-β induces a heart failure phenotype via fibroblasts exosome signaling. Heliyon 2019, 5, e02633. [Google Scholar] [CrossRef]
- Bang, C.; Antoniades, C.; Antonopoulos, A.S.; Eriksson, U.; Franssen, C.; Hamdani, N.; Lehmann, L.; Moessinger, C.; Mongillo, M.; Muhl, L.; et al. Intercellular communication lessons in heart failure. Eur. J. Heart Fail. 2015, 17, 1091–1103. [Google Scholar] [CrossRef]
- Fröhlich, D.; Kuo, W.P.; Frühbeis, C.; Sun, J.J.; Zehendner, C.M.; Luhmann, H.J.; Pinto, S.; Toedling, J.; Trotter, J.; Krämer-Albers, E.M. Multifaceted effects of oligodendroglial exosomes on neurons: Impact on neuronal firing rate, signal transduction and gene regulation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369. [Google Scholar] [CrossRef]
- Christianson, H.C.; Belting, M. Heparan sulfate proteoglycan as a cell-surface endocytosis receptor. Matrix Biol. 2014, 35, 51–55. [Google Scholar] [CrossRef]
- Zeeshan, R.; Mutahir, Z. Cancer metastasis—tricks of the trade. Bosn. J. Basic Med. Sci. 2017, 17, 172–182. [Google Scholar] [CrossRef][Green Version]
- Shtam, T.A.; Kovalev, R.A.; Varfolomeeva, E.Y.; Makarov, E.M.; Kil, Y.V.; Filatov, M.V. Exosomes are natural carriers of exogenous siRNA to human cells in vitro. Cell Commun. Signal. 2013, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367. [Google Scholar] [CrossRef] [PubMed]
- Lakshmi, S.; Essa, M.M.; Hartman, R.E.; Guillemin, G.J.; Sivan, S.; Elumalai, P. Exosomes in Alzheimer’s Disease: Potential Role as Pathological Mediators, Biomarkers and Therapeutic Targets. Neurochem. Res. 2020, 45, 2553–2559. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Ahmad, T.; Sharma, A.; Mabalirajan, U.; Kulshreshtha, A.; Agrawal, A.; Ghosh, B. Let-7 microRNA-mediated regulation of IL-13 and allergic airway inflammation. J. Allergy Clin. Immunol. 2011, 128, 1077–1085. [Google Scholar] [CrossRef] [PubMed]
- Gangadaran, P.; Rajendran, R.L.; Oh, J.M.; Oh, E.J.; Hong, C.M.; Chung, H.Y.; Lee, J.; Ahn, B.C. Identification of Angiogenic Cargo in Extracellular Vesicles Secreted from Human Adipose Tissue-Derived Stem Cells and Induction of Angiogenesis In Vitro and In Vivo. Pharmaceutics 2021, 13, 495. [Google Scholar] [CrossRef]
- Henne, W.M.; Buchkovich, N.J.; Emr, S.D. The ESCRT pathway. Dev. Cell 2011, 21, 77–91. [Google Scholar] [CrossRef]
- Larios, J.; Mercier, V.; Roux, A.; Gruenberg, J. ALIX- and ESCRT-III-dependent sorting of tetraspanins to exosomes. J. Cell Biol. 2020, 219. [Google Scholar] [CrossRef]
- Gesierich, S.; Berezovskiy, I.; Ryschich, E.; Zöller, M. Systemic induction of the angiogenesis switch by the tetraspanin D6.1A/CO-029. Cancer Res. 2006, 66, 7083–7094. [Google Scholar] [CrossRef]
- Hemler, M.E. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 2005, 6, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Loewith, R.; Riezman, H.; Winssinger, N. Sphingolipids and membrane targets for therapeutics. Curr. Opin. Chem. Biol. 2019, 50, 19–28. [Google Scholar] [CrossRef]
- Wortzel, I.; Dror, S.; Kenific, C.M.; Lyden, D. Exosome-Mediated Metastasis: Communication from a Distance. Dev. Cell 2019, 49, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Demory Beckler, M.; Higginbotham, J.N.; Franklin, J.L.; Ham, A.J.; Halvey, P.J.; Imasuen, I.E.; Whitwell, C.; Li, M.; Liebler, D.C.; Coffey, R.J. Proteomic analysis of exosomes from mutant KRAS colon cancer cells identify intercellular transfer of mutant KRAS. Mol. Cell Proteomics 2013, 12, 343–355. [Google Scholar] [CrossRef]
- Nazarenko, I.; Rana, S.; Baumann, A.; McAlear, J.; Hellwig, A.; Trendelenburg, M.; Lochnit, G.; Preissner, K.T.; Zöller, M. Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res. 2010, 70, 1668–1678. [Google Scholar] [CrossRef] [PubMed]
- Shimoda, M.; Principe, S.; Jackson, H.W.; Luga, V.; Fang, H.; Molyneux, S.D.; Shao, Y.W.; Aiken, A.; Waterhouse, P.D.; Karamboulas, C.; et al. Loss of the Timp gene family is sufficient for the acquisition of the CAF-like cell state. Nat. Cell Biol. 2014, 16, 889–901. [Google Scholar] [CrossRef]
- Bunggulawa, E.J.; Wang, W.; Yin, T.; Wang, N.; Durkan, C.; Wang, Y.; Wang, G. Recent advancements in the use of exosomes as drug delivery systems. J. Nanobiotechnol. 2018, 16, 81. [Google Scholar] [CrossRef]
- Umapathi, A.; Nagaraju, N.P.; Madhyastha, H.; Jain, D.; Srinivas, S.P.; Rotello, V.M.; Daima, H.K. Highly efficient and selective antimicrobial isonicotinylhydrazide-coated polyoxometalate-functionalized silver nanoparticles. Colloids Surf. B Biointerfaces 2019, 184, 110522. [Google Scholar] [CrossRef]
- Beatriz, M.; Vilaça, R.; Lopes, C. Exosomes: Innocent Bystanders or Critical Culprits in Neurodegenerative Diseases. Front. Cell Dev. Biol. 2021, 9, 635104. [Google Scholar] [CrossRef]
- Kopkova, A.; Sana, J.; Fadrus, P.; Machackova, T.; Vecera, M.; Vybihal, V.; Juracek, J.; Vychytilova-Faltejskova, P.; Smrcka, M.; Slaby, O. MicroRNA isolation and quantification in cerebrospinal fluid: A comparative methodical study. PLoS ONE 2018, 13, e0208580. [Google Scholar] [CrossRef] [PubMed]
- Tassew, N.G.; Charish, J.; Shabanzadeh, A.P.; Luga, V.; Harada, H.; Farhani, N.; D’Onofrio, P.; Choi, B.; Ellabban, A.; Nickerson, P.E.B.; et al. Exosomes Mediate Mobilization of Autocrine Wnt10b to Promote Axonal Regeneration in the Injured CNS. Cell Rep. 2017, 20, 99–111. [Google Scholar] [CrossRef]
- Brink, P.R.; Valiunas, V.; Gordon, C.; Rosen, M.R.; Cohen, I.S. Can gap junctions deliver? Biochim. Biophys. Acta 2012, 1818, 2076–2081. [Google Scholar] [CrossRef]
- Demonbreun, A.R.; McNally, E.M. Muscle cell communication in development and repair. Curr. Opin. Pharmacol. 2017, 34, 7–14. [Google Scholar] [CrossRef]
- Bellavia, D.; De Luca, A.; Carina, V.; Costa, V.; Raimondi, L.; Salamanna, F.; Alessandro, R.; Fini, M.; Giavaresi, G. Deregulated miRNAs in bone health: Epigenetic roles in osteoporosis. Bone 2019, 122, 52–75. [Google Scholar] [CrossRef] [PubMed]
- Butreddy, A.; Kommineni, N.; Dudhipala, N. Exosomes as Naturally Occurring Vehicles for Delivery of Biopharmaceuticals: Insights from Drug Delivery to Clinical Perspectives. Nanomaterials 2021, 11, 1481. [Google Scholar] [CrossRef] [PubMed]
- Hadjiargyrou, M.; Komatsu, D.E. The Therapeutic Potential of MicroRNAs as Orthobiologics for Skeletal Fractures. J. Bone Miner. Res. 2019, 34, 797–809. [Google Scholar] [CrossRef] [PubMed]
- Hensley, A.P.; McAlinden, A. The role of microRNAs in bone development. Bone 2021, 143, 115760. [Google Scholar] [CrossRef]
- Pethő, A.; Chen, Y.; George, A. Exosomes in Extracellular Matrix Bone Biology. Curr. Osteoporos. Rep. 2018, 16, 58–64. [Google Scholar] [CrossRef]
- Takeda, K.; Akira, S. Role of toll-like receptor in innate immunity. Tanpakushitsu Kakusan Koso 2002, 47, 2097–2102. [Google Scholar] [PubMed]
- Madhyastha, R.; Madhyastha, H.; Nurrahmah, Q.I.; Purbasari, B.; Maruyama, M.; Nakajima, Y. MicroRNA 21 Elicits a Pro-inflammatory Response in Macrophages, with Exosomes Functioning as Delivery Vehicles. Inflammation 2021, 44, 1274–1287. [Google Scholar] [CrossRef]
- Radha, M.; Harishkumar, M.; Nurramah, Q.I.; Purbasari, B.; Nakajima, Y. Pro-inflamatory profile of MIR-21 primed exosomes. Wound Repair Regen. 2021, 29, 1–2. [Google Scholar]
- Simpson, R.J.; Kalra, H.; Mathivanan, S. ExoCarta as a resource for exosomal research. J. Extracell. Vesicles 2012, 1. [Google Scholar] [CrossRef] [PubMed]
- Russo, F.; Di Bella, S.; Nigita, G.; Macca, V.; Laganà, A.; Giugno, R.; Pulvirenti, A.; Ferro, A. miRandola: Extracellular circulating microRNAs database. PLoS ONE 2012, 7, e47786. [Google Scholar] [CrossRef]
- Kalra, H.; Simpson, R.J.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P.; Bond, V.C.; Borràs, F.E.; Breakefield, X.; Budnik, V.; et al. Vesiclepedia: A compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012, 10, e1001450. [Google Scholar] [CrossRef] [PubMed]
- Veziroglu, E.M.; Mias, G.I. Characterizing Extracellular Vesicles and Their Diverse RNA Contents. Front. Genet. 2020, 11, 700. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.Q.; Akhtar, S.; Prabhu, K.S.; Zarif, L.; Khan, R.; Alam, M.; Buddenkotte, J.; Ahmad, A.; Steinhoff, M.; Uddin, S. Exosomes: Emerging Diagnostic and Therapeutic Targets in Cutaneous Diseases. Int. J. Mol. Sci. 2020, 21, 9264. [Google Scholar] [CrossRef]
- Bergqvist, C.; Ezzedine, K. Vitiligo: A focus on pathogenesis and its therapeutic implications. J. Dermatol. 2021, 48, 252–270. [Google Scholar] [CrossRef]
- Barile, L.; Vassalli, G. Exosomes: Therapy delivery tools and biomarkers of diseases. Pharmacol. Ther. 2017, 174, 63–78. [Google Scholar] [CrossRef]
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.N.; Freitas, R.P.; et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010, 12, 19–30. [Google Scholar] [CrossRef]
- Park, J.A.; Sharif, A.S.; Tschumperlin, D.J.; Lau, L.; Limbrey, R.; Howarth, P.; Drazen, J.M. Tissue factor-bearing exosome secretion from human mechanically stimulated bronchial epithelial cells in vitro and in vivo. J. Allergy Clin. Immunol. 2012, 130, 1375–1383. [Google Scholar] [CrossRef]
- Taylor, D.D.; Gercel-Taylor, C. Exosomes/microvesicles: Mediators of cancer-associated immunosuppressive microenvironments. Semin. Immunopathol 2011, 33, 441–454. [Google Scholar] [CrossRef]
- Adriano, B.; Cotto, N.M.; Chauhan, N.; Jaggi, M.; Chauhan, S.C.; Yallapu, M.M. Milk exosomes: Nature’s abundant nanoplatform for theranostic applications. Bioact. Mater. 2021, 6, 2479–2490. [Google Scholar] [CrossRef] [PubMed]
- O’Loughlin, A.J.; Mäger, I.; De Jong, O.G.; Varela, M.A.; Schiffelers, R.M.; El Andaloussi, S.; Wood, M.J.A.; Vader, P. Functional Delivery of Lipid-Conjugated siRNA by Extracellular Vesicles. Mol. Ther. 2017, 25, 1580–1587. [Google Scholar] [CrossRef] [PubMed]
- Thietart, S.; Rautou, P.E. Extracellular vesicles as biomarkers in liver diseases: A clinician’s point of view. J. Hepatol. 2020, 73, 1507–1525. [Google Scholar] [CrossRef]
- Berditchevski, F.; Odintsova, E. Tetraspanins as regulators of protein trafficking. Traffic 2007, 8, 89–96. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- El-Andaloussi, S.; Lee, Y.; Lakhal-Littleton, S.; Li, J.; Seow, Y.; Gardiner, C.; Alvarez-Erviti, L.; Sargent, I.L.; Wood, M.J. Exosome-mediated delivery of siRNA in vitro and in vivo. Nat. Protoc 2012, 7, 2112–2126. [Google Scholar] [CrossRef] [PubMed]
- Lathwal, S.; Yerneni, S.S.; Boye, S.; Muza, U.L.; Takahashi, S.; Sugimoto, N.; Lederer, A.; Das, S.R.; Campbell, P.G.; Matyjaszewski, K. Engineering exosome polymer hybrids by atom transfer radical polymerization. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Sato, Y.T.; Umezaki, K.; Sawada, S.; Mukai, S.A.; Sasaki, Y.; Harada, N.; Shiku, H.; Akiyoshi, K. Engineering hybrid exosomes by membrane fusion with liposomes. Sci. Rep. 2016, 6, 21933. [Google Scholar] [CrossRef] [PubMed]
- Gupta, D.; Liang, X.; Pavlova, S.; Wiklander, O.P.B.; Corso, G.; Zhao, Y.; Saher, O.; Bost, J.; Zickler, A.M.; Piffko, A.; et al. Quantification of extracellular vesicles. J. Extracell. Vesicles 2020, 9, 1800222. [Google Scholar] [CrossRef]
- Mirtaheri, E.; Dolatmoradi, A.; El-Zahab, B. Thermally Assisted Acoustofluidic Separation Based on Membrane Protein Content. Anal. Chem. 2019, 91, 13953–13961. [Google Scholar] [CrossRef] [PubMed]
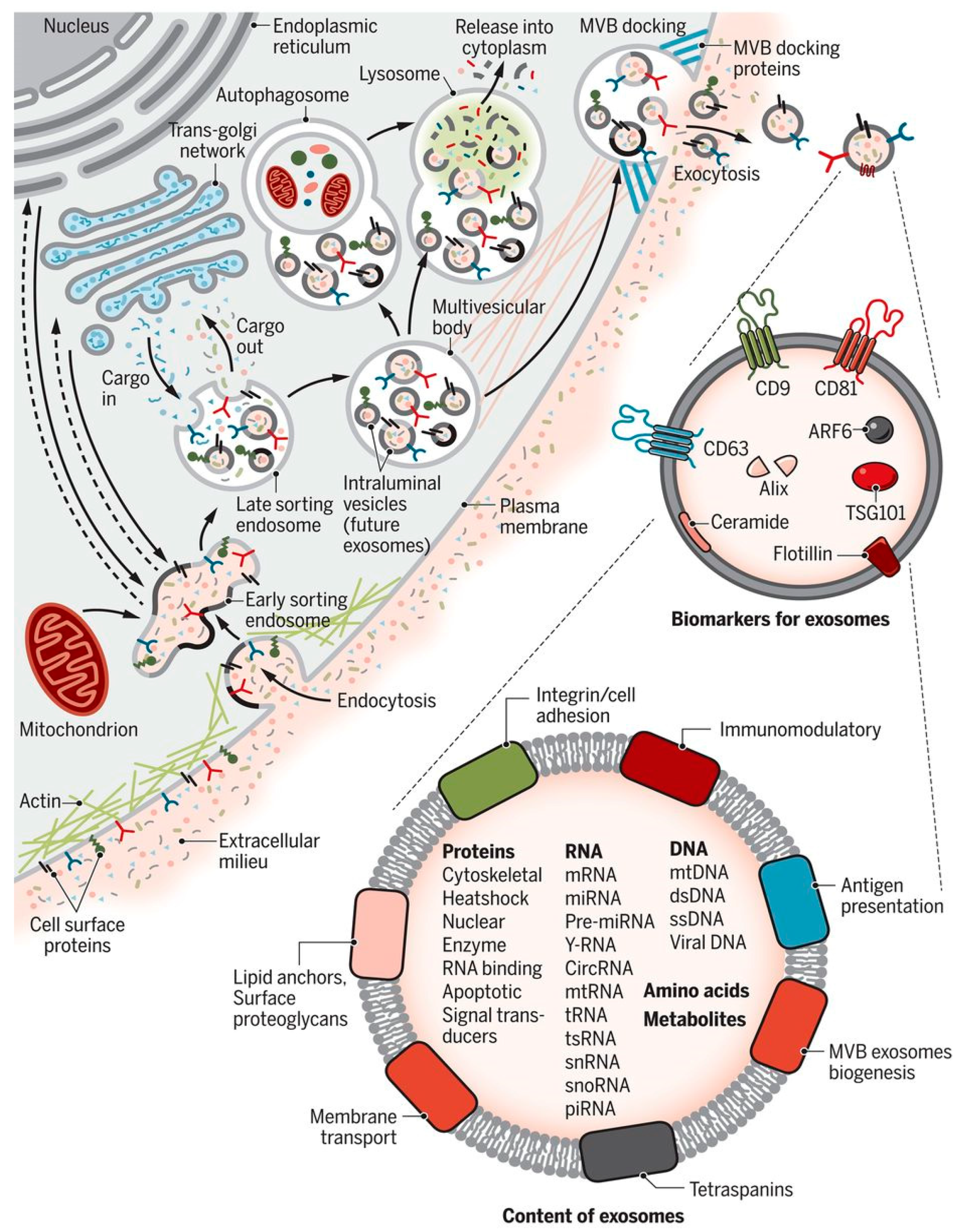
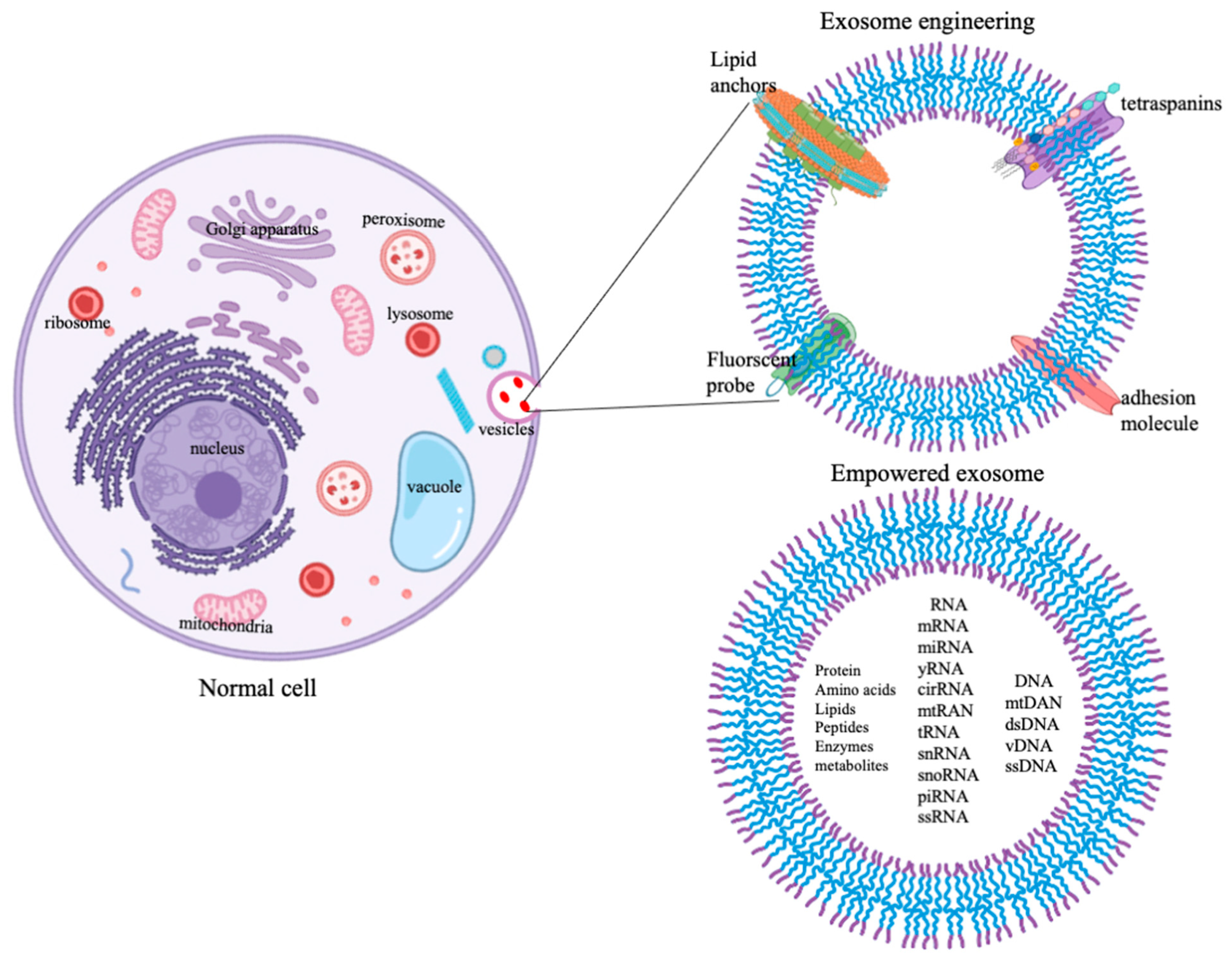
| Cell Type/Tissue/Host Nature | Function | Reference |
|---|---|---|
| Breast cancer-derived | Metastasis protein transport | [14] |
| Virology | COVID-19 infection | [11] |
| Host-pathogen interaction | Toxic transport | [13,15] |
| Fibroblast to cancer cell | Communication | [16] |
| Serum | miRNA transport | [17] |
| Normal cell exosomes | Nanoparticle delivery | [18] |
| Metastasis breast cancer | GTPase and Rab27B delivery | [19,20] |
| Ovarian cancer cell | Lysosomal delivery | [21] |
| Microglia | Neuron signal communication | [22] |
| Cardiomyocytes | Non-coding RNA signals | [23] |
| Osteoblast cells | MiR-31 transport | [24,25,26] |
| Neutrophils | Inflammatory signals | [27,28] |
| Fibroblast | Deviation in TGF beta signaling | [29] |
| Cardiac tissue | Heart failure | [30] |
| Neuronal cells | Neuron diseases | [22,31] |
| Endothelial cell | Endocytosis | [32] |
| Melanoma | miRNA circulation | [17] |
| Alveolar cell | Cancer metastasis | [33] |
| Normal cell | Exogenous siRNA transport | [34,35] |
| Neuronal cell | Alzheimer disease | [36] |
| Oligodendroglia | Cell communication | [31] |
| Mesenchymal stem cell | miR-31 and miR-28 transport | [24,26] |
| Myeloid leukemia | Tumor growth factor communication | [14] |
| Osteoclast | miR-28 loading | [26] |
| Lung airway cell | Let-7 regulation | [37] |
| Adipose-derived stem cells | Angiogenesis | [38] |
| Cargo Types | Technique of Loading | Principle | Merits/Demerits |
|---|---|---|---|
| Synthetic drugs | Incubation | Membrane diffusion | Simple and easy [4] |
| Nucleic acid, peptides, | Transfection | Gene manupulation | Efficiency coefficient to be standardized [78] |
| Drugs, materials | Polymerisation | Issues of membrane pore formation | Very high loading efficiency [79] |
| Nucleic acids, RNA, and Peptides | Freeze-Thaw | Membrane fusion with liposomes | Moderate loading [43] |
| Proteins, Peptides, and Materials | Surfactant | Membrane fusion and nanopore | Very effective and high loading [80] |
| RNAs and DNAs | Dialysis and hybridization | Issues of rapid pH changes in the medium | Easy and less time consuming [18] |
| Nanomaterials | Hybridization | Chemo-biological reaction | Stability [56] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harishkumar, M.; Radha, M.; Yuichi, N.; Muthukalianan, G.K.; Kaoru, O.; Shiomori, K.; Sakai, K.; Nozomi, W. Designer Exosomes: Smart Nano-Communication Tools for Translational Medicine. Bioengineering 2021, 8, 158. https://doi.org/10.3390/bioengineering8110158
Harishkumar M, Radha M, Yuichi N, Muthukalianan GK, Kaoru O, Shiomori K, Sakai K, Nozomi W. Designer Exosomes: Smart Nano-Communication Tools for Translational Medicine. Bioengineering. 2021; 8(11):158. https://doi.org/10.3390/bioengineering8110158
Chicago/Turabian StyleHarishkumar, Madhyastha, Madhyastha Radha, Nakajima Yuichi, Gothandam Kodiveri Muthukalianan, Ohe Kaoru, Koichiro Shiomori, Kentaro Sakai, and Watanabe Nozomi. 2021. "Designer Exosomes: Smart Nano-Communication Tools for Translational Medicine" Bioengineering 8, no. 11: 158. https://doi.org/10.3390/bioengineering8110158
APA StyleHarishkumar, M., Radha, M., Yuichi, N., Muthukalianan, G. K., Kaoru, O., Shiomori, K., Sakai, K., & Nozomi, W. (2021). Designer Exosomes: Smart Nano-Communication Tools for Translational Medicine. Bioengineering, 8(11), 158. https://doi.org/10.3390/bioengineering8110158







