Collagen Mimetic Peptides
Abstract
1. Introduction
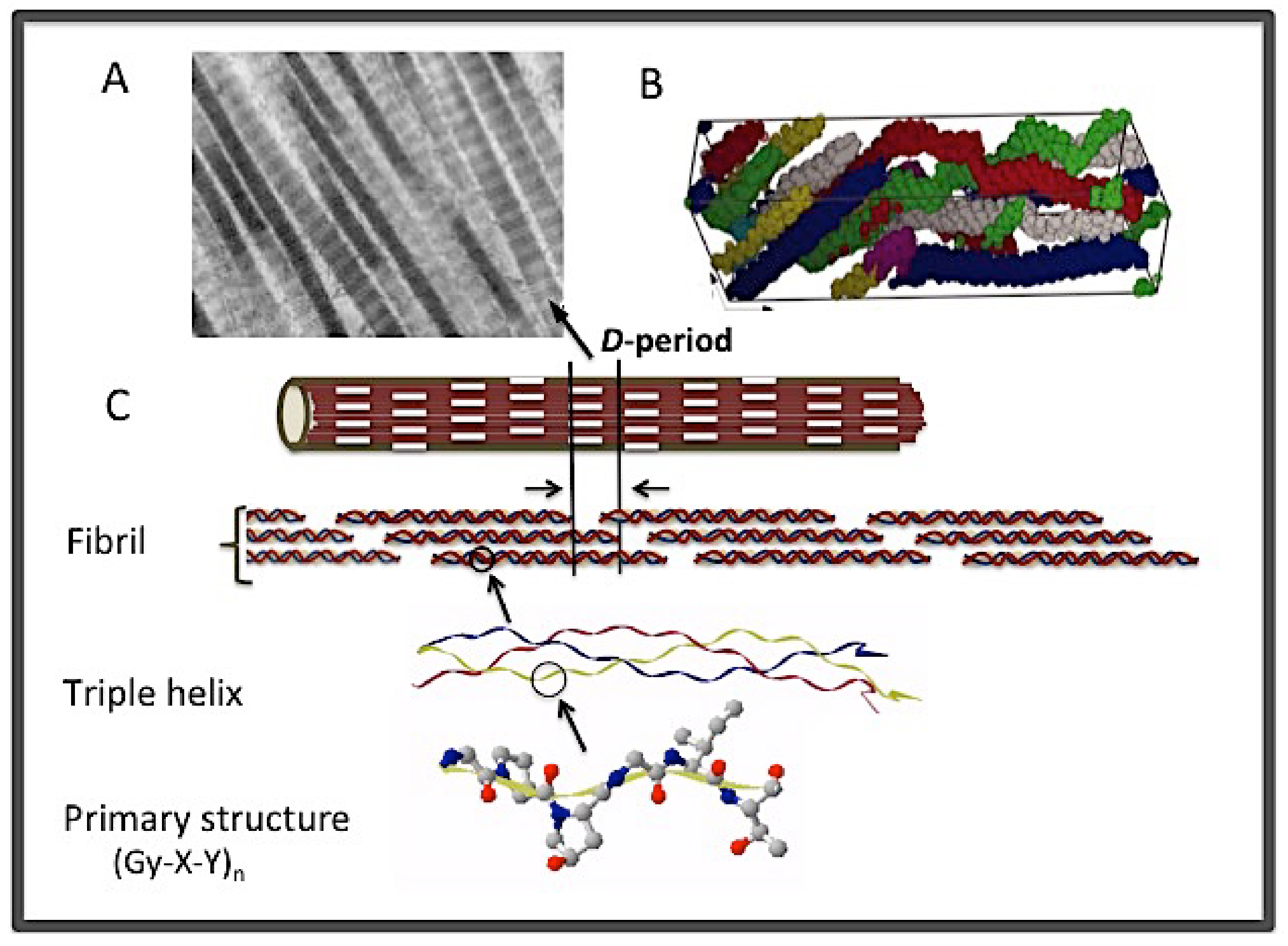
1.1. The Macromolecular Assembly of Collagen
1.2. Collagen-Based Biomaterials
2. Collagen Mimetic Peptides by Chemical Synthesis
2.1. The Homotrimeric CMPs
2.1.1. The Sequence–Structure Relationship
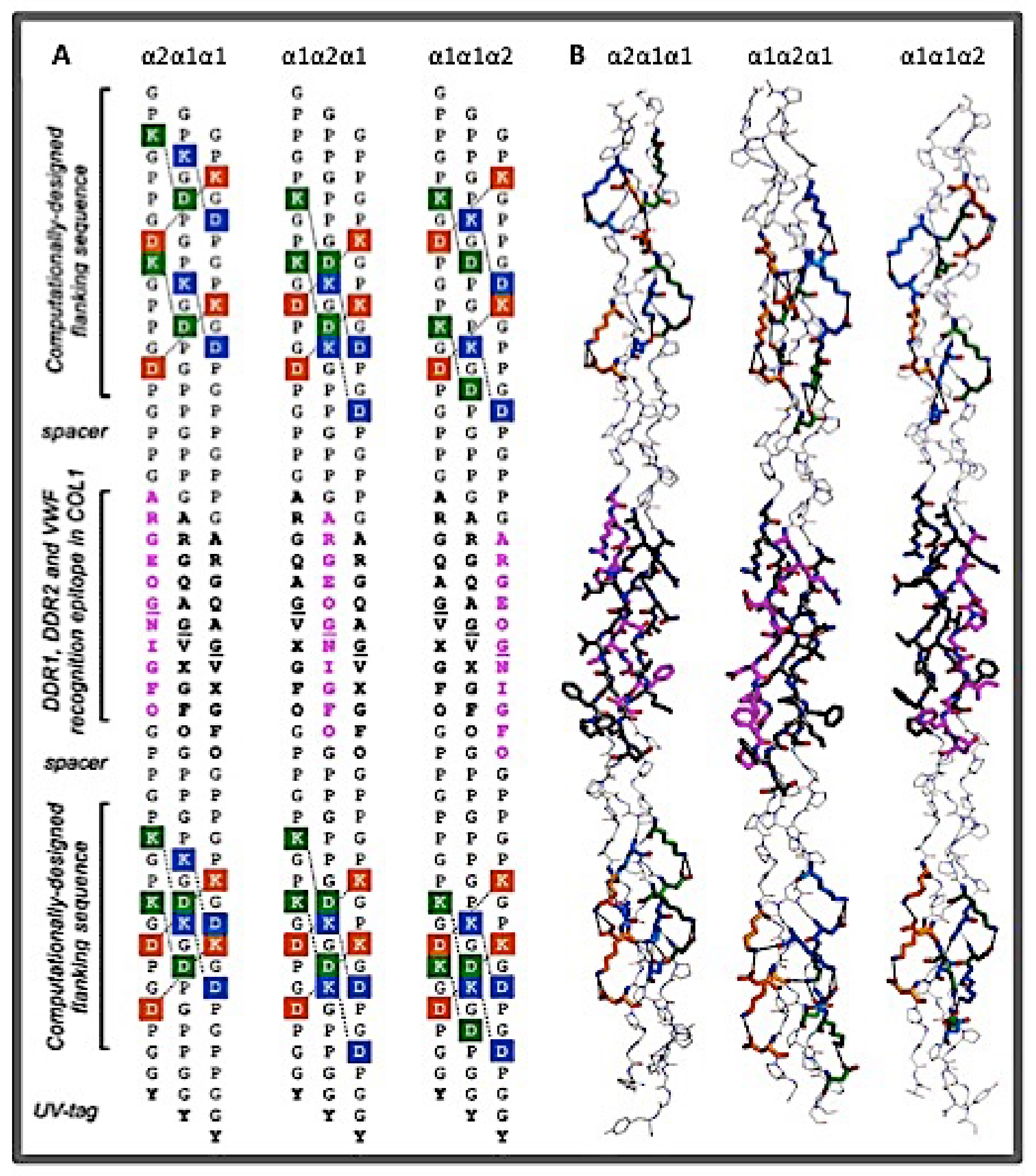
2.1.2. The Binding Sites of Collagen Receptors
2.2. The Heterotrimeric CMPs
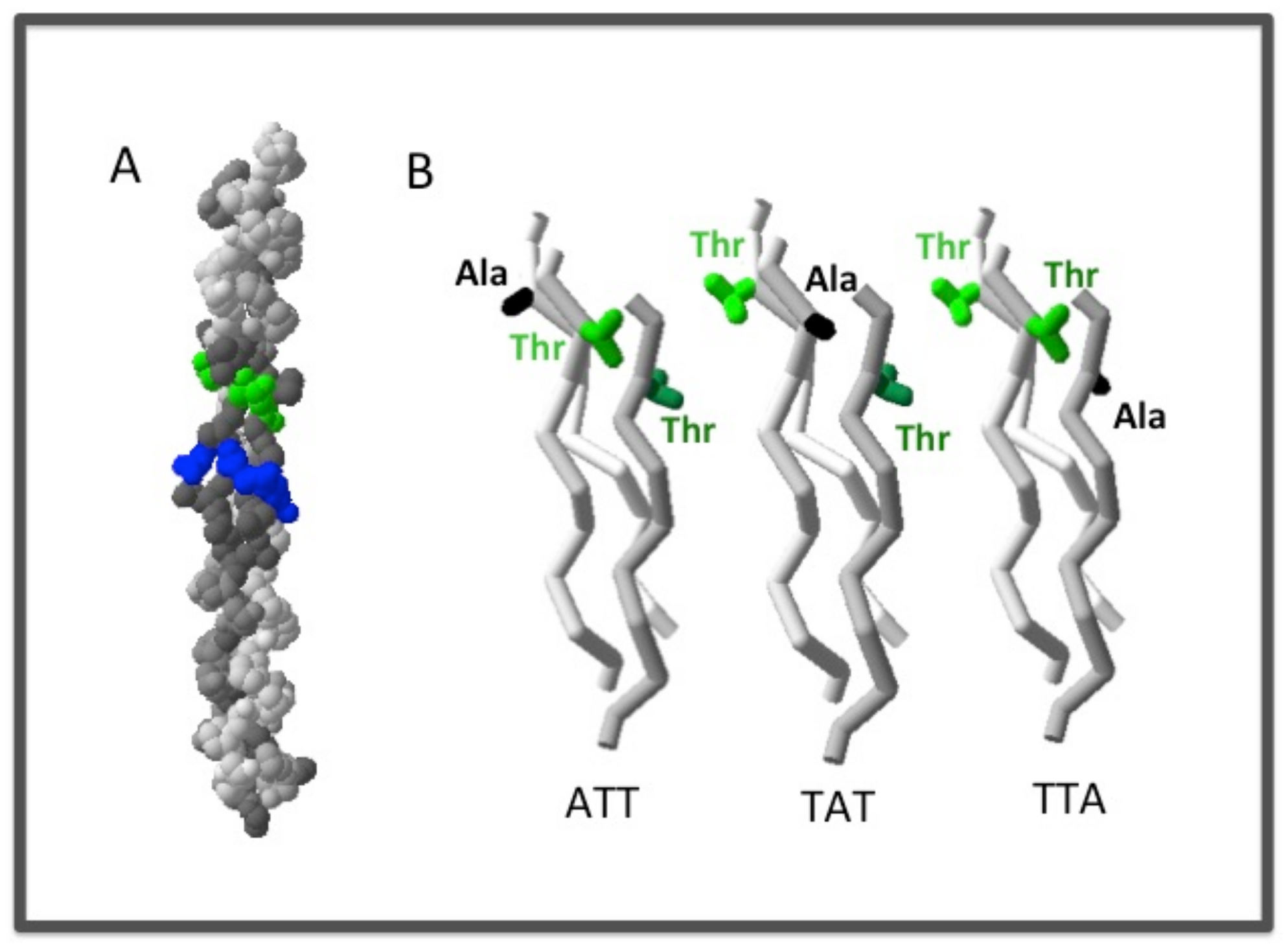
2.2.1. The Chain Register Affects both the Stability and the Binding Affinity of the Triple Helix
2.2.2. The Self-Assembled Heterotrimeric Triple Helix
2.3. Applications of CMPs
2.3.1. Self-Assembled Fibrillar Structures
2.3.2. Interaction of CMPs with Damaged Collagens
3. The Recombinant Collagen Peptides
3.1. The Sequence–Stability Relationship Revisited
3.1.1. Defining the Sequence Requirements of Fibronectin Binding, and of the Proteolysis of MMPs
3.1.2. The Impact of Gly Substitution Mutations
3.2. The Heterotrimeric Recombinant Peptides
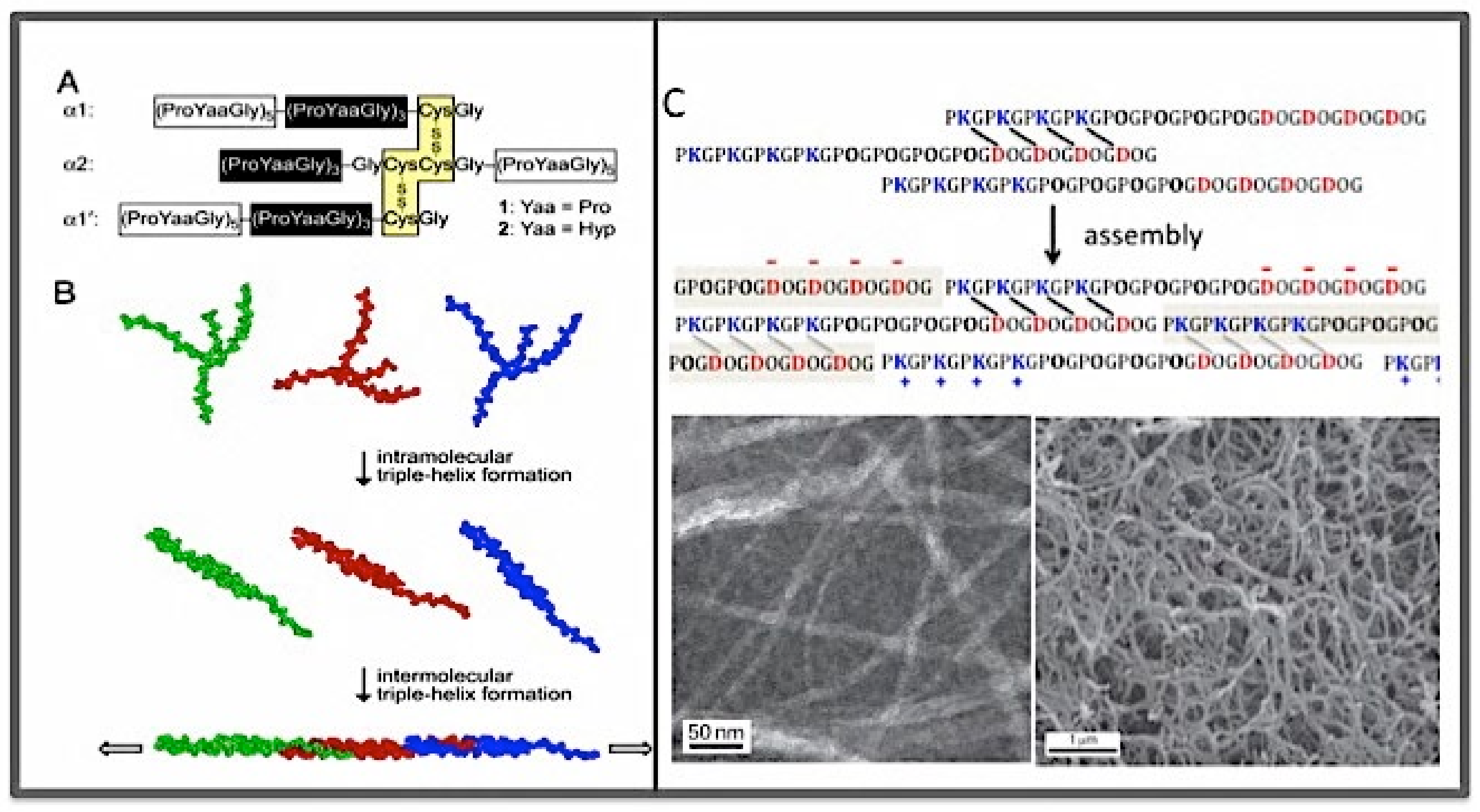
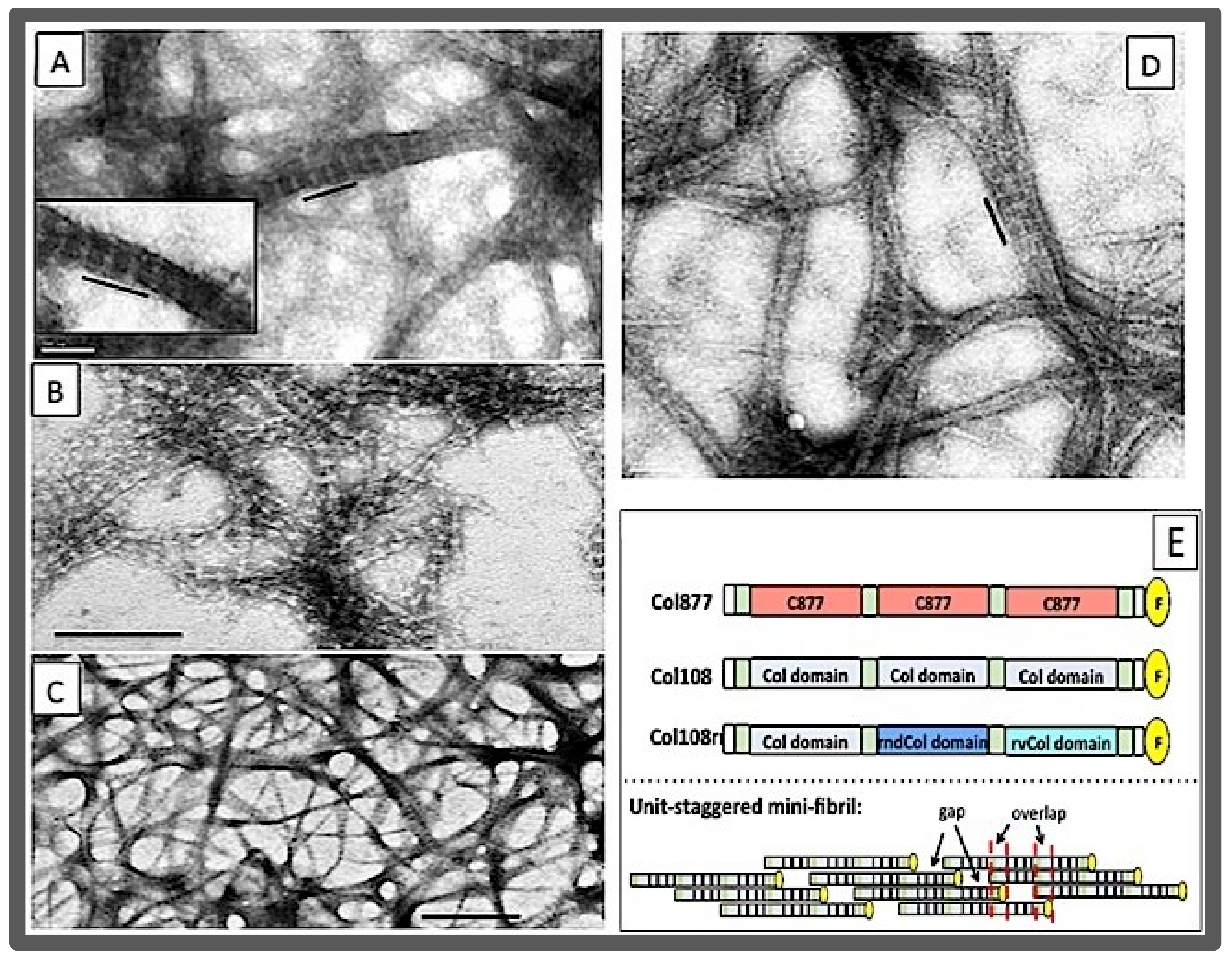
3.3. The Collagen Mimetic Fibrils
- (1)
- The size of the d-period was in a good agreement with the size of the section of triple helix formed by one SU (123 amino acid residues, or 41 GXY triplets) based on the average helical rise of 0.86 nm per GXY tripeptide. Furthermore, the overlap region was in good agreement with the size of the C-terminal overhang;
- (2)
- Another peptide, the 1U108, contained only one SU. This peptide did not have a 123-residue sequence periodicity and did not form fibrils [169];
- (3)
- The pattern of sequence periodicity was the determining factor. In another peptide, peptide Col108rr, the sequences in each of the three SUs of Col108 were shuffled such that while the amino acid composition of this peptide was the same as Col108, the sequence periodicity was lost. As expected, the Col108rr only formed non-specific aggregates [170];
- (4)
- In a new peptide, peptide Col877, the SU of Col108 was replaced by another 123 residues containing residues 877–986 of the α1 chain of human type I collagen [170] Thus, in contrast to Col108rr, Col877 had a very different amino acid composition from that of Col108, but had the same 123-residue sequence periodicity in its primary structure. Remarkably, Col877 can form mini-fibrils with the same 35-nm d-period as Col108. The Col877 mini-fibrils are a clear demonstration that the unit-staggering arrangement is at the foundation of the d-period axial structure.
- (5)
- The Col877 mini-fibril, and the lack of the fibril assembly of Col108rr and 1U108 indicated the foldon domain was not the determining factor of the fibril assembly, since all peptides, including Col108 and 2U108, contained the foldon domain.
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hartgerink, J.D.; Beniash, E.; Stupp, S.I. Self-assembly and mineralization of peptide-amphiphile nanofibers. Science 2001, 294, 1684–1688. [Google Scholar] [CrossRef] [PubMed]
- Hartgerink, J.D.; Beniash, E.; Stupp, S.I. Peptide-amphiphile nanofibers: A versatile scaffold for the preparation of self-assembling materials. Proc. Natl. Acad. Sci. USA 2002, 99, 5133–5138. [Google Scholar] [CrossRef] [PubMed]
- Wojtowicz, A.M.; Shekaran, A.; Oest, M.E.; Dupont, K.M.; Templeman, K.L.; Hutmacher, D.W.; Guldberg, R.E.; Garcia, A.J. Coating of biomaterial scaffolds with the collagen-mimetic peptide GFOGER for bone defect repair. Biomaterials 2010, 31, 2574–2582. [Google Scholar] [CrossRef] [PubMed]
- Lowery, J.L.; Datta, N.; Rutledge, G.C. Effect of fiber diameter, pore size and seeding method on growth of human dermal fibroblasts in electrospun poly(epsilon-caprolactone) fibrous mats. Biomaterials 2010, 31, 491–504. [Google Scholar] [CrossRef]
- Xie, J.; Li, X.; Lipner, J.; Manning, C.N.; Schwartz, A.G.; Thomopoulos, S.; Xia, Y. “Aligned-to-random” nanofiber scaffolds for mimicking the structure of the tendon-to-bone insertion site. Nanoscale 2010, 2, 923–926. [Google Scholar] [CrossRef]
- Ricard-Blum, S. The collagen family. Cold Spring Harb. Perspect Biol. 2011, 3, a004978. [Google Scholar] [CrossRef]
- Ricard-Blum, S.; Ruggiero, F. The collagen superfamily: From the extracellular matrix to the cell membrane. Pathol. Biol. 2005, 53, 430–442. [Google Scholar] [CrossRef]
- Birk, D.E.; Bruckner, P. Collagen suprastructures. Top. Curr. Chem. 2005, 247, 185–205. [Google Scholar] [CrossRef]
- Engel, J.; Bachinger, H.P. Structure, stability and folding of the collagen triple helix. Top. Curr. Chem. 2005, 247, 7–33. [Google Scholar] [CrossRef]
- Brodsky, B.; Thiagarajan, G.; Madhan, B.; Kar, K. Triple-helical peptides: An approach to collagen conformation, stability, and self-association. Biopolymers 2008, 89, 345–353. [Google Scholar] [CrossRef]
- Hulmes, D.J.; Miller, A.; Parry, D.A.; Piez, K.A.; Woodhead-Galloway, J. Analysis of the primary structure of collagen for the origins of molecular packing. J. Mol. Biol. 1973, 79, 137–148. [Google Scholar] [CrossRef]
- Hulmes, D.J.; Miller, A.; Parry, D.A.; Woodhead-Galloway, J. Fundamental periodicities in the amino acid sequence of the collagen alpha1 chain. Biochem. Biophys. Res. Commun. 1977, 77, 574–580. [Google Scholar] [CrossRef]
- Kaur, P.J.; Strawn, R.; Bai, H.; Xu, K.; Ordas, G.; Matsui, H.; Xu, Y. The self-assembly of a mini-fibril with axial periodicity from a designed collagen-mimetic triple helix. J. Biol. Chem. 2015, 290, 9251–9261. [Google Scholar] [CrossRef] [PubMed]
- Hulmes, D.J. Building collagen molecules, fibrils, and suprafibrillar structures. J. Struct. Biol. 2002, 137, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Kramer, R.Z.; Bella, J.; Brodsky, B.; Berman, H.M. The crystal and molecular structure of a collagen-like peptide with a Biol.ogically relevant sequence. J. Mol. Biol. 2001, 311, 131–147. [Google Scholar] [CrossRef]
- Li, Y.; Brodsky, B.; Baum, J. NMR shows hydrophobic interactions replace glycine packing in the triple helix at a natural break in the (Gly-X-Y)n repeat. J. Biol. Chem. 2007, 282, 22699–22706. [Google Scholar] [CrossRef]
- Li, Y.; Kim, S.; Brodsky, B.; Baum, J. Identification of partially disordered peptide intermediates through residue-specific NMR diffusion measurements. J. Am. Chem. Soc. 2005, 127, 10490–10491. [Google Scholar] [CrossRef]
- Fan, P.; Li, M.-H.; Brodsky, B.; Baum, J. Backbone Dynamics of (Pro-Hyp-Gly)10 and a Designed Collagen-like Triple-Helical Peptide by 15N NMR Relaxation and Hydrogen-Exchange Measurements. Biochemistry 1993, 32, 13299–13309. [Google Scholar] [CrossRef]
- Ottl, J.; Battistuta, R.; Pieper, M.; Tschesche, H.; Bode, W.; Kuhn, K.; Moroder, L. Design and synthesis of heterotrimeric collagen peptides with a built-in cystine-knot. Models for collagen catabolism by matrix-metalloproteases. FEBS Lett. 1996, 398, 31–36. [Google Scholar] [CrossRef]
- Piez, K.A.; Trus, B.L. Sequence regularities and packing of collagen molecules. J. Mol. Biol. 1978, 122, 419–432. [Google Scholar] [CrossRef]
- Jalan, A.A.; Sammon, D.; Hartgerink, J.D.; Brear, P.; Stott, K.; Hamaia, S.W.; Hunter, E.J.; Walker, D.R.; Leitinger, B.; Farndale, R.W. Chain alignment of collagen I deciphered using computationally designed heterotrimers. Nat. Chem. Biol. 2020, 16, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Prockop, D.J.; Kivirikko, K.I.; Tuderman, L.; Guzman, N.A. The biosynthesis of collagen and its disorders (first of two parts). N. Engl. J. Med. 1979, 301, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U.; Carrique, L.; Vadon-Le Goff, S.; Mariano, N.; Georges, R.-N.; Delolme, F.; Koivunen, P.; Myllyharju, J.; Moali, C.; Aghajari, N.; et al. Structural basis of homo- and heterotrimerization of collagen I. Nat. Commun. 2017, 8, 14671. [Google Scholar] [CrossRef] [PubMed]
- DiChiara, A.S.; Li, R.C.; Suen, P.H.; Hosseini, A.S.; Taylor, R.J.; Weickhardt, A.F.; Malhotra, D.; McCaslin, D.R. A cysteine-based molecular code informs collagen C-propeptide assembly. Nat. Commun. 2018, 9, 4206. [Google Scholar] [CrossRef] [PubMed]
- Wood, G.C.; Keech, M.K. The formation of fibrils from collagen solutions. 1. The effect of experimental conditions: Kinetic and electron-microscope studies. Biochem. J. 1960, 75, 588–598. [Google Scholar] [CrossRef] [PubMed]
- Piez, K.A. Structure and assembly of the native collagen fibril. Connect. Tissue Res. 1982, 10, 25–36. [Google Scholar] [CrossRef]
- Piez, K.A. Molecular and Aggregate StructuRes. of the Collagens. In Extracellular Matrix Biochemistry; Piez, K.A., Reddi, A.H., Eds.; Elsevier: New York, NY, USA; Amesterdam, The Netherlands; Oxford, UK, 1984. [Google Scholar]
- Piez, K.A.; Miller, A. The structure of collagen fibrils. J. Supramol. Struct. 1974, 2, 121–137. [Google Scholar] [CrossRef]
- Piez, K.A.; Trus, B.L. Microfibrillar structure and packing of collagen: Hydrophobic interactions. J. Mol. Biol. 1977, 110, 701–704. [Google Scholar] [CrossRef]
- Piez, K.A.; Trus, B.L. A new model for packing of type-I collagen molecules in the native fibril. Biosci. Rep. 1981, 1, 801–810. [Google Scholar] [CrossRef]
- Orgel, J.; Sella, I.; Madhurapantula, R.S.; Antipova, O.; Mandelberg, Y.; Kashman, Y.; Benayahu, D.; Benayahu, Y. Molecular and ultrastructural studies of a fibrillar collagen from octocoral (Cnidaria). J. Exp. Biol. 2017, 220, 3327–3335. [Google Scholar] [CrossRef]
- Orgel, J.P.; Antipova, O.; Sagi, I.; Bitler, A.; Qiu, D.; Wang, R.; Xu, Y.; San Antonio, J.D. Collagen fibril surface displays a constellation of sites capable of promoting fibril assembly, stability, and hemostasis. Connect Tissue Res. 2011, 52, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Orgel, J.P.; Irving, T.C.; Miller, A.; Wess, T.J. Microfibrillar structure of type I collagen in situ. Proc. Natl. Acad. Sci. USA 2006, 103, 9001–9005. [Google Scholar] [CrossRef] [PubMed]
- Orgel, J.P.; Persikov, A.V.; Antipova, O. Variation in the helical structure of native collagen. PLoS ONE 2014, 9, e89519. [Google Scholar] [CrossRef] [PubMed]
- Orgel, J.P.; San Antonio, J.D.; Antipova, O. Molecular and structural mapping of collagen fibril interactions. Connect Tissue Res. 2011, 52, 2–17. [Google Scholar] [CrossRef]
- Sweeney, S.M.; Orgel, J.P.; Fertala, A.; McAuliffe, J.D.; Turner, K.R.; Di Lullo, G.A.; Chen, S.; Antipova, O.; Perumal, S.; Ala-Kokko, L.; et al. Candidate cell and matrix interaction domains on the collagen fibril, the predominant protein of vertebrates. J. Biol. Chem. 2008, 283, 21187–21197. [Google Scholar] [CrossRef]
- Herr, A.B.; Farndale, R.W. Structural insights into the interactions between platelet receptors and fibrillar collagen. J. Biol. Chem. 2009, 284, 19781–19785. [Google Scholar] [CrossRef]
- Chang, S.W.; Buehler, M.J. Molecular biomechanics of collagen molecules. Mater. Today 2014, 17, 70–76. [Google Scholar] [CrossRef]
- Buehler, M.J. Nature designs tough collagen: Explaining the nanostructure of collagen fibrils. Proc. Natl. Acad. Sci. USA 2006, 103, 12285–12290. [Google Scholar] [CrossRef] [PubMed]
- Davidenko, N.; Schuster, C.F.; Bax, D.V.; Farndale, R.W.; Hamaia, S.; Best, S.M.; Cameron, R.E. Evaluation of cell binding to collagen and gelatin: A study of the effect of 2D and 3D architecture and surface chemistry. J. Mater. Sci. Mater. Med. 2016, 27, 148. [Google Scholar] [CrossRef] [PubMed]
- Malcor, J.-D.; Hunter, E.J.; Davidenko, N.; Bax, D.V.; Cameron, R.; Best, S.; Sinha, S.; Farndale, R.W. Collagen scaffolds functionalized with triple-helical peptides support 3D HUVEC culture. Regen. Biomater. 2020, 7, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Plant, A.L.; Bhadriraju, K.; Spurlin, T.A.; Elliott, J.T. Cell response to matrix mechanics: Focus on collagen. Biochim. Et Biophys. Acta 2009, 1793, 893–902. [Google Scholar] [CrossRef] [PubMed]
- Sherman, V.R.; Yang, W.; Meyers, M.A. The materials science of collagen. J. Mech. Behav. Biomed. 2015, 52, 22–50. [Google Scholar] [CrossRef]
- Kadler, K.E.; Hill, A.; Canty-Laird, E.G. Collagen fibrillogenesis: Fibronectin, integrins, and minor collagens as organizers and nucleators. Curr. Opin. Cell Biol. 2008, 20, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Kadler, K.E.; Hojima, Y.; Prockop, D.J. Assembly of collagen fibrils de novo by cleavage of the type I pC-collagen with procollagen C-proteinase. Assay of critical concentration demonstrates that collagen self-assembly is a classical example of an entropy-driven process. J. Biol. Chem. 1987, 262, 15696–15701. [Google Scholar] [PubMed]
- Prockop, D.J.; Fertala, A. Inhibition of the self-assembly of collagen I into fibrils with synthetic peptides. Demonstration that assembly is driven by specific binding sites on the monomers. J. Biol. Chem. 1998, 273, 15598–15604. [Google Scholar] [CrossRef]
- Kuznetsova, N.; Leikin, S. Does the triple helical domain of type I collagen encode molecular recognition and fiber assembly while telopeptides serve as catalytic domains? Effect of proteolytic cleavage on fibrillogenesis and on collagen-collagen interaction in fibers. J. Biol. Chem. 1999, 274, 36083–36088. [Google Scholar] [CrossRef]
- Comper, W.D.; Veis, A. The mechanism of nucleation for in vitro collagen fibril formation. Biopolymers 1977, 16, 2113–2131. [Google Scholar] [CrossRef]
- Helseth, D.L., Jr.; Veis, A. Collagen self-assembly in vitro. Differentiating specific telopeptide-dependent interactions using selective enzyme modification and the addition of free amino telopeptide. J. Biol. Chem. 1981, 256, 7118–7128. [Google Scholar]
- Farndale, R.W.; Lisman, T.; Bihan, D.; Hamaia, S.; Smerling, C.S.; Pugh, N.; Konitsiotis, A.; Leitinger, B.; de Groot, P.G.; Jarvis, G.E.; et al. Cell-collagen interactions: The use of peptide Toolkits to investigate collagen-receptor interactions. Biochem. Soc. Trans. 2008, 36, 241–250. [Google Scholar] [CrossRef]
- Di Lullo, G.A.; Sweeney, S.M.; Korkko, J.; Ala-Kokko, L.; San Antonio, J.D. Mapping the ligand-binding sites and disease-associated mutations on the most abundant protein in the human, type I collagen. J. Biol. Chem. 2002, 277, 4223–4231. [Google Scholar] [CrossRef]
- Sweeney, S.M.; Guy, C.A.; Fields, G.B.; San Antonio, J.D. Defining the domains of type I collagen involved in heparin- binding and endothelial tube formation. Proc. Natl. Acad. Sci. USA 1998, 95, 7275–7280. [Google Scholar] [CrossRef] [PubMed]
- Vogel, W.F.; Abdulhussein, R.; Ford, C.E. Sensing extracellular matrix: An update on discoidin domain receptor function. Cell. Signal. 2006, 18, 1108–1116. [Google Scholar] [CrossRef] [PubMed]
- Leitinger, B.; Hohenester, E. Mammalian collagen receptors. Matrix Biol. J. Int. Soc. Matrix Biol. 2007, 26, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Barczyk, M.; Carracedo, S.; Gullberg, D. Integrins. Cell Tissue Res. 2009, 339, 269. [Google Scholar] [CrossRef]
- Banerjee, J.; Azevedo, H.S. Crafting of functional biomaterials by directed molecular self-assembly of triple helical peptide building blocks. Interface Focus 2017, 7, 20160138. [Google Scholar] [CrossRef]
- Amar, S.; Smith, L.; Fields, G.B. Matrix metalloproteinase collagenolysis in health and disease. Biochim. Et Biophys. Acta. Mol. Cell Res. 2017, 1864, 1940–1951. [Google Scholar] [CrossRef]
- Barillari, G. The Impact of Matrix Metalloproteinase-9 on the Sequential Steps of the Metastatic Process. Int. J. Mol. Sci. 2020, 21, 4526. [Google Scholar] [CrossRef]
- Cui, N.; Hu, M.; Khalil, R.A. Biochemical and Biol.ogical Attributes of Matrix Metalloproteinases. Prog. Mol. Biol. Transl. Sci. 2017, 147, 1–73. [Google Scholar] [CrossRef]
- Fallas, J.A.; O’Leary, L.E.; Hartgerink, J.D. Synthetic collagen mimics: Self-assembly of homotrimers, heterotrimers and higher order structures. Chem. Soc Rev. 2010, 39, 3510–3527. [Google Scholar] [CrossRef]
- Lauer-Fields, J.L.; Juska, D.; Fields, G.B. Matrix metalloproteinases and collagen catabolism. Biopolymers 2002, 66, 19–32. [Google Scholar] [CrossRef]
- Lauer-Fields, J.L.; Tuzinski, K.A.; Shimokawa, K.; Nagase, H.; Fields, G.B. Hydrolysis of triple-helical collagen peptide models by matrix metalloproteinases. J. Biol. Chem. 2000, 275, 13282–13290. [Google Scholar] [CrossRef] [PubMed]
- Pedchenko, V.; Kitching, A.R.; Hudson, B.G. Goodpasture’s autoimmune disease—A collagen IV disorder. Matrix Biol. J. Int. Soc. Matrix Biol. 2018, 71—72, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Miyake, M.; Hori, S.; Morizawa, Y.; Tatsumi, Y.; Toritsuka, M.; Ohnishi, S.; Shimada, K.; Furuya, H.; Khadka, V.S.; Deng, Y.; et al. Collagen type IV alpha 1 (COL4A1) and collagen type XIII alpha 1 (COL13A1) produced in cancer cells promote tumor budding at the invasion front in human urothelial carcinoma of the bladder. Oncotarget 2017, 8, 36099–36114. [Google Scholar] [CrossRef] [PubMed]
- Vaniotis, G.; Rayes, R.F.; Qi, S.; Milette, S.; Wang, N.; Perrino, S.; Bourdeau, F.; Nyström, H.; He, Y.; Lamarche-Vane, N.; et al. Collagen IV-conveyed signals can regulate chemokine production and promote liver metastasis. Oncogene 2018, 37, 3790–3805. [Google Scholar] [CrossRef]
- Jayadev, R.; Chi, Q.; Keeley, D.P.; Hastie, E.L.; Kelley, L.C.; Sherwood, D.R. α-Integrins dictate distinct modes of type IV collagen recruitment to basement membranes. J. Cell Biol. 2019, 218, 3098–3116. [Google Scholar] [CrossRef]
- Ramshaw, J.A.; Werkmeister, J.A.; Glattauer, V. Collagen-based biomaterials. Biotechnol. Genet. Eng. Rev. 1996, 13, 335–382. [Google Scholar] [CrossRef]
- Kumar, V.A.; Shi, S.; Wang, B.K.; Li, I.C.; Jalan, A.A.; Sarkar, B.; Wickremasinghe, N.C.; Hartgerink, J.D. Drug-triggered and cross-linked self-assembling nanofibrous hydrogels. J. Am. Chem. Soc. 2015, 137, 4823–4830. [Google Scholar] [CrossRef]
- Kumar, V.A.; Taylor, N.L.; Jalan, A.A.; Hwang, L.K.; Wang, B.K.; Hartgerink, J.D. A nanostructured synthetic collagen mimic for hemostasis. Biomacromolecules 2014, 15, 1484–1490. [Google Scholar] [CrossRef]
- Fields, C.G.; Lovdahl, C.M.; Miles, A.J.; Hagen, V.L.; Fields, G.B. Solid-phase synthesis and stability of triple-helical peptides incorporating native collagen sequences. Biopolymers 1993, 33, 1695–1707. [Google Scholar] [CrossRef]
- Heidemann, E.; Roth, W. Synthesis and Investigation of Collagen Model Peptides. Adv. Polym. Sci. 1982, 43, 143–203. [Google Scholar]
- Sakakibara, S.; Inouye, K.; Shudo, K.; Kishida, Y.; Kobayashi, Y.; Prockop, D.J. Synthesis of (Pro-Hyp-Gly) n of defined molecular weights. Evidence for the stabilization of collagen triple helix by hydroxypyroline. Biochim. Biophys. Acta 1973, 303, 198–202. [Google Scholar] [CrossRef]
- Suto, K.; Noda, H. Conformational change of the triple-helical structure. IV. Kinetics of the helix-folding of (Pro-Pro-Gly)n (n equals 10, 12, and 15). Biopolymers 1974, 13, 2477–2488. [Google Scholar] [CrossRef] [PubMed]
- Fields, G.D.; Prockop, D.J. Perspectives on Synthesis and Applications of Triple-Helical Collagen Model Peptides. Biopolymers 1996, 40, 345. [Google Scholar] [CrossRef]
- Brodsky, B.; Persikov, A.V. Molecular structure of the collagen triple helix. Adv. Protein Chem. 2005, 70, 301–339. [Google Scholar] [CrossRef] [PubMed]
- Baum, J.; Brodsky, B. Folding of peptide models of collagen and misfolding diseases. Curre. Opin. Struct. Biol. 1999, 9, 122–128. [Google Scholar] [CrossRef]
- Buevich, A.V.; Silva, T.; Brodsky, B.; Baum, J. Transformation of the mechanism of triple-helix peptide folding in the absence of a C-terminal nucleation domain and its implications for mutations in collagen disorders. J. Biol. Chem. 2004, 279, 46890–46895. [Google Scholar] [CrossRef] [PubMed]
- Hyde, T.J.; Bryan, M.A.; Brodsky, B.; Baum, J. Sequence dependence of renucleation after a Gly mutation in model collagen peptides. J. Biol. Chem. 2006, 281, 36937–36943. [Google Scholar] [CrossRef]
- Xu, Y.; Bhate, M.; Brodsky, B. Characterization of the Nucleation Step and Folding of a Collagen Triple-Helix Peptide. Biochemistry 2002, 41, 8143–8151. [Google Scholar] [CrossRef]
- Persikov, A.V.; Xu, Y.; Brodsky, B. Equilibrium thermal transitions of collagen model peptides. Protein Sci. 2004, 13, 893–902. [Google Scholar] [CrossRef]
- Persikov, A.V.; Ramshaw, J.A.M.; Brodsky, B. Prediction of Collagen Stability from Amino Acid Sequence. J. Biol. Chem. 2005, 280, 19343–19349. [Google Scholar] [CrossRef]
- Bella, J.; Eaton, M.; Brodsky, B.; Berman, H.M. Crystal and Molecular Structure of a Collagen-Like Peptide at 1.9 A Resolution. Science 1994, 266, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Okuyama, K.; Okuyama, K.; Arnott, S.; Takayanagi, M.; Kakudo, M. Crystal and molecular structure of a collagen-like polypeptide (Pro-Pro-Gly)10. J. Mol. Biol. 1981, 152, 427–443. [Google Scholar] [CrossRef]
- Nagarajan, V.; Kamitori, S.; Okuyama, K. Structure analysis of a collagen-model peptide with a (Pro-Hyp-Gly) sequence repeat. J. Biochem. 1999, 125, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Persikov, A.V.; Ramshaw, J.A.; Kirkpatrick, A.; Brodsky, B. Triple-helix propensity of hydroxyproline and fluoroproline: Comparison of host-guest and repeating tripeptide collagen models. J. Am. Chem. Soc. 2003, 125, 11500–11501. [Google Scholar] [CrossRef] [PubMed]
- Persikov, A.V.; Ramshaw, J.A.; Kirkpatrick, A.; Brodsky, B. Electrostatic interactions involving lysine make major contributions to collagen triple-helix stability. Biochemistry 2005, 44, 1414–1422. [Google Scholar] [CrossRef]
- Persikov, A.V.; Ramshaw, J.A.M.; Kirkpatrick, A.; Brodsky, B. Amino Acid Propensities for the Collagen Triple-Helix. Biochemistry 2000, 39, 14960–14967. [Google Scholar] [CrossRef]
- Persikov, A.V.; Ramshaw, J.A.M.; Kirkpatrick, A.; Brodsky, B. Peptide investigations of pairwise interactions in the collagen triple-helix. J. Mol. Biol. 2002, 316, 385–394. [Google Scholar] [CrossRef]
- Knight, C.G.; Morton, L.F.; Onley, D.J.; Peachey, A.R.; Messent, A.J.; Smethurst, P.A.; Tuckwell, D.S.; Farndale, R.W.; Barnes, M.J. Identification in collagen type I of an integrin alpha2 beta1-binding site containing an essential GER sequence. J. Biol. Chem. 1998, 273, 33287–33294. [Google Scholar] [CrossRef]
- Knight, C.G.; Morton, L.F.; Peachey, A.R.; Tuckwell, D.S.; Farndale, R.W.; Barnes, M.J. The collagen-binding A-domains of integrins alpha(1)beta(1) and alpha(2)beta(1) recognize the same specific amino acid sequence, GFOGER, in native (triple-helical) collagens. J. Biol. Chem. 2000, 275, 35–40. [Google Scholar] [CrossRef]
- Tuckwell, D.; Calderwood, D.A.; Green, L.J.; Humphries, M.J. Integrin alpha 2 I-domain is a binding site for collagens. J. Cell Sci. 1995, 108, 1629–1637. [Google Scholar]
- Michishita, M.; Videm, V.; Arnaout, M.A. A novel divalent cation-binding site in the A domain of the beta 2 integrin CR3 (CD11b/CD18) is essential for ligand binding. Cell 1993, 72, 857–867. [Google Scholar] [CrossRef]
- Kamata, T.; Liddington, R.C.; Takada, Y. Interaction between collagen and the alpha(2) I-domain of integrin alpha(2)beta(1). Critical role of conserved residues in the metal ion-dependent adhesion site (MIDAS) region. J. Biol. Chem. 1999, 274, 32108–32111. [Google Scholar] [CrossRef] [PubMed]
- Emsley, J.; Knight, C.G.; Farndale, R.W.; Barnes, M.J. Structural basis of collagen recognition by integrin alpha2beta1. Cell 2000, 101, 47–56. [Google Scholar] [CrossRef]
- Raynal, N.; Hamaia, S.W.; Siljander, P.R.; Maddox, B.; Peachey, A.R.; Fernandez, R.; Foley, L.J.; Slatter, D.A.; Jarvis, G.E. Use of synthetic peptides to locate novel integrin alpha2beta1-binding motifs in human collagen III. J. Biol. Chem. 2006, 281, 3821–3831. [Google Scholar] [CrossRef]
- Slatter, D.A.; Foley, L.A.; Peachey, A.R.; Nietlispach, D.; Farndale, R.W. Rapid synthesis of a register-specific heterotrimeric type I collagen helix encompassing the integrin alpha2beta1 binding site. J. Mol. Biol. 2006, 359, 289–298. [Google Scholar] [CrossRef]
- Lisman, T.; Raynal, N.; Groeneveld, D.; Maddox, B.; Peachey, A.R.; Huizinga, E.G.; de Groot, P.G.; Farndale, R.W. A single high-affinity binding site for von Willebrand factor in collagen III, identified using synthetic triple-helical peptides. Blood 2006, 108, 3753–3756. [Google Scholar] [CrossRef]
- Knight, C.G.; Morton, L.F.; Onley, D.J.; Peachey, A.R.; Ichinohe, T.; Okuma, M.; Farndale, R.W.; Barnes, M.J. Collagen-platelet interaction: Gly-Pro-Hyp is uniquely specific for platelet Gp VI and mediates platelet activation by collagen. Cardiovasc. Res. 1999, 41, 450–457. [Google Scholar] [CrossRef]
- Xu, H.; Raynal, N.; Stathopoulos, S.; Myllyharju, J.; Farndale, R.W.; Leitinger, B. Collagen binding specificity of the discoidin domain receptors: Binding sites on collagens II and III and molecular determinants for collagen IV recognition by DDR1. Matrix Biol. J. Int. Soc. Matrix Biol. 2011, 30, 16–26. [Google Scholar] [CrossRef]
- Konitsiotis, A.D.; Raynal, N.; Bihan, D.; Hohenester, E.; Farndale, R.W.; Leitinger, B. Characterization of high affinity binding motifs for the discoidin domain receptor DDR2 in collagen. J. Biol. Chem. 2008, 283, 6861–6868. [Google Scholar] [CrossRef]
- Jarvis, G.E.; Raynal, N.; Langford, J.P.; Onley, D.J.; Andrews, A.; Smethurst, P.A.; Farndale, R.W. Identification of a major GpVI-binding locus in human type III collagen. Blood 2008, 111, 4986–4996. [Google Scholar] [CrossRef]
- Lebbink, R.J.; Raynal, N.; de Ruiter, T.; Bihan, D.G.; Farndale, R.W.; Meyaard, L. Identification of multiple potent binding sites for human leukocyte associated Ig-like receptor LAIR on collagens II and III. Matrix Biol. J. Int. Soc. Matrix Biol. 2009, 28, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Hinerman, J.M.; Blaszczyk, M.; Miller, J.L.; Conrady, D.G.; Barrow, A.D.; Chirgadze, D.Y.; Bihan, D.; Farndale, R.W.; Herr, A.B. Structural basis for collagen recognition by the immune receptor OSCAR. Blood 2016, 127, 529–537. [Google Scholar] [CrossRef] [PubMed]
- Perumal, S.; Antipova, O.; Orgel, J.P. Collagen fibril architecture, domain organization, and triple-helical conformation govern its proteolysis. Proc. Natl. Acad. Sci. USA 2008, 105, 2824–2829. [Google Scholar] [CrossRef]
- Ottl, J.; Gabriel, D.; Murphy, G.; Knauper, V.; Tominaga, Y.; Nagase, H.; Kroger, M.; Tschesche, H.; Bode, W.; Moroder, L. Recognition and catabolism of synthetic heterotrimeric collagen peptides by matrix metalloproteinases. Chem. Biol. 2000, 7, 119–132. [Google Scholar] [CrossRef]
- Boudko, S.P.; Bachinger, H.P. The NC2 domain of type IX collagen determines the chain register of the triple helix. J. Biol. Chem. 2012, 287, 44536–44545. [Google Scholar] [CrossRef]
- Boudko, S.P.; Engel, J.; Okuyama, K.; Mizuno, K.; Bachinger, H.P.; Schumacher, M.A. Crystal structure of human type III collagen Gly991-Gly1032 cystine knot-containing peptide shows both 7/2 and 10/3 triple helical symmetries. J. Biol. Chem. 2008, 283, 32580–32589. [Google Scholar] [CrossRef]
- Sacca, B.; Renner, C.; Moroder, L. The Chain Register in Heterotrimeric Collagen Peptides Affects Triple Helix Stability and Folding Kinetics. J. Mol. Biol. 2002, 324, 309–318. [Google Scholar] [CrossRef]
- Saccà, B.; Sinner, E.K.; Kaiser, J.; Lübken, C.; Eble, J.A.; Moroder, L. Binding and docking of synthetic heterotrimeric collagen type IV peptides with alpha1beta1 integrin. Chembiochem. Eur. J. Chem. Biol. 2002, 3, 904–907. [Google Scholar] [CrossRef]
- Xu, F.; Zahid, S.; Silva, T.; Nanda, V. Computational design of a collagen A:B:C-type heterotrimer. J. Am. Chem. Soc. 2011, 133, 15260–15263. [Google Scholar] [CrossRef]
- Xu, F.; Zhang, L.; Koder, R.L.; Nanda, V. De novo self-assembling collagen heterotrimers using explicit positive and negative design. Biochemistry 2010, 49, 2307–2316. [Google Scholar] [CrossRef]
- Zheng, H.; Lu, C.; Lan, J.; Fan, S.; Nanda, V.; Xu, F. How electrostatic networks modulate specificity and stability of collagen. Proc. Natl. Acad. Sci. USA 2018, 115, 6207–6212. [Google Scholar] [CrossRef]
- Fallas, J.A.; Hartgerink, J.D. Computational design of self-assembling register-specific collagen heterotrimers. Nat. Commun 2012, 3, 1087. [Google Scholar] [CrossRef] [PubMed]
- Fallas, J.A.; Lee, M.A.; Jalan, A.A.; Hartgerink, J.D. Rational design of single-composition ABC collagen heterotrimers. J. Am. Chem. Soc. 2012, 134, 1430–1433. [Google Scholar] [CrossRef] [PubMed]
- Gauba, V.; Hartgerink, J.D. Surprisingly high stability of collagen ABC heterotrimer: Evaluation of side chain charge pairs. J. Am. Chem. Soc. 2007, 129, 15034–15041. [Google Scholar] [CrossRef] [PubMed]
- Gauba, V.; Hartgerink, J.D. Self-assembled heterotrimeric collagen triple helices directed through electrostatic interactions. J. Am. Chem. Soc. 2007, 129, 2683–2690. [Google Scholar] [CrossRef] [PubMed]
- Jalan, A.A.; Demeler, B.; Hartgerink, J.D. Hydroxyproline-free single composition ABC collagen heterotrimer. J. Am. Chem. Soc. 2013, 135, 6014–6017. [Google Scholar] [CrossRef]
- Jalan, A.A.; Hartgerink, J.D. Pairwise interactions in collagen and the design of heterotrimeric helices. Curr. Opin. Chem. Biol. 2013, 17, 960–967. [Google Scholar] [CrossRef]
- Jalan, A.A.; Hartgerink, J.D. Simultaneous control of composition and register of an AAB-type collagen heterotrimer. Biomacromolecules 2013, 14, 179–185. [Google Scholar] [CrossRef]
- O’Leary, L.E.; Fallas, J.A.; Hartgerink, J.D. Positive and negative design leads to compositional control in AAB collagen heterotrimers. J. Am. Chem. Soc. 2011, 133, 5432–5443. [Google Scholar] [CrossRef]
- Russell, L.E.; Fallas, J.A.; Hartgerink, J.D. Selective assembly of a high stability AAB collagen heterotrimer. J. Am. Chem. Soc. 2010, 132, 3242–3243. [Google Scholar] [CrossRef]
- Tanrikulu, I.C.; Forticaux, A.; Jin, S.; Raines, R.T. Peptide tessellation yields micrometre-scale collagen triple helices. Nat. Chem. 2016, 8, 1008–1014. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, L.E.; Fallas, J.A.; Bakota, E.L.; Kang, M.K.; Hartgerink, J.D. Multi-hierarchical self-assembly of a collagen mimetic peptide from triple helix to nanofibre and hydrogel. Nat. Chem. 2011, 3, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Kotch, F.W.; Raines, R.T. Self-assembly of synthetic collagen triple helices. Proc. Natl. Acad. Sci. USA 2006, 103, 3028–3033. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, C.M.; Kadoya, Y.; Hozumi, K.; Okano-Kosugi, H.; Asada, S.; Kitagawa, K.; Nomizu, M.; Koide, T. A collagen-mimetic triple helical supramolecule that evokes integrin-dependent cell responses. Biomaterials 2010, 31, 1925–1934. [Google Scholar] [CrossRef]
- Koide, T.; Homma, D.L.; Asada, S.; Kitagawa, K. Self-complementary peptides for the formation of collagen-like triple helical supramolecules. Bioorg. Med. Chem. Lett. 2005, 15, 5230–5233. [Google Scholar] [CrossRef]
- Sarkar, B.; O’Leary, L.E.; Hartgerink, J.D. Self-assembly of fiber-forming collagen mimetic peptides controlled by triple-helical nucleation. J. Am. Chem. Soc. 2014, 136, 14417–14424. [Google Scholar] [CrossRef]
- Rele, S.; Song, Y.; Apkarian, R.P.; Qu, Z.; Conticello, V.P.; Chaikof, E.L. D-periodic collagen-mimetic microfibers. J. Am. Chem. Soc. 2007, 129, 14780–14787. [Google Scholar] [CrossRef]
- Bai, H.; Xu, K.; Xu, Y.; Matsui, H. Fabrication of Au nanowire in uniform length and diameter using a new monodisperse and rigid biomolecular template, collagen triple helix. Angewante Chem. 2007, 46, 3319–3322. [Google Scholar] [CrossRef]
- Kaur, P.; Maeda, Y.; Mutter, A.C.; Matsunaga, T.; Xu, Y.; Matsui, H. 3D Self-Assembly of Triple Helix Peptide NanowiRes. into Micron-Sized Crystalline Cubes with Joint Hubs of Ligand-Conjugated Au Nanoparticles. Nature 2009, 49, 8375–8378. [Google Scholar]
- Kar, K.; Amin, P.; Bryan, M.A.; Persikov, A.V.; Mohs, A.; Wang, Y.H.; Brodsky, B. Self-association of collagen triple helic peptides into higher order structures. J. Biol. Chem. 2006, 281, 33283–33290. [Google Scholar] [CrossRef]
- Kar, K.; Ibrar, S.; Nanda, V.; Getz, T.M.; Kunapuli, S.P.; Brodsky, B. Aromatic interactions promote self-association of collagen triple-helical peptides to higher-order structures. Biochemistry 2009, 48, 7959–7968. [Google Scholar] [CrossRef] [PubMed]
- Kar, K.; Wang, Y.H.; Brodsky, B. Sequence dependence of kinetics and morphology of collagen model peptide self-assembly into higher order structures. Protein Sci. Publ. Protein Soc. 2008, 17, 1086–1095. [Google Scholar] [CrossRef]
- McGuinness, K.; Khan, I.J.; Nanda, V. Morphological diversity and polymorphism of self-assembling collagen peptides controlled by length of hydrophobic domains. ACS Nano 2014, 8, 12514–12523. [Google Scholar] [CrossRef]
- Parmar, A.S.; James, J.K.; Grisham, D.R.; Pike, D.H.; Nanda, V. Dissecting Electrostatic Contributions to Folding and Self-Assembly Using Designed Multicomponent Peptide Systems. J. Am. Chem. Soc. 2016, 138, 4362–4367. [Google Scholar] [CrossRef] [PubMed]
- Cejas, M.A.; Kinney, W.A.; Chen, C.; Vinter, J.G.; Almond, H.R., Jr.; Balss, K.M.; Maryanoff, C.A.; Schmidt, U.; Breslav, M.; Mahan, A.; et al. Thrombogenic collagen-mimetic peptides: Self-assembly of triple helix-based fibrils driven by hydrophobic interactions. Proc. Natl. Acad. Sci. USA 2008, 105, 8513–8518. [Google Scholar] [CrossRef] [PubMed]
- Cejas, M.A.; Kinney, W.A.; Chen, C.; Leo, G.C.; Tounge, B.A.; Vinter, J.G.; Joshi, P.P.; Maryanoff, B.E. Collagen-related peptides: Self-assembly of short, single strands into a functional biomaterial of micrometer scale. J. Am. Chem. Soc. 2007, 129, 2202–2203. [Google Scholar] [CrossRef] [PubMed]
- Przybyla, D.E.; Chmielewski, J. Metal-triggered radial self-assembly of collagen peptide fibers. J. Am. Chem. Soc. 2008, 130, 12610–12611. [Google Scholar] [CrossRef] [PubMed]
- Przybyla, D.E.; Chmielewski, J. Metal-triggered collagen peptide disk formation. J. Am. Chem. Soc. 2010, 132, 7866–7867. [Google Scholar] [CrossRef]
- Li, Y.; Yu, S.M. Targeting and mimicking collagens via triple helical peptide assembly. Curr. Opin. Chem. Biol. 2013, 17, 968–975. [Google Scholar] [CrossRef]
- Li, Y.; Ho, D.; Meng, H.; Chan, T.R.; An, B.; Yu, H.; Brodsky, B.; Jun, A.S.; Michael Yu, S. Direct detection of collagenous proteins by fluorescently labeled collagen mimetic peptides. Bioconjug. Chem. 2013, 24, 9–16. [Google Scholar] [CrossRef]
- Li, Y.; Foss, C.A.; Summerfield, D.D.; Doyle, J.J.; Torok, C.M.; Dietz, H.C.; Pomper, M.G.; Yu, S.M. Targeting collagen strands by photo-triggered triple-helix hybridization. Proc. Natl. Acad. Sci. USA 2012, 109, 14767–14772. [Google Scholar] [CrossRef]
- Hwang, J.; San, B.H.; Turner, N.J.; White, L.J.; Faulk, D.M.; Badylak, S.F.; Li, Y.; Yu, S.M. Molecular assessment of collagen denaturation in decellularized tissues using a collagen hybridizing peptide. Acta Biomater. 2017, 53, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Hodges, J.A.; Raines, R.T. Stereoelectronic and steric effects in the collagen triple helix: Toward a code for strand association. J. Am. Chem. Soc. 2005, 127, 15923–15932. [Google Scholar] [CrossRef]
- Barth, D.; Milbradt, A.G.; Renner, C.; Moroder, L. A (4R)- or a (4S)-fluoroproline residue in position Xaa of the (Xaa-Yaa-Gly) collagen repeat severely affects triple-helix formation. Chembiochem. Eur. J. Chem. Biol. 2004, 5, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Zitnay, J.L.; Li, Y.; Qin, Z.; San, B.H.; Depalle, B.; Reese, S.P.; Buehler, M.J.; Yu, S.M.; Weiss, J.A. Molecular level detection and localization of mechanical damage in collagen enabled by collagen hybridizing peptides. Nat. Commun. 2017, 8, 14913. [Google Scholar] [CrossRef]
- Wahyudi, H.; Reynolds, A.A.; Li, Y.; Owen, S.C.; Yu, S.M. Targeting collagen for diagnostic imaging and therapeutic delivery. J. Control. Release Off. J. Control. Release Soc. 2016, 240, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Ellison, A.J.; Dempwolff, F.; Kearns, D.B.; Raines, R.T. Role for Cell-Surface Collagen of Streptococcus pyogenes in Infections. ACS Infect. Dis. 2020, 6, 1836–1843. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, A.B.; Karim, A.; Ocotl, E.; Dones, J.M.; Chacko, J.V.; Liu, A.; Raines, R.T.; Gibson, A.L.F.; Eliceiri, K.W. Optical imaging of collagen fiber damage to assess thermally injured human skin. Wound Repair Regen. Off. Publ. Wound Health Soc. Eur. Tissue Repair Soc. 2020, 28, 848–855. [Google Scholar] [CrossRef] [PubMed]
- Ellison, A.J.; Tanrikulu, I.C.; Dones, J.M.; Raines, R.T. Cyclic Peptide Mimetic of Damaged Collagen. Biomacromolecules 2020, 21, 1539–1547. [Google Scholar] [CrossRef]
- Dones, J.M.; Tanrikulu, I.C.; Chacko, J.V.; Schroeder, A.B.; Hoang, T.T.; Gibson, A.L.F.; Eliceiri, K.W.; Raines, R.T. Optimization of interstrand interactions enables burn detection with a collagen-mimetic peptide. Org. Biomol. Chem. 2019, 17, 9906–9912. [Google Scholar] [CrossRef]
- Song, J.Y.; Pineault, K.M.; Dones, J.M.; Raines, R.T.; Wellik, D.M. Hox genes maintain critical roles in the adult skeleton. Proc. Natl. Acad. Sci. USA 2020, 117, 7296–7304. [Google Scholar] [CrossRef]
- Sivashanmugam, A.; Murray, V.; Cui, C.; Zhang, Y.; Wang, J.; Li, Q. Practical protocols for production of very high yields of recombinant proteins using Escherichia coli. Protein Sci. Publ. Protein Soc. 2009, 18, 936–948. [Google Scholar] [CrossRef] [PubMed]
- An, B.; Brodsky, B. Collagen binding to OSCAR: The odd couple. Blood 2016, 127, 521–522. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Brodsky, B.; Inouye, M. Dissecting a bacterial collagen domain from Streptococcus pyogenes: Sequence and length-dependent variations in triple helix stability and folding. J. Biol. Chem. 2011, 286, 18960–18968. [Google Scholar] [CrossRef] [PubMed]
- Perret, S.; Merle, C.; Bernocco, S.; Berland, P.; Garrone, R.; Hulmes, D.J.; Theisen, M.; Ruggiero, F. Unhydroxylated triple helical collagen I produced in transgenic plants provides new clues on the role of hydroxyproline in collagen folding and fibril formation. J. Biol. Chem. 2001, 276, 43693–43698. [Google Scholar] [CrossRef] [PubMed]
- Olsen, D.R.; Leigh, S.D.; Chang, R.; McMullin, H.; Ong, W.; Tai, E.; Chisholm, G.; Birk, D.E.; Berg, R.A.; Hitzeman, R.A.; et al. Production of human type I collagen in yeast reveals unexpected new insights into the molecular assembly of collagen trimers. J. Biol. Chem. 2001, 276, 24038–24043. [Google Scholar] [CrossRef]
- Mohs, A.; Silva, T.; Yoshida, T.; Amin, R.; Lukomski, S.; Inouye, M.; Brodsky, B. Mechanism of stabilization of a bacterial collagen triple helix in the absence of hydroxyproline. J. Biol. Chem. 2007, 282, 29757–29765. [Google Scholar] [CrossRef] [PubMed]
- Yoshizumi, A.; Yu, Z.; Silva, T.; Thiagarajan, G.; Ramshaw, J.A.; Inouye, M.; Brodsky, B. Self-association of streptococcus pyogenes collagen-like constructs into higher order structures. Protein Sci. Publ. Protein Soc. 2009, 18, 1241–1251. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; An, B.; Ramshaw, J.A.; Brodsky, B. Bacterial collagen-like proteins that form triple-helical structures. J. Struct. Biol. 2014, 186, 451–461. [Google Scholar] [CrossRef]
- An, B.; Abbonante, V.; Yigit, S.; Balduini, A.; Kaplan, D.L.; Brodsky, B. Definition of the native and denatured type II collagen binding site for fibronectin using a recombinant collagen system. J. Biol. Chem. 2014, 289, 4941–4951. [Google Scholar] [CrossRef]
- An, B.; Abbonante, V.; Xu, H.; Gavriilidou, D.; Yoshizumi, A.; Bihan, D.; Farndale, R.W.; Kaplan, D.L.; Balduini, A.; Leitinger, B.; et al. Recombinant Collagen Engineered to Bind to Discoidin Domain Receptor Functions as a Receptor Inhibitor. J. Biol. Chem. 2016, 291, 4343–4355. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Visse, R.; Inouye, M.; Nagase, H.; Brodsky, B. Defining requirements for collagenase cleavage in collagen type III using a bacterial collagen system. J. Biol. Chem. 2012, 287, 22988–22997. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Nowak, I.; Kirchner, M.; Xu, Y. Recombinant collagen studies link the severe conformational changes induced by osteogenesis imperfecta mutations to the disruption of a set of interchain salt bridges. J. Biol. Chem. 2008, 283, 34337–34344. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Rashid, S.; Yu, Z.; Yoshizumi, A.; Hwang, E.; Brodsky, B. Location of glycine mutations within a bacterial collagen protein affects degree of disruption of triple-helix folding and conformation. J. Biol. Chem. 2011, 286, 2041–2046. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Cheng, H.; Silva, T.; Baum, J.; Brodsky, B. Osteogenesis imperfecta missense mutations in collagen: Structural consequences of a glycine to alanine replacement at a highly charged site. Biochemistry 2011, 50, 10771–10780. [Google Scholar] [CrossRef]
- Boudko, S.P.; Zientek, K.D.; Vance, J.; Hacker, J.L.; Engel, J.; Bachinger, H.P. The NC2 domain of collagen IX provides chain selection and heterotrimerization. J. Biol. Chem. 2010, 285, 23721–23731. [Google Scholar] [CrossRef]
- Boudko, S.P.; Engel, J.; Bachinger, H.P. The crucial role of trimerization domains in collagen folding. Int J. Biochem Cell Biol. 2012, 44, 21–32. [Google Scholar] [CrossRef]
- Strawn, R.; Chen, F.; Jeet Haven, P.; Wong, S.; Park-Arias, A.; De Leeuw, M.; Xu, Y. To achieve self-assembled collagen mimetic fibrils using designed peptides. Biopolymers 2018, 109, e23226. [Google Scholar] [CrossRef]
- Chen, F.; Strawn, R.; Xu, Y. The predominant roles of the sequence periodicity in the self-assembly of collagen-mimetic mini-fibrils. Protein Sci. Publ. Protein Soc. 2019, 28, 1640–1651. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Kirchner, M. Collagen Mimetic Peptides. Bioengineering 2021, 8, 5. https://doi.org/10.3390/bioengineering8010005
Xu Y, Kirchner M. Collagen Mimetic Peptides. Bioengineering. 2021; 8(1):5. https://doi.org/10.3390/bioengineering8010005
Chicago/Turabian StyleXu, Yujia, and Michele Kirchner. 2021. "Collagen Mimetic Peptides" Bioengineering 8, no. 1: 5. https://doi.org/10.3390/bioengineering8010005
APA StyleXu, Y., & Kirchner, M. (2021). Collagen Mimetic Peptides. Bioengineering, 8(1), 5. https://doi.org/10.3390/bioengineering8010005



