Advancements in Radiomics-Based AI for Pancreatic Ductal Adenocarcinoma
Abstract
1. Introduction
2. Related Works and Contributions
3. Research Methodology
- Studies that implemented radiomics or deep learning-based radiomic analyses.
- Investigations covering uni- or multi-modality imaging approaches (e.g., CT, MRI, and PET).
- Fusion models integrating radiomics with machine learning or deep learning techniques.
- Research exploring single- or multi-omics (e.g., radiogenomics).
- English-language publications that reported human subjects’ data on pancreatic cancer.
4. Comprehensive Review of the Literature by Clinical Application
4.1. Disease Classification
4.2. Disease Detection
4.3. Survival Prediction
4.4. Treatment Response
4.5. Radiogenomics
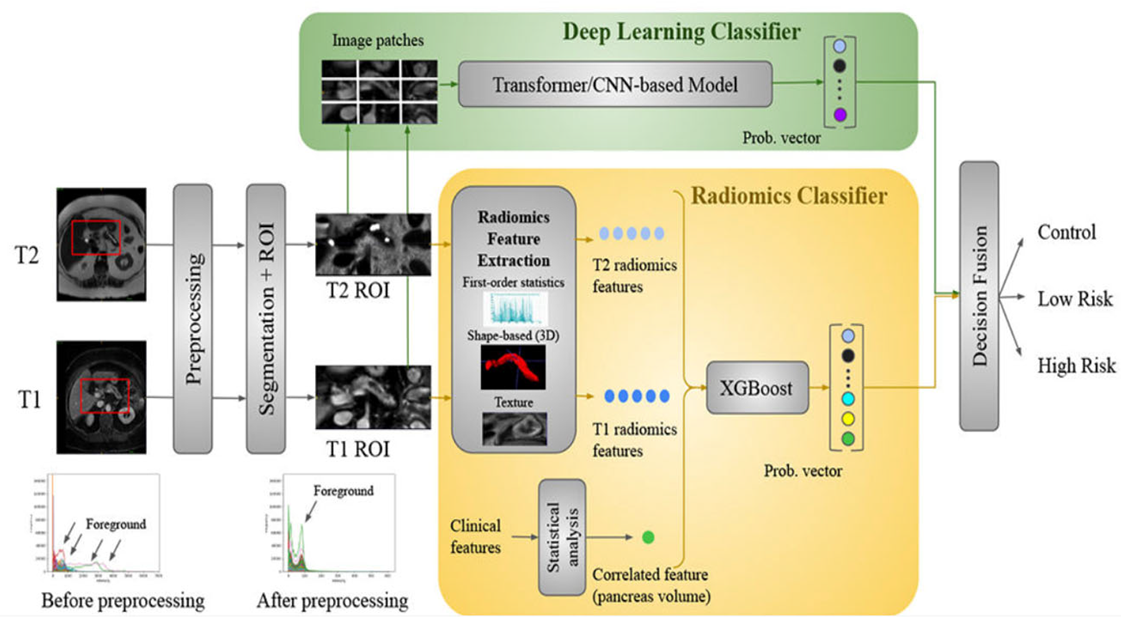
4.6. Deep Radiomics Fusion Models
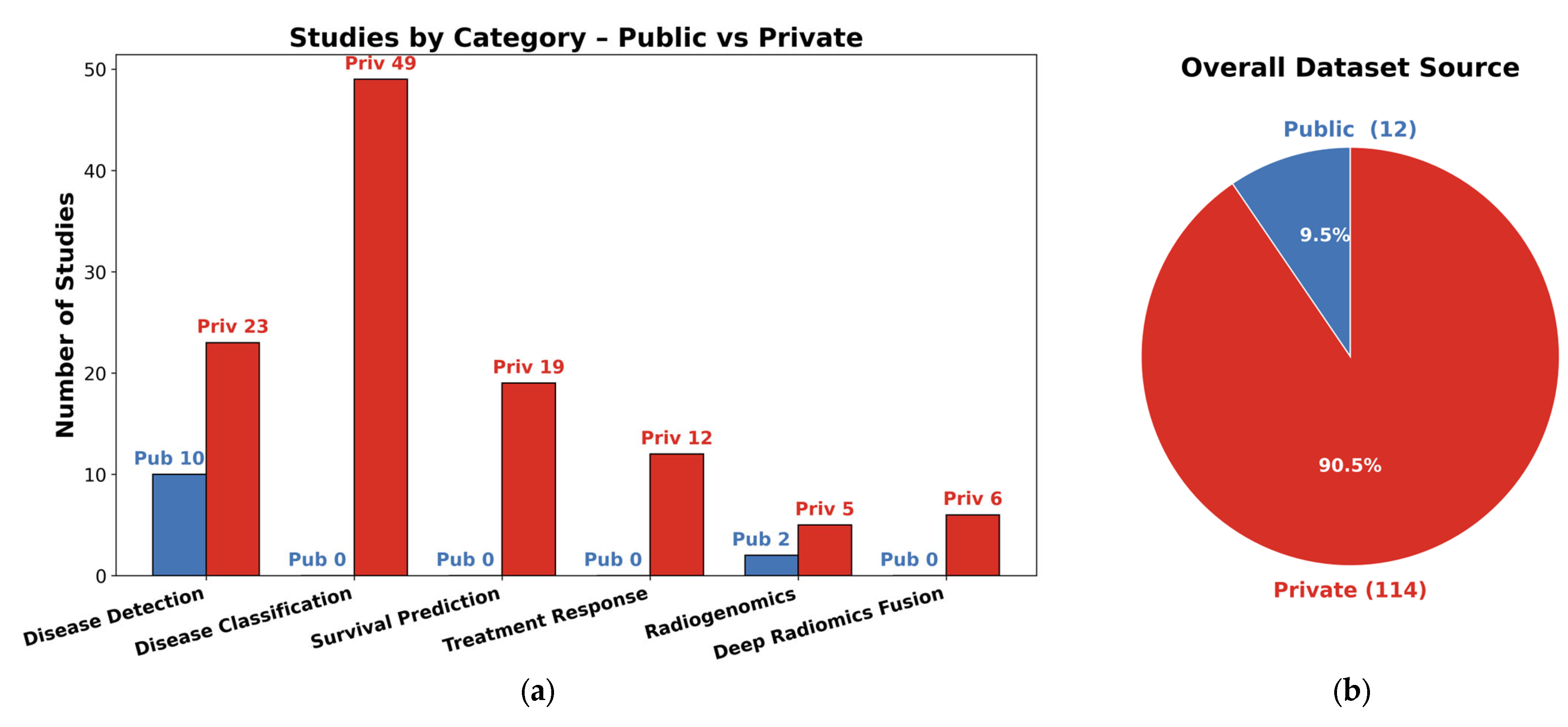
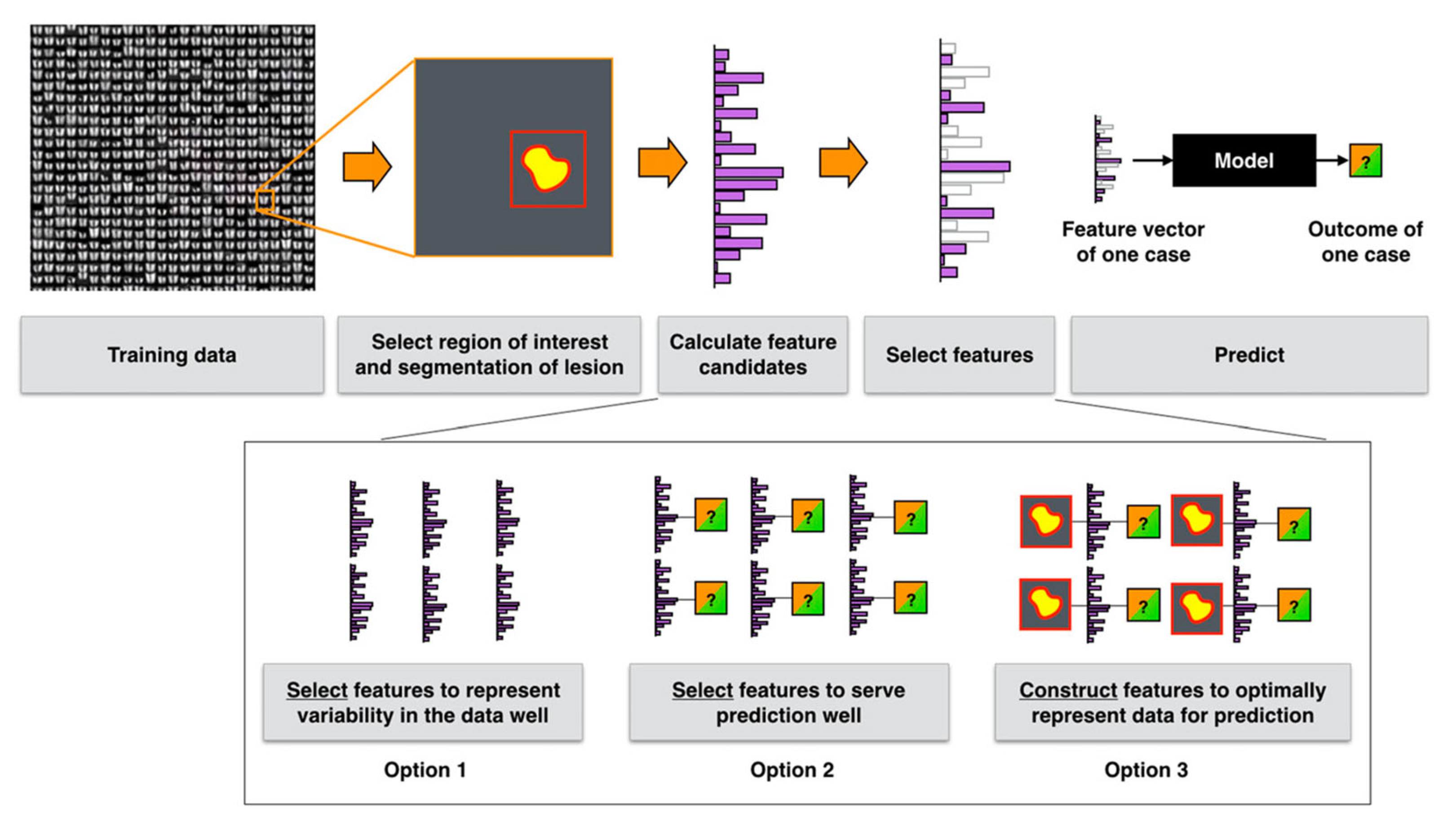
5. Datasets, Features, and Methods
5.1. Datasets
5.2. Features and Methods
5.3. Radiomics and Deep Radiomics Comparison
6. Discussion
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer Statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef]
- Hidalgo, M. Pancreatic Cancer. N. Engl. J. Med. 2010, 362, 1605–1617. [Google Scholar] [CrossRef] [PubMed]
- Mayerhoefer, M.E.; Materka, A.; Langs, G.; Häggström, I.; Szczypiński, P.; Gibbs, P.; Cook, G. Introduction to Radiomics. J. Nucl. Med. 2020, 61, 488–495. [Google Scholar] [CrossRef] [PubMed]
- Nougaret, S.; Tibermacine, H.; Tardieu, M.; Sala, E. Radiomics: An Introductory Guide to What It May Foretell. Curr. Oncol. Rep. 2019, 21, 70. [Google Scholar] [CrossRef] [PubMed]
- Kocak, B.; Durmaz, E.S.; Ates, E.; Kilickesmez, O. Radiomics with Artificial Intelligence: A Practical Guide for Beginners. Diagn. Interv. Radiol. 2019, 25, 485–495. [Google Scholar] [CrossRef]
- Giger, M.L. Machine Learning in Medical Imaging. J. Am. Coll. Radiol. 2018, 15, 512–520. [Google Scholar] [CrossRef]
- Bizzego, A.; Bussola, N.; Salvalai, D.; Chierici, M.; Maggio, V.; Jurman, G.; Furlanello, C. Integrating Deep and Radiomics Features in Cancer Bioimaging. In Proceedings of the 2019 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), Siena, Italy, 9–11 July 2019; pp. 1–8. [Google Scholar]
- Casà, C.; Piras, A.; D’Aviero, A.; Preziosi, F.; Mariani, S.; Cusumano, D.; Romano, A.; Boskoski, I.; Lenkowicz, J.; Dinapoli, N.; et al. The Impact of Radiomics in Diagnosis and Staging of Pancreatic Cancer. Ther. Adv. Gastrointest. Endosc. 2022, 15, 26317745221081596. [Google Scholar] [CrossRef]
- Castiglioni, I.; Rundo, L.; Codari, M.; Di Leo, G.; Salvatore, C.; Interlenghi, M.; Gallivanone, F.; Cozzi, A.; D’Amico, N.C.; Sardanelli, F. AI Applications to Medical Images: From Machine Learning to Deep Learning. Phys. Medica 2021, 83, 9–24. [Google Scholar] [CrossRef]
- Guiot, J.; Vaidyanathan, A.; Deprez, L.; Zerka, F.; Danthine, D.; Frix, A.; Lambin, P.; Bottari, F.; Tsoutzidis, N.; Miraglio, B.; et al. A Review in Radiomics: Making Personalized Medicine a Reality via Routine Imaging. Med. Res. Rev. 2022, 42, 426–440. [Google Scholar] [CrossRef]
- Yao, L.; Zhang, Z.; Keles, E.; Yazici, C.; Tirkes, T.; Bagci, U. A Review of Deep Learning and Radiomics Approaches for Pancreatic Cancer Diagnosis from Medical Imaging. Curr. Opin. Gastroenterol. 2023, 39, 436–447. [Google Scholar] [CrossRef]
- Huang, B.; Huang, H.; Zhang, S.; Zhang, D.; Shi, Q.; Liu, J.; Guo, J. Artificial Intelligence in Pancreatic Cancer. Theranostics 2022, 12, 6931–6954. [Google Scholar] [CrossRef]
- Hayashi, H.; Uemura, N.; Matsumura, K.; Zhao, L.; Sato, H.; Shiraishi, Y.; Yamashita, Y.; Baba, H. Recent Advances in Artificial Intelligence for Pancreatic Ductal Adenocarcinoma. World J. Gastroenterol. 2021, 27, 7480–7496. [Google Scholar] [CrossRef] [PubMed]
- Abunahel, B.M.; Pontre, B.; Kumar, H.; Petrov, M.S. Pancreas Image Mining: A Systematic Review of Radiomics. Eur. Radiol. 2021, 31, 3447–3467. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, T.M.; Kawamoto, S.; Hruban, R.H.; Fishman, E.K.; Soyer, P.; Chu, L.C. A Primer on Artificial Intelligence in Pancreatic Imaging. Diagn. Interv. Imaging 2023, 104, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Avanzo, M.; Stancanello, J.; El Naqa, I. Beyond Imaging: The Promise of Radiomics. Phys. Medica 2017, 38, 122–139. [Google Scholar] [CrossRef]
- He, M.; Liu, Z.; Lin, Y.; Wan, J.; Li, J.; Xu, K.; Wang, Y.; Jin, Z.; Tian, J.; Xue, H. Differentiation of Atypical Non-Functional Pancreatic Neuroendocrine Tumor and Pancreatic Ductal Adenocarcinoma Using CT Based Radiomics. Eur. J. Radiol. 2019, 117, 102–111. [Google Scholar] [CrossRef]
- Xie, H.; Ma, S.; Guo, X.; Zhang, X.; Wang, X. Preoperative Differentiation of Pancreatic Mucinous Cystic Neoplasm from Macrocystic Serous Cystic Adenoma Using Radiomics: Preliminary Findings and Comparison with Radiological Model. Eur. J. Radiol. 2020, 122, 108747. [Google Scholar] [CrossRef]
- Mashayekhi, R.; Parekh, V.S.; Faghih, M.; Singh, V.K.; Jacobs, M.A.; Zaheer, A. Radiomic Features of the Pancreas on CT Imaging Accurately Differentiate Functional Abdominal Pain, Recurrent Acute Pancreatitis, and Chronic Pancreatitis. Eur. J. Radiol. 2020, 123, 108778. [Google Scholar] [CrossRef]
- Attiyeh, M.A.; Chakraborty, J.; Gazit, L.; Langdon-Embry, L.; Gonen, M.; Balachandran, V.P.; D’Angelica, M.I.; DeMatteo, R.P.; Jarnagin, W.R.; Kingham, T.P.; et al. Preoperative Risk Prediction for Intraductal Papillary Mucinous Neoplasms by Quantitative CT Image Analysis. HPB 2019, 21, 212–218. [Google Scholar] [CrossRef]
- Liu, J.; Hu, L.; Zhou, B.; Wu, C.; Cheng, Y. Development and Validation of a Novel Model Incorporating MRI-Based Radiomics Signature with Clinical Biomarkers for Distinguishing Pancreatic Carcinoma from Mass-Forming Chronic Pancreatitis. Transl. Oncol. 2022, 18, 101357. [Google Scholar] [CrossRef]
- Kulali, F.; Semiz-Oysu, A.; Demir, M.; Segmen-Yilmaz, M.; Bukte, Y. Role of Diffusion-Weighted MR Imaging in Predicting the Grade of Nonfunctional Pancreatic Neuroendocrine Tumors. Diagn. Interv. Imaging 2018, 99, 301–309. [Google Scholar] [CrossRef]
- Park, S.; Chu, L.C.; Hruban, R.H.; Vogelstein, B.; Kinzler, K.W.; Yuille, A.L.; Fouladi, D.F.; Shayesteh, S.; Ghandili, S.; Wolfgang, C.L.; et al. Differentiating Autoimmune Pancreatitis from Pancreatic Ductal Adenocarcinoma with CT Radiomics Features. Diagn. Interv. Imaging 2020, 101, 555–564. [Google Scholar] [CrossRef]
- Wei, R.; Lin, K.; Yan, W.; Guo, Y.; Wang, Y.; Li, J.; Zhu, J. Computer-Aided Diagnosis of Pancreas Serous Cystic Neoplasms: A Radiomics Method on Preoperative MDCT Images. Technol. Cancer Res. Treat. 2019, 18, 1533033818824339. [Google Scholar] [CrossRef]
- Reinert, C.P.; Baumgartner, K.; Hepp, T.; Bitzer, M.; Horger, M. Complementary Role of Computed Tomography Texture Analysis for Differentiation of Pancreatic Ductal Adenocarcinoma from Pancreatic Neuroendocrine Tumors in the Portal-Venous Enhancement Phase. Abdom. Radiol. 2020, 45, 750–758. [Google Scholar] [CrossRef] [PubMed]
- Polk, S.L.; Choi, J.W.; McGettigan, M.J.; Rose, T.; Ahmed, A.; Kim, J.; Jiang, K.; Balagurunathan, Y.; Qi, J.; Farah, P.T.; et al. Multiphase Computed Tomography Radiomics of Pancreatic Intraductal Papillary Mucinous Neoplasms to Predict Malignancy. World J. Gastroenterol. 2020, 26, 3458–3471. [Google Scholar] [CrossRef] [PubMed]
- Flammia, F.; Innocenti, T.; Galluzzo, A.; Danti, G.; Chiti, G.; Grazzini, G.; Bettarini, S.; Tortoli, P.; Busoni, S.; Dragoni, G.; et al. Branch Duct-Intraductal Papillary Mucinous Neoplasms (BD-IPMNs): An MRI-Based Radiomic Model to Determine the Malignant Degeneration Potential. Radiol. Med. 2023, 128, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, G.; Mori, M.; Panzeri, M.M.; Barbera, M.; Palumbo, D.; Sini, C.; Muffatti, F.; Andreasi, V.; Steidler, S.; Doglioni, C.; et al. CT-Derived Radiomic Features to Discriminate Histologic Characteristics of Pancreatic Neuroendocrine Tumors. Radiol. Med. 2021, 126, 745–760. [Google Scholar] [CrossRef]
- Tikhonova, V.S.; Karmazanovsky, G.G.; Kondratyev, E.V.; Gruzdev, I.S.; Mikhaylyuk, K.A.; Sinelnikov, M.Y.; Revishvili, A.S. Radiomics Model–Based Algorithm for Preoperative Prediction of Pancreatic Ductal Adenocarcinoma Grade. Eur. Radiol. 2022, 33, 1152–1161. [Google Scholar] [CrossRef]
- Zhang, H.; Meng, Y.; Li, Q.; Yu, J.; Liu, F.; Fang, X.; Li, J.; Feng, X.; Zhou, J.; Zhu, M.; et al. Two Nomograms for Differentiating Mass-Forming Chronic Pancreatitis from Pancreatic Ductal Adenocarcinoma in Patients with Chronic Pancreatitis. Eur. Radiol. 2022, 32, 6336–6347. [Google Scholar] [CrossRef]
- Kim, D.W.; Kim, H.J.; Kim, K.W.; Byun, J.H.; Song, K.B.; Kim, J.H.; Hong, S.-M. Neuroendocrine Neoplasms of the Pancreas at Dynamic Enhanced CT: Comparison between Grade 3 Neuroendocrine Carcinoma and Grade 1/2 Neuroendocrine Tumour. Eur. Radiol. 2015, 25, 1375–1383. [Google Scholar] [CrossRef]
- Li, Q.; Zhou, Z.; Chen, Y.; Yu, J.; Zhang, H.; Meng, Y.; Zhu, M.; Li, N.; Zhou, J.; Liu, F.; et al. Fully Automated Magnetic Resonance Imaging-Based Radiomics Analysis for Differentiating Pancreatic Adenosquamous Carcinoma from Pancreatic Ductal Adenocarcinoma. Abdom. Radiol. 2023, 48, 2074–2084. [Google Scholar] [CrossRef]
- Chu, L.C.; Park, S.; Soleimani, S.; Fouladi, D.F.; Shayesteh, S.; He, J.; Javed, A.A.; Wolfgang, C.L.; Vogelstein, B.; Kinzler, K.W.; et al. Classification of Pancreatic Cystic Neoplasms Using Radiomic Feature Analysis Is Equivalent to an Experienced Academic Radiologist: A Step toward Computer-Augmented Diagnostics for Radiologists. Abdom. Radiol. 2022, 47, 4139–4150. [Google Scholar] [CrossRef]
- Yang, R.; Chen, Y.; Sa, G.; Li, K.; Hu, H.; Zhou, J.; Guan, Q.; Chen, F. CT Classification Model of Pancreatic Serous Cystic Neoplasms and Mucinous Cystic Neoplasms Based on a Deep Neural Network. Abdom. Radiol. 2022, 47, 232–241. [Google Scholar] [CrossRef]
- Chen, S.; Ren, S.; Guo, K.; Daniels, M.J.; Wang, Z.; Chen, R. Preoperative Differentiation of Serous Cystic Neoplasms from Mucin-Producing Pancreatic Cystic Neoplasms Using a CT-Based Radiomics Nomogram. Abdom. Radiol. 2021, 46, 2637–2646. [Google Scholar] [CrossRef] [PubMed]
- Bian, Y.; Li, J.; Cao, K.; Fang, X.; Jiang, H.; Ma, C.; Jin, G.; Lu, J.; Wang, L. Magnetic Resonance Imaging Radiomic Analysis Can Preoperatively Predict G1 and G2/3 Grades in Patients with NF-PNETs. Abdom. Radiol. 2021, 46, 667–680. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Zhao, R.; Zhang, J.; Guo, K.; Gu, X.; Duan, S.; Wang, Z.; Chen, R. Diagnostic Accuracy of Unenhanced CT Texture Analysis to Differentiate Mass-Forming Pancreatitis from Pancreatic Ductal Adenocarcinoma. Abdom. Radiol. 2020, 45, 1524–1533. [Google Scholar] [CrossRef] [PubMed]
- van der Pol, C.B.; Lee, S.; Tsai, S.; Larocque, N.; Alayed, A.; Williams, P.; Schieda, N. Differentiation of Pancreatic Neuroendocrine Tumors from Pancreas Renal Cell Carcinoma Metastases on CT Using Qualitative and Quantitative Features. Abdom. Radiol. 2019, 44, 992–999. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, J.; He, J.; Xu, S. Preoperative Differentiation of Pancreatic Cystic Neoplasm Subtypes on Computed Tomography Radiomics. Quant. Imaging Med. Surg. 2023, 13, 6395–6411. [Google Scholar] [CrossRef]
- Chang, N.; Cui, L.; Luo, Y.; Chang, Z.; Yu, B.; Liu, Z. Development and Multicenter Validation of a CT-Based Radiomics Signature for Discriminating Histological Grades of Pancreatic Ductal Adenocarcinoma. Quant. Imaging Med. Surg. 2020, 10, 692–702. [Google Scholar] [CrossRef]
- Zhu, M.; Xu, C.; Yu, J.; Wu, Y.; Li, C.; Zhang, M.; Jin, Z.; Li, Z. Differentiation of Pancreatic Cancer and Chronic Pancreatitis Using Computer-Aided Diagnosis of Endoscopic Ultrasound (EUS) Images: A Diagnostic Test. PLoS ONE 2013, 8, e63820. [Google Scholar] [CrossRef]
- Săftoiu, A.; Vilmann, P.; Gorunescu, F.; Janssen, J.; Hocke, M.; Larsen, M.; Iglesias–Garcia, J.; Arcidiacono, P.; Will, U.; Giovannini, M.; et al. Efficacy of an Artificial Neural Network–Based Approach to Endoscopic Ultrasound Elastography in Diagnosis of Focal Pancreatic Masses. Clin. Gastroenterol. Hepatol. 2012, 10, 84–90.e1. [Google Scholar] [CrossRef] [PubMed]
- Kang, T.W.; Kim, S.H.; Lee, J.; Kim, A.Y.; Jang, K.M.; Choi, D.; Kim, M.J. Differentiation between Pancreatic Metastases from Renal Cell Carcinoma and Hypervascular Neuroendocrine Tumour: Use of Relative Percentage Washout Value and Its Clinical Implication. Eur. J. Radiol. 2015, 84, 2089–2096. [Google Scholar] [CrossRef] [PubMed]
- Hanania, A.N.; Bantis, L.E.; Feng, Z.; Wang, H.; Tamm, E.P.; Katz, M.H.; Maitra, A.; Koay, E.J. Quantitative Imaging to Evaluate Malignant Potential of IPMNs. Oncotarget 2016, 7, 85776–85784. [Google Scholar] [CrossRef] [PubMed]
- Salanitri, F.P.; Bellitto, G.; Palazzo, S.; Irmakci, I.; Wallace, M.; Bolan, C.; Engels, M.; Hoogenboom, S.; Aldinucci, M.; Bagci, U.; et al. Neural Transformers for Intraductal Papillary Mucosal Neoplasms (IPMN) Classification in MRI Images. In Proceedings of the 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Scotland, UK, 11–15 July 2022; pp. 475–479. [Google Scholar]
- Gai, T.; Thai, T.; Jones, M.; Jo, J.; Zheng, B. Applying a Radiomics-Based CAD Scheme to Classify between Malignant and Benign Pancreatic Tumors Using CT Images. J. X-Ray Sci. Technol. 2022, 30, 377–388. [Google Scholar] [CrossRef]
- Pawlik, T.M.; Laheru, D.; Hruban, R.H.; Coleman, J.; Wolfgang, C.L.; Campbell, K.; Ali, S.; Fishman, E.K.; Schulick, R.D.; Herman, J.M. Evaluating the Impact of a Single-Day Multidisciplinary Clinic on the Management of Pancreatic Cancer. Ann. Surg. Oncol. 2008, 15, 2081–2088. [Google Scholar] [CrossRef]
- Chakraborty, J.; Midya, A.; Gazit, L.; Attiyeh, M.; Langdon-Embry, L.; Allen, P.J.; Do, R.K.G.; Simpson, A.L. CT Radiomics to Predict High-risk Intraductal Papillary Mucinous Neoplasms of the Pancreas. Med. Phys. 2018, 45, 5019–5029. [Google Scholar] [CrossRef]
- Zhang, T.; Xiang, Y.; Wang, H.; Yun, H.; Liu, Y.; Wang, X.; Zhang, H. Radiomics Combined with Multiple Machine Learning Algorithms in Differentiating Pancreatic Ductal Adenocarcinoma from Pancreatic Neuroendocrine Tumor: More Hands Produce a Stronger Flame. J. Clin. Med. 2022, 11, 6789. [Google Scholar] [CrossRef]
- Ma, X.; Wang, Y.-R.; Zhuo, L.-Y.; Yin, X.-P.; Ren, J.-L.; Li, C.-Y.; Xing, L.-H.; Zheng, T.-T. Retrospective Analysis of the Value of Enhanced CT Radiomics Analysis in the Differential Diagnosis Between Pancreatic Cancer and Chronic Pancreatitis. Int. J. Gen. Med. 2022, 15, 233–241. [Google Scholar] [CrossRef]
- Vaiyapuri, T.; Dutta, A.K.; Punithavathi, I.S.H.; Duraipandy, P.; Alotaibi, S.S.; Alsolai, H.; Mohamed, A.; Mahgoub, H. Intelligent Deep-Learning-Enabled Decision-Making Medical System for Pancreatic Tumor Classification on CT Images. Healthcare 2022, 10, 677. [Google Scholar] [CrossRef]
- Wang, X.; Qiu, J.-J.; Tan, C.-L.; Chen, Y.-H.; Tan, Q.-Q.; Ren, S.-J.; Yang, F.; Yao, W.-Q.; Cao, D.; Ke, N.-W.; et al. Development and Validation of a Novel Radiomics-Based Nomogram With Machine Learning to Preoperatively Predict Histologic Grade in Pancreatic Neuroendocrine Tumors. Front. Oncol. 2022, 12, 843376. [Google Scholar] [CrossRef]
- Shi, Y.-J.; Zhu, H.-T.; Liu, Y.-L.; Wei, Y.-Y.; Qin, X.-B.; Zhang, X.-Y.; Li, X.-T.; Sun, Y.-S. Radiomics Analysis Based on Diffusion Kurtosis Imaging and T2 Weighted Imaging for Differentiation of Pancreatic Neuroendocrine Tumors From Solid Pseudopapillary Tumors. Front. Oncol. 2020, 10, 1624. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Zhang, J.; Chen, J.; Cui, W.; Zhao, R.; Qiu, W.; Duan, S.; Chen, R.; Chen, X.; Wang, Z. Evaluation of Texture Analysis for the Differential Diagnosis of Mass-Forming Pancreatitis From Pancreatic Ductal Adenocarcinoma on Contrast-Enhanced CT Images. Front. Oncol. 2019, 9, 1171. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Guo, X.; Ou, X.; Zhang, W.; Ma, X. Discrimination of Pancreatic Serous Cystadenomas From Mucinous Cystadenomas With CT Textural Features: Based on Machine Learning. Front. Oncol. 2019, 9, 494. [Google Scholar] [CrossRef]
- Li, H.; Shi, K.; Reichert, M.; Lin, K.; Tselousov, N.; Braren, R.; Fu, D.; Schmid, R.; Li, J.; Menze, B. Differential Diagnosis for Pancreatic Cysts in CT Scans Using Densely-Connected Convolutional Networks. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 2095–2098. [Google Scholar]
- Bevilacqua, A.; Calabrò, D.; Malavasi, S.; Ricci, C.; Casadei, R.; Campana, D.; Baiocco, S.; Fanti, S.; Ambrosini, V. A [68Ga]Ga-DOTANOC PET/CT Radiomic Model for Non-Invasive Prediction of Tumour Grade in Pancreatic Neuroendocrine Tumours. Diagnostics 2021, 11, 870. [Google Scholar] [CrossRef]
- Gu, D.; Hu, Y.; Ding, H.; Wei, J.; Chen, K.; Liu, H.; Zeng, M.; Tian, J. CT Radiomics May Predict the Grade of Pancreatic Neuroendocrine Tumors: A Multicenter Study. Eur. Radiol. 2019, 29, 6880–6890. [Google Scholar] [CrossRef]
- Kuwahara, T.; Hara, K.; Mizuno, N.; Okuno, N.; Matsumoto, S.; Obata, M.; Kurita, Y.; Koda, H.; Toriyama, K.; Onishi, S.; et al. Usefulness of Deep Learning Analysis for the Diagnosis of Malignancy in Intraductal Papillary Mucinous Neoplasms of the Pancreas. Clin. Transl. Gastroenterol. 2019, 10, e00045. [Google Scholar] [CrossRef]
- Tobaly, D.; Santinha, J.; Sartoris, R.; Dioguardi Burgio, M.; Matos, C.; Cros, J.; Couvelard, A.; Rebours, V.; Sauvanet, A.; Ronot, M.; et al. CT-Based Radiomics Analysis to Predict Malignancy in Patients with Intraductal Papillary Mucinous Neoplasm (IPMN) of the Pancreas. Cancers 2020, 12, 3089. [Google Scholar] [CrossRef]
- Li, J.; Lu, J.; Liang, P.; Li, A.; Hu, Y.; Shen, Y.; Hu, D.; Li, Z. Differentiation of Atypical Pancreatic Neuroendocrine Tumors from Pancreatic Ductal Adenocarcinomas: Using Whole-tumor CT Texture Analysis as Quantitative Biomarkers. Cancer Med. 2018, 7, 4924–4931. [Google Scholar] [CrossRef]
- Hernandez-Barco, Y.G.; Daye, D.; Fernandez-del Castillo, C.F.; Parker, R.F.; Casey, B.W.; Warshaw, A.L.; Ferrone, C.R.; Lillemoe, K.D.; Qadan, M. IPMN-LEARN: A Linear Support Vector Machine Learning Model for Predicting Low-Grade Intraductal Papillary Mucinous Neoplasms. Ann. Hepato-Biliary-Pancreat. Surg. 2023, 27, 195–200. [Google Scholar] [CrossRef]
- Cui, S.; Tang, T.; Su, Q.; Wang, Y.; Shu, Z.; Yang, W.; Gong, X. Radiomic Nomogram Based on MRI to Predict Grade of Branching Type Intraductal Papillary Mucinous Neoplasms of the Pancreas: A Multicenter Study. Cancer Imaging 2021, 21, 26. [Google Scholar] [CrossRef]
- Guo, C.; Zhuge, X.; Wang, Q.; Xiao, W.; Wang, Z.; Wang, Z.; Feng, Z.; Chen, X. The Differentiation of Pancreatic Neuroendocrine Carcinoma from Pancreatic Ductal Adenocarcinoma: The Values of CT Imaging Features and Texture Analysis. Cancer Imaging 2018, 18, 37. [Google Scholar] [CrossRef]
- Tong, T.; Gu, J.; Xu, D.; Song, L.; Zhao, Q.; Cheng, F.; Yuan, Z.; Tian, S.; Yang, X.; Tian, J.; et al. Deep Learning Radiomics Based on Contrast-Enhanced Ultrasound Images for Assisted Diagnosis of Pancreatic Ductal Adenocarcinoma and Chronic Pancreatitis. BMC Med. 2022, 20, 74. [Google Scholar] [CrossRef]
- Liang, W.; Tian, W.; Wang, Y.; Wang, P.; Wang, Y.; Zhang, H.; Ruan, S.; Shao, J.; Zhang, X.; Huang, D.; et al. Classification Prediction of Pancreatic Cystic Neoplasms Based on Radiomics Deep Learning Models. BMC Cancer 2022, 22, 1237. [Google Scholar] [CrossRef]
- Korfiatis, P.; Suman, G.; Patnam, N.G.; Trivedi, K.H.; Karbhari, A.; Mukherjee, S.; Cook, C.; Klug, J.R.; Patra, A.; Khasawneh, H.; et al. Automated Artificial Intelligence Model Trained on a Large Data Set Can Detect Pancreas Cancer on Diagnostic Computed Tomography Scans as Well as Visually Occult Preinvasive Cancer on Prediagnostic Computed Tomography Scans. Gastroenterology 2023, 165, 1533–1546.e4. [Google Scholar] [CrossRef]
- Alizadeh Savareh, B.; Asadzadeh Aghdaie, H.; Behmanesh, A.; Bashiri, A.; Sadeghi, A.; Zali, M.; Shams, R. A Machine Learning Approach Identified a Diagnostic Model for Pancreatic Cancer through Using Circulating MicroRNA Signatures. Pancreatology 2020, 20, 1195–1204. [Google Scholar] [CrossRef]
- D’Onofrio, M.; Tedesco, G.; Cardobi, N.; De Robertis, R.; Sarno, A.; Capelli, P.; Martini, P.T.; Giannotti, G.; Beleù, A.; Marchegiani, G.; et al. Magnetic Resonance (MR) for Mural Nodule Detection Studying Intraductal Papillary Mucinous Neoplasms (IPMN) of Pancreas: Imaging-Pathologic Correlation. Pancreatology 2021, 21, 180–187. [Google Scholar] [CrossRef]
- Xia, Y.; Yu, Q.; Chu, L.; Kawamoto, S.; Park, S.; Liu, F.; Chen, J.; Zhu, Z.; Li, B.; Zhou, Z.; et al. The FELIX Project: Deep Networks To Detect Pancreatic Neoplasms. medRxiv 2022. [Google Scholar] [CrossRef]
- Chen, J.; Xia, Y.; Yao, J.; Yan, K.; Zhang, J.; Lu, L.; Wang, F.; Zhou, B.; Qiu, M.; Yu, Q.; et al. CancerUniT: Towards a Single Unified Model for Effective Detection, Segmentation, and Diagnosis of Eight Major Cancers Using a Large Collection of CT Scans. arXiv 2023, arXiv:2301.12291. [Google Scholar]
- Zhang, Z.; Li, S.; Wang, Z.; Lu, Y. A Novel and Efficient Tumor Detection Framework for Pancreatic Cancer via CT Images. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 1160–1164. [Google Scholar]
- Chen, P.-T.; Chang, D.; Yen, H.; Liu, K.-L.; Huang, S.-Y.; Roth, H.; Wu, M.-S.; Liao, W.-C.; Wang, W. Radiomic Features at CT Can Distinguish Pancreatic Cancer from Noncancerous Pancreas. Radiol. Imaging Cancer 2021, 3, e210010. [Google Scholar] [CrossRef]
- Chen, P.-T.; Wu, T.; Wang, P.; Chang, D.; Liu, K.-L.; Wu, M.-S.; Roth, H.R.; Lee, P.-C.; Liao, W.-C.; Wang, W. Pancreatic Cancer Detection on CT Scans with Deep Learning: A Nationwide Population-Based Study. Radiology 2023, 306, 172–182. [Google Scholar] [CrossRef]
- Chu, L.C.; Park, S.; Kawamoto, S.; Fouladi, D.F.; Shayesteh, S.; Zinreich, E.S.; Graves, J.S.; Horton, K.M.; Hruban, R.H.; Yuille, A.L.; et al. Utility of CT Radiomics Features in Differentiation of Pancreatic Ductal Adenocarcinoma From Normal Pancreatic Tissue. Am. J. Roentgenol. 2019, 213, 349–357. [Google Scholar] [CrossRef]
- Liu, S.-L.; Li, S.; Guo, Y.-T.; Zhou, Y.-P.; Zhang, Z.-D.; Li, S.; Lu, Y. Establishment and Application of an Artificial Intelligence Diagnosis System for Pancreatic Cancer with a Faster Region-Based Convolutional Neural Network. Chin. Med. J. 2019, 132, 2795–2803. [Google Scholar] [CrossRef]
- Abel, L.; Wasserthal, J.; Weikert, T.; Sauter, A.W.; Nesic, I.; Obradovic, M.; Yang, S.; Manneck, S.; Glessgen, C.; Ospel, J.M.; et al. Automated Detection of Pancreatic Cystic Lesions on CT Using Deep Learning. Diagnostics 2021, 11, 901. [Google Scholar] [CrossRef]
- Ozkan, M.; Cakiroglu, M.; Kocaman, O.; Kurt, M.; Yilmaz, B.; Can, G.; Korkmaz, U.; Dandil, E.; Eksi, Z. Age-Based Computer-Aided Diagnosis Approach for Pancreatic Cancer on Endoscopic Ultrasound Images. Endosc. Ultrasound 2016, 5, 101. [Google Scholar] [CrossRef]
- Zhang, Z.-M.; Wang, J.-S.; Zulfiqar, H.; Lv, H.; Dao, F.-Y.; Lin, H. Early Diagnosis of Pancreatic Ductal Adenocarcinoma by Combining Relative Expression Orderings With Machine-Learning Method. Front. Cell Dev. Biol. 2020, 8, 582864. [Google Scholar] [CrossRef]
- Deng, Y.; Ming, B.; Zhou, T.; Wu, J.; Chen, Y.; Liu, P.; Zhang, J.; Zhang, S.; Chen, T.; Zhang, X.-M. Radiomics Model Based on MR Images to Discriminate Pancreatic Ductal Adenocarcinoma and Mass-Forming Chronic Pancreatitis Lesions. Front. Oncol. 2021, 11, 620981. [Google Scholar] [CrossRef]
- Javed, S.; Qureshi, T.A.; Gaddam, S.; Wang, L.; Azab, L.; Wachsman, A.M.; Chen, W.; Asadpour, V.; Jeon, C.Y.; Wu, B.; et al. Risk Prediction of Pancreatic Cancer Using AI Analysis of Pancreatic Subregions in Computed Tomography Images. Front. Oncol. 2022, 12, 1007990. [Google Scholar] [CrossRef]
- Qureshi, T.A.; Gaddam, S.; Wachsman, A.M.; Wang, L.; Azab, L.; Asadpour, V.; Chen, W.; Xie, Y.; Wu, B.; Pandol, S.J.; et al. Predicting Pancreatic Ductal Adenocarcinoma Using Artificial Intelligence Analysis of Pre-Diagnostic Computed Tomography Images. Cancer Biomark. 2022, 33, 211–217. [Google Scholar] [CrossRef]
- Park, H.J.; Shin, K.; You, M.-W.; Kyung, S.-G.; Kim, S.Y.; Park, S.H.; Byun, J.H.; Kim, N.; Kim, H.J. Deep Learning–Based Detection of Solid and Cystic Pancreatic Neoplasms at Contrast-Enhanced CT. Radiology 2023, 306, 140–149. [Google Scholar] [CrossRef]
- Mukherjee, S.; Patra, A.; Khasawneh, H.; Korfiatis, P.; Rajamohan, N.; Suman, G.; Majumder, S.; Panda, A.; Johnson, M.P.; Larson, N.B.; et al. Radiomics-Based Machine-Learning Models Can Detect Pancreatic Cancer on Prediagnostic Computed Tomography Scans at a Substantial Lead Time Before Clinical Diagnosis. Gastroenterology 2022, 163, 1435–1446.e3. [Google Scholar] [CrossRef]
- Chen, W.; Zhou, Y.; Asadpour, V.; Parker, R.A.; Puttock, E.J.; Lustigova, E.; Wu, B.U. Quantitative Radiomic Features From Computed Tomography Can Predict Pancreatic Cancer up to 36 Months Before Diagnosis. Clin. Transl. Gastroenterol. 2023, 14, e00548. [Google Scholar] [CrossRef]
- Frøkjær, J.B.; Lisitskaya, M.V.; Jørgensen, A.S.; Østergaard, L.R.; Hansen, T.M.; Drewes, A.M.; Olesen, S.S. Pancreatic Magnetic Resonance Imaging Texture Analysis in Chronic Pancreatitis: A Feasibility and Validation Study. Abdom. Radiol. 2020, 45, 1497–1506. [Google Scholar] [CrossRef] [PubMed]
- Gonoi, W.; Hayashi, T.Y.; Okuma, H.; Akahane, M.; Nakai, Y.; Mizuno, S.; Tateishi, R.; Isayama, H.; Koike, K.; Ohtomo, K. Development of Pancreatic Cancer Is Predictable Well in Advance Using Contrast-Enhanced CT: A Case–Cohort Study. Eur. Radiol. 2017, 27, 4941–4950. [Google Scholar] [CrossRef] [PubMed]
- Si, K.; Xue, Y.; Yu, X.; Zhu, X.; Li, Q.; Gong, W.; Liang, T.; Duan, S. Fully End-to-End Deep-Learning-Based Diagnosis of Pancreatic Tumors. Theranostics 2021, 11, 1982–1990. [Google Scholar] [CrossRef]
- Ma, H.; Liu, Z.-X.; Zhang, J.-J.; Wu, F.-T.; Xu, C.-F.; Shen, Z.; Yu, C.-H.; Li, Y.-M. Construction of a Convolutional Neural Network Classifier Developed by Computed Tomography Images for Pancreatic Cancer Diagnosis. World J. Gastroenterol. 2020, 26, 5156–5168. [Google Scholar] [CrossRef]
- Hsieh, M.H.; Sun, L.-M.; Lin, C.-L.; Hsieh, M.-J.; Hsu, C.-Y.; Kao, C.-H. Development of a Prediction Model for Pancreatic Cancer in Patients with Type 2 Diabetes Using Logistic Regression and Artificial Neural Network Models. Cancer Manag. Res. 2018, 10, 6317–6324. [Google Scholar] [CrossRef]
- Muhammad, W.; Hart, G.R.; Nartowt, B.; Farrell, J.J.; Johung, K.; Liang, Y.; Deng, J. Pancreatic Cancer Prediction Through an Artificial Neural Network. Front. Artif. Intell. 2019, 2, 2. [Google Scholar] [CrossRef]
- Boursi, B.; Finkelman, B.; Giantonio, B.J.; Haynes, K.; Rustgi, A.K.; Rhim, A.D.; Mamtani, R.; Yang, Y.-X. A Clinical Prediction Model to Assess Risk for Pancreatic Cancer Among Patients With New-Onset Diabetes. Gastroenterology 2017, 152, 840–850.e3. [Google Scholar] [CrossRef]
- Appelbaum, L.; Cambronero, J.P.; Stevens, J.P.; Horng, S.; Pollick, K.; Silva, G.; Haneuse, S.; Piatkowski, G.; Benhaga, N.; Duey, S.; et al. Development and Validation of a Pancreatic Cancer Risk Model for the General Population Using Electronic Health Records: An Observational Study. Eur. J. Cancer 2021, 143, 19–30. [Google Scholar] [CrossRef]
- Das, A.; Nguyen, C.C.; Li, F.; Li, B. Digital Image Analysis of EUS Images Accurately Differentiates Pancreatic Cancer from Chronic Pancreatitis and Normal Tissue. Gastrointest. Endosc. 2008, 67, 861–867. [Google Scholar] [CrossRef]
- Urman, J.M.; Herranz, J.M.; Uriarte, I.; Rullán, M.; Oyón, D.; González, B.; Fernandez-Urién, I.; Carrascosa, J.; Bolado, F.; Zabalza, L.; et al. Pilot Multi-Omic Analysis of Human Bile from Benign and Malignant Biliary Strictures: A Machine-Learning Approach. Cancers 2020, 12, 1644. [Google Scholar] [CrossRef]
- Liu, K.-L.; Wu, T.; Chen, P.-T.; Tsai, Y.M.; Roth, H.; Wu, M.-S.; Liao, W.-C.; Wang, W. Deep Learning to Distinguish Pancreatic Cancer Tissue from Non-Cancerous Pancreatic Tissue: A Retrospective Study with Cross-Racial External Validation. Lancet Digit. Health 2020, 2, e303–e313. [Google Scholar] [CrossRef]
- Săftoiu, A.; Vilmann, P.; Gorunescu, F.; Gheonea, D.I.; Gorunescu, M.; Ciurea, T.; Popescu, G.L.; Iordache, A.; Hassan, H.; Iordache, S. Neural Network Analysis of Dynamic Sequences of EUS Elastography Used for the Differential Diagnosis of Chronic Pancreatitis and Pancreatic Cancer. Gastrointest. Endosc. 2008, 68, 1086–1094. [Google Scholar] [CrossRef]
- Cheng, S.-H.; Cheng, Y.-J.; Jin, Z.-Y.; Xue, H.-D. Unresectable Pancreatic Ductal Adenocarcinoma: Role of CT Quantitative Imaging Biomarkers for Predicting Outcomes of Patients Treated with Chemotherapy. Eur. J. Radiol. 2019, 113, 188–197. [Google Scholar] [CrossRef]
- Khalvati, F.; Zhang, Y.; Baig, S.; Lobo-Mueller, E.M.; Karanicolas, P.; Gallinger, S.; Haider, M.A. Prognostic Value of CT Radiomic Features in Resectable Pancreatic Ductal Adenocarcinoma. Sci. Rep. 2019, 9, 5449. [Google Scholar] [CrossRef] [PubMed]
- Yun, G.; Kim, Y.H.; Lee, Y.J.; Kim, B.; Hwang, J.-H.; Choi, D.J. Tumor Heterogeneity of Pancreas Head Cancer Assessed by CT Texture Analysis: Association with Survival Outcomes after Curative Resection. Sci. Rep. 2018, 8, 7226. [Google Scholar] [CrossRef] [PubMed]
- Eilaghi, A.; Baig, S.; Zhang, Y.; Zhang, J.; Karanicolas, P.; Gallinger, S.; Khalvati, F.; Haider, M.A. CT Texture Features Are Associated with Overall Survival in Pancreatic Ductal Adenocarcinoma–A Quantitative Analysis. BMC Med. Imaging 2017, 17, 38. [Google Scholar] [CrossRef] [PubMed]
- Miyata, T.; Hayashi, H.; Yamashita, Y.; Matsumura, K.; Nakao, Y.; Itoyama, R.; Yamao, T.; Tsukamoto, M.; Okabe, H.; Imai, K.; et al. Prognostic Value of the Preoperative Tumor Marker Index in Resected Pancreatic Ductal Adenocarcinoma: A Retrospective Single-Institution Study. Ann. Surg. Oncol. 2021, 28, 1572–1580. [Google Scholar] [CrossRef]
- Healy, G.M.; Salinas-Miranda, E.; Jain, R.; Dong, X.; Deniffel, D.; Borgida, A.; Hosni, A.; Ryan, D.T.; Njeze, N.; McGuire, A.; et al. Pre-Operative Radiomics Model for Prognostication in Resectable Pancreatic Adenocarcinoma with External Validation. Eur. Radiol. 2022, 32, 2492–2505. [Google Scholar] [CrossRef]
- Attiyeh, M.A.; Chakraborty, J.; Doussot, A.; Langdon-Embry, L.; Mainarich, S.; Gönen, M.; Balachandran, V.P.; D’Angelica, M.I.; DeMatteo, R.P.; Jarnagin, W.R.; et al. Survival Prediction in Pancreatic Ductal Adenocarcinoma by Quantitative Computed Tomography Image Analysis. Ann. Surg. Oncol. 2018, 25, 1034–1042. [Google Scholar] [CrossRef]
- Xie, T.; Wang, X.; Li, M.; Tong, T.; Yu, X.; Zhou, Z. Pancreatic Ductal Adenocarcinoma: A Radiomics Nomogram Outperforms Clinical Model and TNM Staging for Survival Estimation after Curative Resection. Eur. Radiol. 2020, 30, 2513–2524. [Google Scholar] [CrossRef]
- Kim, B.R.; Kim, J.H.; Ahn, S.J.; Joo, I.; Choi, S.-Y.; Park, S.J.; Han, J.K. CT Prediction of Resectability and Prognosis in Patients with Pancreatic Ductal Adenocarcinoma after Neoadjuvant Treatment Using Image Findings and Texture Analysis. Eur. Radiol. 2019, 29, 362–372. [Google Scholar] [CrossRef]
- Choi, M.H.; Lee, Y.J.; Yoon, S.B.; Choi, J.-I.; Jung, S.E.; Rha, S.E. MRI of Pancreatic Ductal Adenocarcinoma: Texture Analysis of T2-Weighted Images for Predicting Long-Term Outcome. Abdom. Radiol. 2019, 44, 122–130. [Google Scholar] [CrossRef]
- Parr, E.; Du, Q.; Zhang, C.; Lin, C.; Kamal, A.; McAlister, J.; Liang, X.; Bavitz, K.; Rux, G.; Hollingsworth, M.; et al. Radiomics-Based Outcome Prediction for Pancreatic Cancer Following Stereotactic Body Radiotherapy. Cancers 2020, 12, 1051. [Google Scholar] [CrossRef] [PubMed]
- Cozzi, L.; Comito, T.; Fogliata, A.; Franzese, C.; Franceschini, D.; Bonifacio, C.; Tozzi, A.; Di Brina, L.; Clerici, E.; Tomatis, S.; et al. Computed Tomography Based Radiomic Signature as Predictive of Survival and Local Control after Stereotactic Body Radiation Therapy in Pancreatic Carcinoma. PLoS ONE 2019, 14, e0210758. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Li, X.; Zhang, Q.; Guo, C.; Zhang, X.; Lao, M.; Shen, Y.; Xiao, W.; Ying, S.; Sun, K.; et al. Development of a Novel Multiparametric MRI Radiomic Nomogram for Preoperative Evaluation of Early Recurrence in Resectable Pancreatic Cancer. J. Magn. Reson. Imaging 2020, 52, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhao, Y.; Xu, J.; Shao, S.; Yu, D. Development and External Validation of a Radiomics Combined with Clinical Nomogram for Preoperative Prediction Prognosis of Resectable Pancreatic Ductal Adenocarcinoma Patients. Front. Oncol. 2022, 12, 1037672. [Google Scholar] [CrossRef]
- Chakraborty, J.; Langdon-Embry, L.; Cunanan, K.M.; Escalon, J.G.; Allen, P.J.; Lowery, M.A.; O’Reilly, E.M.; Gönen, M.; Do, R.G.; Simpson, A.L. Preliminary Study of Tumor Heterogeneity in Imaging Predicts Two Year Survival in Pancreatic Cancer Patients. PLoS ONE 2017, 12, e0188022. [Google Scholar] [CrossRef]
- Kaissis, G.; Ziegelmayer, S.; Lohöfer, F.; Algül, H.; Eiber, M.; Weichert, W.; Schmid, R.; Friess, H.; Rummeny, E.; Ankerst, D.; et al. A Machine Learning Model for the Prediction of Survival and Tumor Subtype in Pancreatic Ductal Adenocarcinoma from Preoperative Diffusion-Weighted Imaging. Eur. Radiol. Exp. 2019, 3, 41. [Google Scholar] [CrossRef]
- Zhang, Y.; Lobo-Mueller, E.M.; Karanicolas, P.; Gallinger, S.; Haider, M.A.; Khalvati, F. CNN-Based Survival Model for Pancreatic Ductal Adenocarcinoma in Medical Imaging. BMC Med. Imaging 2020, 20, 11. [Google Scholar] [CrossRef]
- Shi, H.; Wei, Y.; Cheng, S.; Lu, Z.; Zhang, K.; Jiang, K.; Xu, Q. Survival Prediction after Upfront Surgery in Patients with Pancreatic Ductal Adenocarcinoma: Radiomic, Clinic-Pathologic and Body Composition Analysis. Pancreatology 2021, 21, 731–737. [Google Scholar] [CrossRef]
- Rezaee, N.; Barbon, C.; Zaki, A.; He, J.; Salman, B.; Hruban, R.H.; Cameron, J.L.; Herman, J.M.; Ahuja, N.; Lennon, A.M.; et al. Intraductal Papillary Mucinous Neoplasm (IPMN) with High-Grade Dysplasia Is a Risk Factor for the Subsequent Development of Pancreatic Ductal Adenocarcinoma. HPB 2016, 18, 236–246. [Google Scholar] [CrossRef]
- Abraham, J.P.; Magee, D.; Cremolini, C.; Antoniotti, C.; Halbert, D.D.; Xiu, J.; Stafford, P.; Berry, D.A.; Oberley, M.J.; Shields, A.F.; et al. Clinical Validation of a Machine-Learning–Derived Signature Predictive of Outcomes from First-Line Oxaliplatin-Based Chemotherapy in Advanced Colorectal Cancer. Clin. Cancer Res. 2021, 27, 1174–1183. [Google Scholar] [CrossRef]
- Ciaravino, V.; Cardobi, N.; de Robertis, R.; Capelli, P.; Melisi, D.; Simionato, F.; Marchegiani, G.; Salvia, R.; D’Onofrio, M. CT Texture Analysis of Ductal Adenocarcinoma Downstaged After Chemotherapy. Anticancer Res. 2018, 38, 4889–4895. [Google Scholar] [CrossRef] [PubMed]
- Mu, W.; Liu, C.; Gao, F.; Qi, Y.; Lu, H.; Liu, Z.; Zhang, X.; Cai, X.; Ji, R.Y.; Hou, Y.; et al. Prediction of Clinically Relevant Pancreatico-Enteric Anastomotic Fistulas after Pancreatoduodenectomy Using Deep Learning of Preoperative Computed Tomography. Theranostics 2020, 10, 9779–9788. [Google Scholar] [CrossRef] [PubMed]
- Nasief, H.; Zheng, C.; Schott, D.; Hall, W.; Tsai, S.; Erickson, B.; Allen Li, X. A Machine Learning Based Delta-Radiomics Process for Early Prediction of Treatment Response of Pancreatic Cancer. npj Precis. Oncol. 2019, 3, 25. [Google Scholar] [CrossRef]
- Nasief, H.; Hall, W.; Zheng, C.; Tsai, S.; Wang, L.; Erickson, B.; Li, X.A. Improving Treatment Response Prediction for Chemoradiation Therapy of Pancreatic Cancer Using a Combination of Delta-Radiomics and the Clinical Biomarker CA19-9. Front. Oncol. 2020, 9, 1464. [Google Scholar] [CrossRef]
- McClaine, R.J.; Lowy, A.M.; Sussman, J.J.; Schmulewitz, N.; Grisell, D.L.; Ahmad, S.A. Neoadjuvant Therapy May Lead to Successful Surgical Resection and Improved Survival in Patients with Borderline Resectable Pancreatic Cancer. HPB 2010, 12, 73–79. [Google Scholar] [CrossRef]
- Yue, Y.; Osipov, A.; Fraass, B.; Sandler, H.; Zhang, X.; Nissen, N.; Hendifar, A.; Tuli, R. Identifying Prognostic Intratumor Heterogeneity Using Pre- and Post-Radiotherapy 18F-FDG PET Images for Pancreatic Cancer Patients. J. Gastrointest. Oncol. 2017, 8, 127–138. [Google Scholar] [CrossRef]
- Cassinotto, C.; Cortade, J.; Belleannée, G.; Lapuyade, B.; Terrebonne, E.; Vendrely, V.; Laurent, C.; Sa-Cunha, A. An Evaluation of the Accuracy of CT When Determining Resectability of Pancreatic Head Adenocarcinoma after Neoadjuvant Treatment. Eur. J. Radiol. 2013, 82, 589–593. [Google Scholar] [CrossRef]
- Chen, X.; Oshima, K.; Schott, D.; Wu, H.; Hall, W.; Song, Y.; Tao, Y.; Li, D.; Zheng, C.; Knechtges, P.; et al. Assessment of Treatment Response during Chemoradiation Therapy for Pancreatic Cancer Based on Quantitative Radiomic Analysis of Daily CTs: An Exploratory Study. PLoS ONE 2017, 12, e0178961. [Google Scholar] [CrossRef]
- Rigiroli, F.; Hoye, J.; Lerebours, R.; Lafata, K.J.; Li, C.; Meyer, M.; Lyu, P.; Ding, Y.; Schwartz, F.R.; Mettu, N.B.; et al. CT Radiomic Features of Superior Mesenteric Artery Involvement in Pancreatic Ductal Adenocarcinoma: A Pilot Study. Radiology 2021, 301, 610–622. [Google Scholar] [CrossRef]
- Bian, Y.; Jiang, H.; Ma, C.; Cao, K.; Fang, X.; Li, J.; Wang, L.; Zheng, J.; Lu, J. Performance of CT-Based Radiomics in Diagnosis of Superior Mesenteric Vein Resection Margin in Patients with Pancreatic Head Cancer. Abdom. Radiol. 2020, 45, 759–773. [Google Scholar] [CrossRef] [PubMed]
- Gregucci, F.; Fiorentino, A.; Mazzola, R.; Ricchetti, F.; Bonaparte, I.; Surgo, A.; Figlia, V.; Carbonara, R.; Caliandro, M.; Ciliberti, M.P.; et al. Radiomic Analysis to Predict Local Response in Locally Advanced Pancreatic Cancer Treated with Stereotactic Body Radiation Therapy. Radiol. Med. 2022, 127, 100–107. [Google Scholar] [CrossRef] [PubMed]
- McGovern, J.M.; Singhi, A.D.; Borhani, A.A.; Furlan, A.; McGrath, K.M.; Zeh, H.J.; Bahary, N.; Dasyam, A.K. CT Radiogenomic Characterization of the Alternative Lengthening of Telomeres Phenotype in Pancreatic Neuroendocrine Tumors. Am. J. Roentgenol. 2018, 211, 1020–1025. [Google Scholar] [CrossRef] [PubMed]
- Attiyeh, M.A.; Chakraborty, J.; McIntyre, C.A.; Kappagantula, R.; Chou, Y.; Askan, G.; Seier, K.; Gonen, M.; Basturk, O.; Balachandran, V.P.; et al. CT Radiomics Associations with Genotype and Stromal Content in Pancreatic Ductal Adenocarcinoma. Abdom. Radiol. 2019, 44, 3148–3157. [Google Scholar] [CrossRef]
- Lim, C.H.; Cho, Y.S.; Choi, J.Y.; Lee, K.-H.; Lee, J.K.; Min, J.H.; Hyun, S.H. Imaging Phenotype Using 18F-Fluorodeoxyglucose Positron Emission Tomography–Based Radiomics and Genetic Alterations of Pancreatic Ductal Adenocarcinoma. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 2113–2122. [Google Scholar] [CrossRef]
- Iwatate, Y.; Hoshino, I.; Yokota, H.; Ishige, F.; Itami, M.; Mori, Y.; Chiba, S.; Arimitsu, H.; Yanagibashi, H.; Nagase, H.; et al. Radiogenomics for Predicting P53 Status, PD-L1 Expression, and Prognosis with Machine Learning in Pancreatic Cancer. Br. J. Cancer 2020, 123, 1253–1261. [Google Scholar] [CrossRef]
- Tang, Y.; Su, Y.; Zheng, J.; Zhuo, M.; Qian, Q.; Shen, Q.; Lin, P.; Chen, Z. Radiogenomic Analysis for Predicting Lymph Node Metastasis and Molecular Annotation of Radiomic Features in Pancreatic Cancer. J. Transl. Med. 2024, 22, 690. [Google Scholar] [CrossRef]
- Hinzpeter, R.; Kulanthaivelu, R.; Kohan, A.; Avery, L.; Pham, N.-A.; Ortega, C.; Metser, U.; Haider, M.; Veit-Haibach, P. CT Radiomics and Whole Genome Sequencing in Patients with Pancreatic Ductal Adenocarcinoma: Predictive Radiogenomics Modeling. Cancers 2022, 14, 6224. [Google Scholar] [CrossRef]
- Iwatate, Y.; Yokota, H.; Hoshino, I.; Ishige, F.; Kuwayama, N.; Itami, M.; Mori, Y.; Chiba, S.; Arimitsu, H.; Yanagibashi, H.; et al. Machine Learning with Imaging Features to Predict the Expression of ITGAV, Which Is a Poor Prognostic Factor Derived from Transcriptome Analysis in Pancreatic Cancer. Int. J. Oncol. 2022, 60, 60. [Google Scholar] [CrossRef] [PubMed]
- Dmitriev, K.; Kaufman, A.E.; Javed, A.A.; Hruban, R.H.; Fishman, E.K.; Lennon, A.M.; Saltz, J.H. Classification of Pancreatic Cysts in Computed Tomography Images Using a Random Forest and Convolutional Neural Network Ensemble. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer International Publishing: Cham, Switzerland, 2017; pp. 150–158. [Google Scholar]
- Ziegelmayer, S.; Kaissis, G.; Harder, F.; Jungmann, F.; Müller, T.; Makowski, M.; Braren, R. Deep Convolutional Neural Network-Assisted Feature Extraction for Diagnostic Discrimination and Feature Visualization in Pancreatic Ductal Adenocarcinoma (PDAC) versus Autoimmune Pancreatitis (AIP). J. Clin. Med. 2020, 9, 4013. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lobo-Mueller, E.M.; Karanicolas, P.; Gallinger, S.; Haider, M.A.; Khalvati, F. Improving Prognostic Performance in Resectable Pancreatic Ductal Adenocarcinoma Using Radiomics and Deep Learning Features Fusion in CT Images. Sci. Rep. 2021, 11, 1378. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Jia, G.; Wu, Z.; Wang, T.; Wang, H.; Wei, K.; Cheng, C.; Liu, Z.; Zuo, C. A Multidomain Fusion Model of Radiomics and Deep Learning to Discriminate between PDAC and AIP Based on 18F-FDG PET/CT Images. Jpn J. Radiol. 2023, 41, 417–427. [Google Scholar] [CrossRef]
- Yao, L.; Zhang, Z.; Demir, U.; Keles, E.; Vendrami, C.; Agarunov, E.; Bolan, C.; Schoots, I.; Bruno, M.; Keswani, R.; et al. Radiomics Boosts Deep Learning Model for IPMN Classification. In International Workshop on Machine Learning in Medical Imaging; Springer Nature: Cham, Switzerland, 2024; pp. 134–143. [Google Scholar]
- Vétil, R.; Abi-Nader, C.; Bône, A.; Vullierme, M.-P.; Rohé, M.-M.; Gori, P.; Bloch, I. Non-Redundant Combination of Hand-Crafted and Deep Learning Radiomics: Application to the Early Detection of Pancreatic Cancer. In MICCAI Workshop on Cancer Prevention through Early Detection; Springer Nature: Cham, Switzerland, 2023; pp. 68–82. [Google Scholar]
- Amaral, M.J.; Oliveira, R.C.; Donato, P.; Tralhão, J.G. Pancreatic Cancer Biomarkers: Oncogenic Mutations, Tissue and Liquid Biopsies, and Radiomics—A Review. Dig. Dis. Sci. 2023, 68, 2811–2823. [Google Scholar] [CrossRef]
- Sollini, M.; Antunovic, L.; Chiti, A.; Kirienko, M. Towards Clinical Application of Image Mining: A Systematic Review on Artificial Intelligence and Radiomics. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 2656–2672. [Google Scholar] [CrossRef]


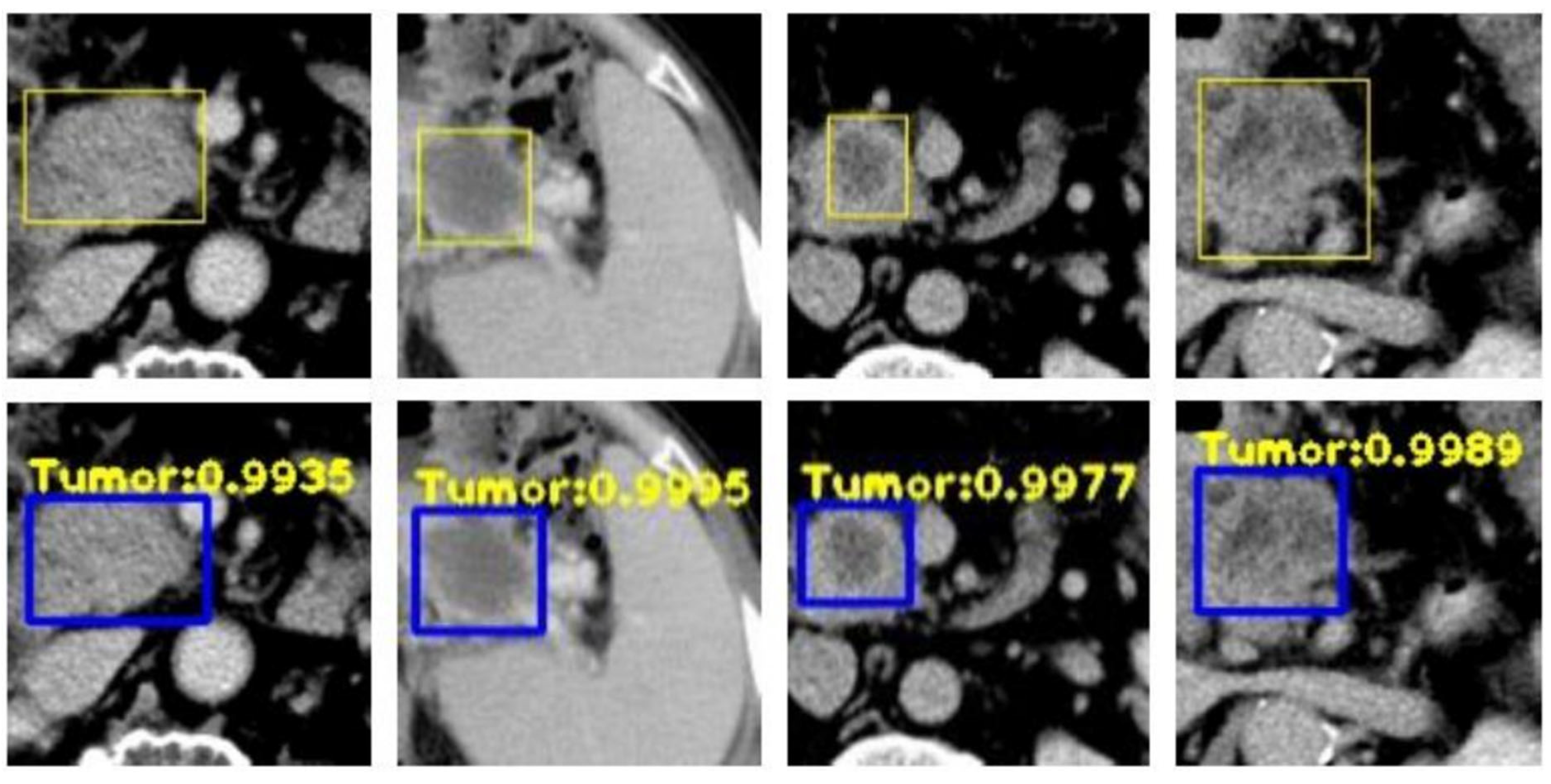
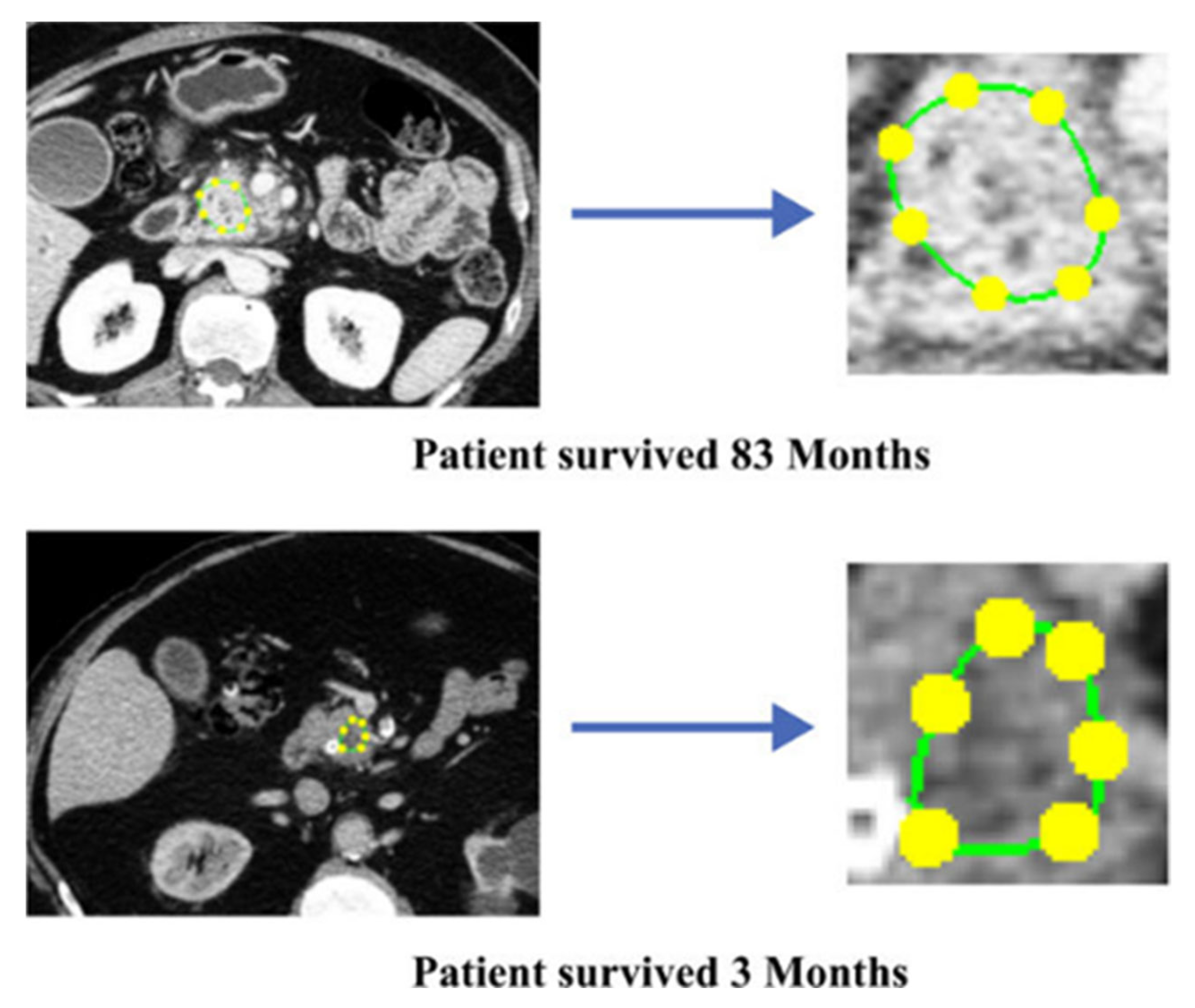




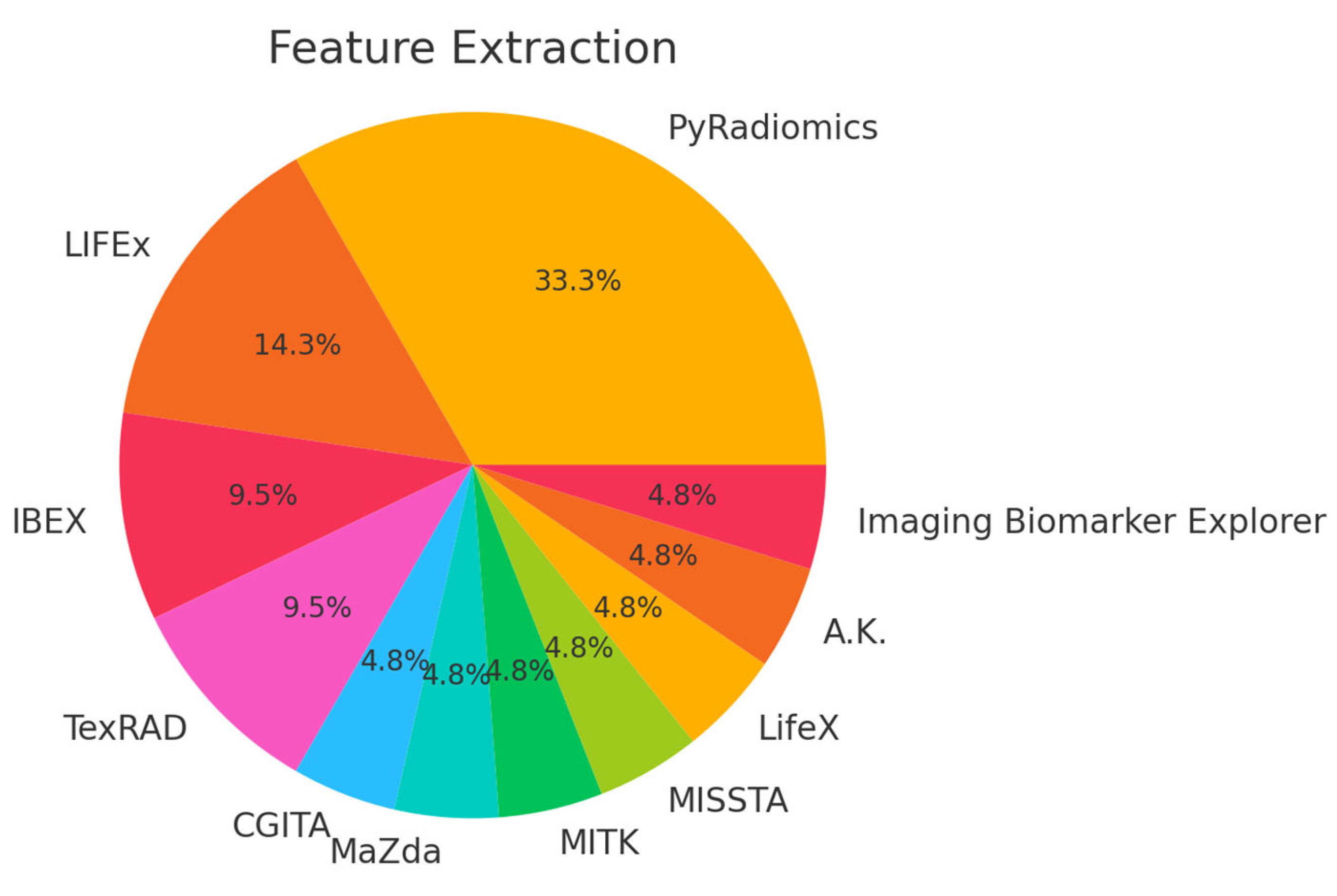

| Ref. | Dataset | Software/Tool/Prog. Lang. * | Features | ML/DL Model | Results |
|---|---|---|---|---|---|
| He et al. (2019) [17] | 147 patients (80 PDAC, 67 NF-pNET) | ITK-SNAP, MATLAB, R | 7 | SVM, Random Forest | Integrated model AUC = 0.884 |
| Xie et al. (2020) [18] | 57 patients (MCN vs. MaSCA) | MRIcron | 1942 | Logistic Regression | AUC = 0.994, Acc. = 98.2% |
| Mashayekhi et al. (2020) [19] | 56 patients (FAP, RAP, and CP) | N/A ** | 54 | IsoSVM | Acc. = 82.1%, AUC = 0.77–0.95 |
| Attiyeh et al. (2018) [20] | 103 patients (BD-IPMNs) | N/A | Quantitative imaging features | Random Forest | AUC = 0.79 |
| Liu et al. (2022) [21] | 102 patients (PC vs. MFCP) | Pyradiomics | 6 | LASSO Regression | AUC = 0.973 (train), 0.960 (validation) |
| Kulali et al. (2018) [22] | 30 patients (NF-pNETs and hepatic metastases) | N/A | N/A | N/A | Lower ADC values correlated with high Ki-67 index, MRI predictive tool |
| Park et al. (2020) [23] | 182 patients (89 AIP, 93 PDAC) | N/A | 431 | Random Forest | AUC = 0.975, Acc. = 95.2% |
| Wei et al. (2019) [24] | 260 patients (SCNs vs. PCNs) | N/A | 409 | SVM | AUC = 0.837 (validation) |
| Reinert et al. (2020) [25] | 95 patients (53 PDAC, 42 PNENs) | Pyradiomics | 92 | Logistic Regression | AUC = 0.79, Acc. = 75.8% |
| Polk et al. (2020) [26] | 51 patients (IPMNs) | Healthmyne | 39 | Logistic Regression | AUC = 0.93 (with ICG criteria) |
| Flammia et al. (2023) [27] | 50 patients (BD-IPMNs) | 3D Slicer | 107 | LASSO Regression | AUC = 0.80–0.99 |
| Benedetti et al. (2021) [28] | 39 patients (pancreatic neuroendocrine tumors) | CGITA (MATLAB) | 69 | N/A | Sphericity AUC = 0.79, tumor volume AUC = 0.79, and voxel-alignment AUC = 0.80–0.85 |
| Tikhonova et al. (2022) [29] | 91 patients (PDAC grading) | LifEx | 5 | LASSO Regression | AUC = 0.75 (grade ≥ 2), AUC = 0.66 (grade 3) |
| Zhang et al. (2022) [30] | 138 patients (MFCP vs. PDAC) | Pyradiomics | LASSO selected features | Logistic Regression | AUC = 0.91 (train), 0.93 (validation) |
| Kim et al. (2015) [31] | 167 lesions (161 patients, pancreatic neuroendocrine neoplasms) | SPSS 18 | N/A | N/A | Portal enhancement ratio < 1.1 achieved 92.3% sensitivity, 80.5% specificity |
| Li et al. (2023) [32] | 512 patients (PASC vs. PDAC) | Pyradiomics | N/A | LDA | AUC = 0.94 (validation), sensitivity = 67.57%, and specificity = 97.44% |
| Chu et al. (2022) [33] | 214 patients (PCNs) | N/A | 488 | Random Forest | AUC = 0.940 |
| Yang et al. (2022) [34] | 110 patients (SCNs vs. MCNs) | N/A | N/A | MMRF-ResNet | AUC = 0.98, Acc. = 92.69% |
| Chen et al. (2021) [35] | 89 patients (SCNs vs. PCNs) | N/A | 710 | Logistic Regression | AUC = 0.960 (train), 0.817 (validation) |
| Bian et al. (2021) [36] | 157 patients (NF-pNETs grading) | N/A | 7 | LASSO Regression | AUC = 0.775 |
| Ren et al. (2020) [37] | 109 patients (MFP vs. PDAC) | N/A | 396 | Random Forest | Acc. = 93.3%, sensitivity = 92.2%, and specificity = 94.2% |
| Van der Pol et al. (2019) [38] | 71 patients (PNETs vs. RCC metastases) | N/A | Entropy and tumor size | Logistic Regression | AUC = 0.77, sensitivity = 71.4%, and specificity = 79.1% |
| Zhang et al. (2023) [39] | 143 patients (PCNs subtypes) | N/A | 1218 | Random Forest | Acc. = 80.4% (train), 70.7% (test), and binary models AUC = 0.914–0.926 |
| Chang et al. (2020) [40] | 301 patients (PDAC grading) | IBEX | LASSO selected features | SVM | AUC = 0.961 (train), 0.910 (test), and 0.770 (external validation) |
| Zhu et al. (2013) [41] | 388 patients (PC vs. CP) | N/A | 105 | SVM | Acc. = 94.26%, sensitivity = 96.25%, and specificity = 93.38% |
| Săftoiu et al. (2012) [42] | 258 patients (pancreatic cancer vs. CP) | N/A | Hue histogram features | MLP Neural Network | AUC = 0.94, Acc. = 91.14% (train), and 84.27% (test) |
| Kang et al. (2015) [43] | 44 patients (pRCC vs. pNETs) | N/A | Relative Percentage Washout (RPW) | Threshold-based classification | Acc. = 83.8%, sensitivity = 83.8%, and specificity = 83.9% |
| Hanania et al. (2016) [44] | 53 patients (34 HG-IPMNs, 19 LG-IPMNs) | N/A | 360 | Logistic Regression | AUC = 0.96, sensitivity = 97%, and specificity = 88% |
| Proietto Salanitri et al. (2022) [45] | 139 patients (normal, LGD, HGD, and adenocarcinoma) | N4 Bias Correction, Gaussian Smoothing, and TensorFlow | N/A | Vision Transformer (ViT) | Acc. = 70%, precision = 67%, and recall = 64% |
| Gai et al. (2022) [46] | 77 patients (33 malignant, 44 benign) | MaZda | 1267, reduced to 12 | SVM | AUC = 0.750, sensitivity = 60.6%, specificity = 81.8%, and Acc. = 72.7% |
| Pawlik et al. (2008) [47] | 203 patients (multidisciplinary pancreatic cancer review) | N/A | N/A | N/A | Treatment plan changed in 23.6% of cases |
| Chakraborty et al. (2018) [48] | 103 patients (BD-IPMNs) | Scout Liver (Analogic Corp.) | Radiographically inspired (RiFs) + texture features | Random Forest | AUC = 0.81 (with clinical variables), AUC = 0.77 (radiomics alone) |
| Zhang et al. (2022) [49] | 238 patients (156 PDAC, 82 pNET) | LIFEx | 48 | Gradient Boosting Decision Tree (GBDT) + Random Forest | AUC = 0.971 (train), 0.930 (validation), sensitivity = 0.804, and specificity = 0.973 |
| Ma et al. (2022) [50] | 175 patients (151 PC, 24 CP) | MITK, PyRadiomics | 1037 | LASSO + Logistic Regression | AUC = 0.980, sensitivity = 94.7%, and specificity = 91.7% |
| Vaiyapuri et al. (2022) [51] | 500 CT images (250 tumor, 250 non-tumor) | TensorFlow | N/A | MobileNet + Autoencoder + Emperor Penguin Optimizer (EPO) | Acc. = 99.35%, sensitivity = 99.35%, and specificity = 98.84% |
| Wang et al. (2022) [52] | 139 patients (PNETs grading) | N/A | 1133 | SVM | AUC = 0.919 (train), 0.875 (validation) |
| Shi et al. (2020) [53] | 66 patients (31 PNETs, 35 SPTs) | ITK-SNAP | 195 | Logistic Regression | AUC = 0.97 (train), 0.86 (validation), sensitivity = 95%, and specificity = 91.67% |
| Ren et al. (2019) [54] | 109 patients (30 MFP, 79 PDAC) | AnalysisKit (GE Healthcare) | 396 | Logistic Regression | AUC = 0.98, sensitivity = 94%, and specificity = 92% |
| Yang et al. (2019) [55] | 78 patients (53 SCAs, 25 MCAs) | LIFEx | 22 (2 mm slices)/18 (5 mm slices) | Random Forest | AUC = 0.77 (train, 2 mm), 0.66 (validation, 2 mm), and 0.75 (validation, 5 mm) |
| Li et al. (2019) [56] | 206 patients (64 IPMNs, 35 MCNs, 66 SCNs, and 41 SPTs) | TensorFlow | N/A | DenseNet CNN | Acc. = 72.8% (outperforms manual reading at 48.1%) |
| Bevilacqua et al. (2021) [57] | 51 patients (PanNETs, G1 vs. G2) | ImageJ 1.53f | N/A | Logistic Regression | AUC = 0.90 (best model), sensitivity = 88%, and specificity = 89% |
| Gu et al. (2019) [58] | 138 patients (PNETs, G1 vs. G2/3) | N/A | 853 | Random Forest | AUC = 0.974 (train), 0.902 (validation) |
| Kuwahara et al. (2019) [59] | 206 patients (50 for deep learning analysis, 3970 EUS images) | TensorFlow | N/A | ResNet50 CNN | AUC = 0.98, sensitivity = 95.7%, specificity = 92.6%, and Acc. = 94.0% |
| Tobaly et al. (2020) [60] | 408 patients (181 LGD, 128 HGD, and 99 invasive carcinoma) | MedSeg, PyRadiomics | 107 | LASSO + Logistic Regression | AUC = 0.84 (train), 0.71 (validation) |
| Li et al. (2018) [61] | 127 patients (50 PDAC, 77 pNET) | FireVoxel | Histogram-based texture features | Threshold-Based Classification | AUC = 0.887, sensitivity = 90%, and specificity = 80% |
| Hernandez-Barco et al. (2023) [62] | 575 patients (IPMN surgical cases) | N/A | 18 clinical and imaging variables | Linear SVM | AUC = 0.82, Acc. = 77.4%, sensitivity = 83%, and specificity = 72% |
| Cui et al. (2021) [63] | 202 patients (BD-IPMN grading) | ITK-SNAP, MITK | 1312 | LASSO + Logistic Regression | AUC = 0.903 (train), 0.884 (validation 1), and 0.876 (validation 2) |
| Guo et al. (2018) [64] | 42 patients (28 PDAC, 14 PNEC) | MATLAB | Contrast ratio + texture features | Threshold-Based Classification | AUC = 0.98–0.99 (contrast ratio), 0.71–0.72 (texture features) |
| Tong et al. (2022) [65] | 558 patients (PDAC vs. CP) | ResNet-50 (DL model) | N/A | Deep Learning (CNN) | AUC = 0.986 (train), 0.978 (internal validation), and 0.953 (external validation) |
| Liang et al. (2022) [66] | 193 patients (99 SCA, 55 MCA, and 39 IPMN) | ITK-SNAP | 1067 | SVM, CNN (Hybrid Model) | AUC = 0.916 (SCA), 0.973 (MCA vs. IPMN) |
| Ref. | Dataset | Software/Tool/Prog. Lang. * | Features | ML/DL Model | Results |
|---|---|---|---|---|---|
| Korfiatis et al. (2023) [67] | 696 PC, 1080 control CTs | TensorFlow 2.3.1 | N/A ** | Modified ResNet + Attention Modules | AUROC = 0.97, Acc. = 92% |
| Alizadeh Savareh et al. (2020) [68] | 671 miRNA profiles | MATLAB 2019 | N/A | ANN + PSO + NCA | Acc. = 93%, Sensitivity = 93%, and Specificity = 92% |
| D’Onofrio et al. (2021) [69] | 91 MRI scans | MeVisLab, MATLAB | ADC Histogram (Entropy-based) | N/A | Acc. = 89.01%, Sensitivity = 90.77%, and Specificity = 84.62% |
| Xia et al. (2023) [70] | 662 PDAC, 450 PanNETs, 458 cysts, and 846 normal | N/A | N/A | 3D U-Net | Sensitivity = 97%, Specificity = 99%, and DSC = 87% |
| Chen et al. (2023) [71] | 10,673 patients (8 cancers + 1055 controls) | nnUNet, CTLabler, and ITK-SNAP | N/A | CancerUniT Transformer | Sensitivity = 93.3%, Specificity = 81.7%, and DSC = 62.8% |
| Zhang et al. (2020) [72] | 2890 pancreatic CTs | TensorFlow | N/A | Faster R-CNN + AFPN | AUC = 0.9455, Acc. = 90.18%, Sensitivity = 83.76%, and Specificity = 91.79% |
| Chen et al. (2021) [73] | 436 PDAC, 479 control CTs | PyRadiomics | 88 features | XGBoost 2.1.0 | Acc. = 95.0%, Sensitivity = 94.7%, and Specificity = 95.4% |
| Chen et al. (2022) [74] | 546 PC, 733 control | TensorFlow | N/A | Ensemble CNNs | AUC = 0.96, Sensitivity = 89.9%, and Specificity = 95.9% |
| Chu et al. (2019) [75] | 190 PDAC, 190 controls | Velocity 3.2.0 | 478 features | Random Forest | Acc. = 99.2%, AUC = 99.9%, Sensitivity = 100%, and Specificity = 98.5% |
| Liu et al. (2019) [76] | 238 PC, 4385 CTs | N/A | N/A | Faster R-CNN + VGG16 | AUC = 0.9632 |
| Abel et al. (2021) [77] | 221 CTs, 543 cysts | SPSS Statistics, nnUNet | N/A | CNN (2-step nnU-Net) | Sensitivity = 78.8%, Specificity = 96.2% |
| Ozkan et al. (2016) [78] | 332 EUS images (202 PC, 130 non-PC) | MATLAB | 122 features | ANN | Acc. = 87.5%, Sensitivity = 83.3%, and Specificity = 93.3% |
| Zhang et al. (2020) [79] | 573 PDAC, 153 adjacent normal, 10 pancreatitis, and 74 normal | LibSVM v3.23 | N/A | SVM | Acc. = 98.77%, Sensitivity = 98.65%, and Specificity = 100% |
| Deng et al. (2021) [80] | 119 MRI scans (PDAC vs. MFCP) | IBEX | N/A | SVM | AUC = 0.997 (Training), 0.962 (Validation) |
| Javed et al. (2022) [81] | 108 CTs | ITK-SNAP | N/A | Naïve Bayes + RFE | Acc. = 89.3%, Sensitivity = 86%, and Specificity = 93% |
| Qureshi et al. (2022) [82] | 108 CTs (36 pre-diagnostic PDAC, 36 PC, and 36 control) | ITK-SNAP | 4000 features | Naïve Bayes | Acc. = 86% |
| Park et al. (2022) [83] | 852 training, 603 and 589 test patients | nnU-Net | N/A | 3D CNN | AUC = 0.91 |
| Mukherjee et al. (2022) [84] | 155 pre-diagnostic CTs, 265 normal | 3D Slicer, PyRadiomics | 88 features | SVM | Acc. = 92.2%, AUC = 0.98 |
| Chen et al. (2023) [85] | 227 non-CP, 70 CP | MATLAB | 111 features | SVM | AUC = 0.99 |
| Frøkjær et al. (2020) [86] | 77 CP, 22 controls | 3D Slicer | 851 features | Bayes Classifier | Acc. = 98%, Sensitivity = 97%, and Specificity = 100% |
| Gonoi et al. (2017) [87] | 9 PDAC, 103 controls | N/A | N/A | Kaplan–Meier survival analysis | Identified Early Imaging Markers |
| Si et al. (2021) [88] | 143,945 CT images (319 patients), 107,036 test images (347 patients) | TensorFlow | N/A | ResNet18 (pancreas detection), U-Net32 (segmentation), and ResNet34 (classification) | AUC = 0.871, Acc. = 82.7%, and F1-score = 88.5% |
| Ma et al. (2020) [89] | 7245 CT images (412 patients) | N/A | N/A | CNN | Acc. = 95.47% (Plain Scan), 95.76% (Arterial Phase), Sensitivity = 91.58%, and Specificity = 98.27% |
| Hsieh et al. (2018) [90] | 1,358,634 patients (3092 pancreatic cancer cases) | Python 3.7 (scikit-learn), TensorFlow | 22 clinical variables | Logistic Regression, ANN | AUC = 0.727 (LR), 0.605 (ANN), and F1-score = 0.997 |
| Muhammad et al. (2019) [91] | 800,114 respondents (NHIS and PLCO datasets), 898 pancreatic cancer cases | N/A | 18 personal health features | Artificial Neural Network (ANN) | AUC = 0.86 (Training), 0.85 (Testing), Sensitivity = 87.3%, and Specificity = 80.7% |
| Boursi et al. (2017) [92] | 109,385 new-onset diabetes patients (390 diagnosed with PDAC) | N/A | N/A | Logistic Regression | AUC = 0.82, Specificity = 94%, and Sensitivity = 44.7% |
| Appelbaum et al. (2021) [93] | 594 PDAC cases, 100,787 controls (training), 408 PDAC cases, and 160,185 controls (validation) | L2-regularized logistic regression, neural network | ICD codes, comorbidities, and medication history | Logistic Regression, Neural Network | AUC = 0.71 (training), 0.68 (validation) |
| Das et al. (2008) [94] | 110 normal pancreas, 99 CP, and 110 PC (EUS images) | ImageJ | 228 features reduced to 11 | Artificial Neural Network (ANN) | AUC = 0.93, Sensitivity = 93%, and Specificity = 92% |
| Urman et al. (2020) [95] | 129 bile samples (57 PDAC, 36 CCA, and 36 benign) | UHPLC-MS, HPLC-MS/MS | N/A | Neural Network | AUC = 1.00 |
| Liu et al. (2020) [96] | 370 PC, 320 controls | N/A | N/A | CNN | AUC = 1.00 (Local), AUC = 0.83 (External) |
| Săftoiu et al. (2008) [97] | 68 patients (32 PC, 11 CP, 22 normal, and 3 PNET) | ImageJ | 228 features reduced to 11 | Multilayer Perceptron (MLP) Neural Network | AUC = 0.932, Sensitivity = 91.4%, Specificity = 87.9%, and Acc. = 89.7% |
| Ref. | Dataset | Software/Tool/Prog. Lang. * | Features | ML/DL Model | Results |
|---|---|---|---|---|---|
| Cheng et al. (2019) [98] | 41 patients (unresectable PDAC, contrast-enhanced CT) | TexRAD | Mean intensity, entropy, skewness, kurtosis, and SD | None | Higher SD associated with longer OS (p = 0.04) |
| Khalvati et al. (2019) [99] | 98 patients (resectable PDAC, contrast-enhanced CT) | PyRadiomics v2.0.1 | 410 extracted, 277 robust | Cox proportional-hazards regression | HR = 1.56, p = 0.005 |
| Yun et al. (2018) [100] | 88 patients (pancreatic head cancer, contrast-enhanced CT) | In-house software | Histogram and GLCM texture features | None | Lower SD and contrast are associated with poor DFS |
| Eilaghi et al. (2017) [101] | 30 patients (resectable PDAC, contrast-enhanced CT) | MATLAB (R2015a) | 5 GLCM texture features | None | Dissimilarity (p = 0.045) and IDN (p = 0.046) significant for OS |
| Miyata et al. (2020) [102] | 183 patients (resected PDAC, tumor markers) | JMP v12 (SAS Institute) | None (clinical markers only) | None | High Pre-TI associated with worse OS (HR = 2.27, p < 0.0001) |
| Healy et al. (2022) [103] | 352 training, 215 validation (resectable PDAC, contrast-enhanced CT) | PyRadiomics v3.0 | IBSI-compliant radiomics features | LASSO Cox regression | C-index = 0.545 (radiomics), 0.497 (clinical) |
| Attiyeh et al. (2018) [104] | 161 patients (resectable PDAC, contrast-enhanced CT) | MATLAB (R2015a) | CT texture features | Cox proportional-hazards regression | C-index = 0.69 (radiomics), 0.74 (clinical) |
| Xie et al. (2020) [105] | 220 patients (resectable PDAC, contrast-enhanced CT) | R software | 300 radiomics features | LASSO regression | AUC = 0.87 (training), 0.85 (validation) |
| Kim et al. (2019) [106] | 45 patients (PDAC post-neoadjuvant therapy, contrast-enhanced CT) | MISSTA | GLCM texture features | None | Higher entropy (HR = 0.159, p = 0.005) predicted longer OS |
| Choi et al. (2019) [107] | 66 patients (PDAC, MRI T2-weighted imaging) | TexRAD | Histogram and GLCM features | None | Higher entropy (p = 0.002) correlated with worse OS |
| Parr et al. (2020) [108] | 74 patients (PDAC, SBRT, and contrast-enhanced CT) | 3D Slicer | 800+ radiomics features | None | Radiomics model outperformed clinical (C-index = 0.66) |
| Cozzi et al. (2019) [109] | 100 patients (PDAC, SBRT, and contrast-enhanced CT) | LifeX | Radiomics features | Cox regression | C-index = 0.73–0.75 for OS prediction |
| Tang et al. (2019) [110] | 303 patients (resectable PDAC, and MRI multiparametric) | ITK-SNAP, A.K. | 328 radiomics features | LASSO logistic regression | AUC = 0.87 (training), 0.85 (validation) |
| Wang et al. (2022) [111] | 184 patients (resectable PDAC, contrast-enhanced CT) | PyRadiomics | 1409 extracted, LASSO selected | Cox regression | C-index = 0.74 (radiomics), 0.68 (clinical) |
| Chakraborty et al. (2017) [112] | 35 patients (PDAC, contrast-enhanced CT) | MATLAB (R2015a) | 255 texture features | Naïve Bayes classifier | AUC = 0.90 (leave-one-out), 0.80 (3-fold CV), and Acc. = 82.86% |
| Kaissis et al. (2019) [113] | 102 training, 30 validation (PDAC, diffusion-weighted MRI) | PyRadiomics | ADC-based radiomic features | Random Forest | AUC = 0.90 (survival prediction), 89% acc. for tumor subtype classification |
| Zhang et al. (2020) [114] | 68 training, 30 validation (resectable PDAC, contrast-enhanced CT) | None | None | CNN (6-layer) | C-index = 0.651, IPA = 11.81% |
| Shi et al. (2021) [115] | 299 patients (resectable PDAC, contrast-enhanced CT) | A.K. (GE Healthcare), ITK-SNAP | 1409 extracted, LASSO selected | Cox regression | C-index = 0.74 (radiomics), 0.68 (clinical) |
| Rezaee et al. (2016) [116] | 616 patients (IPMN, pancreatic resection) | None | None | None | High-grade dysplasia linked to increased PDAC risk, median OS = 92 months |
| Ref. | Dataset | Software/Tool/Prog. Lang. * | Features | ML/DL Model | Results |
|---|---|---|---|---|---|
| Abraham et al. (2021) [117] | 517 patients (105 training, 412 validation, and 55 FOLFIRI control) | N/A ** | 67 gene signatures | Bayesian Regularization Neural Network | OS HR = 0.629 (p = 0.04) for FOLFOX, 0.483 (p = 0.02) for FOLFOXIRI |
| Ciaravino et al. (2018) [118] | 31 patients (17 downstaged, 14 progression) | MaZda | Histogram, texture, and kurtosis | N/A | Kurtosis change (p = 0.0046) is significant in responders |
| Mu et al. (2020) [119] | 583 patients (513 training, 70 validation) | 3D Slicer, Keras, Python | N/A | CNN | AUC = 0.85 (train), 0.81 (val), and 0.89 (test) |
| Nasief et al. (2019) [120] | 90 patients, 2520 daily CT scans | IBEX | 1300+ features | Bayesian Regularization Neural Network | AUC = 0.94 |
| Nasief et al. (2020) [121] | 24 patients (672 CT datasets) | IBEX | 1300+ features | Regression Model | C-index = 0.87, HR = 0.58 |
| McClaine et al. (2010) [122] | 29 patients (26 neoadjuvant, 12 resected) | N/A | N/A | N/A | Median survival: 15.5 months (unresected) vs. 23.3 months (resected), p = 0.015 |
| Yue et al. (2017) [123] | 26 PA patients (19 external-beam RT, 7 SBRT) | N/A | Texture features from PET | Lasso Regression, Cox Model | OS = 29.3 months (low-risk) vs. 17.7 months (high-risk) |
| Cassinotto et al. (2013) [124] | 80 patients (38 neoadjuvant) | N/A | N/A | N/A | CT acc. lower after neoadjuvant (58% vs. 83%, p = 0.039) |
| Chen et al. (2017) [125] | 20 patients, daily CT scans | N/A | Mean CT number, volume, and skewness | N/A | MCTN decrease (−4.7 HU, p < 0.001) correlated with response |
| Rigiroli et al. (2021) [126] | 194 PDAC patients (148 neoadjuvant) | Siemens SyngoVia Frontier Radiomics | 1695 features | Logistic Regression | AUC = 0.71, sensitivity = 62%, and specificity = 77% |
| Bian et al. (2020) [127] | 181 PDAC patients | N/A | 1029 features (portal phase CT) | Logistic Regression | AUC = 0.75, sensitivity = 64.8%, and specificity = 74% |
| Gregucci et al. (2022) [128] | 37 locally advanced PDAC patients | Imaging Biomarker Explorer | 27 radiomic features | Logistic Regression | AUC = 0.851 |
| Ref. | Dataset | Software/Tool/Prog. Lang. * | Features | ML/DL Model | Results |
|---|---|---|---|---|---|
| McGovern et al. (2018) [129] | 121 PanNET patients | N/A ** | Tumor size, shape, necrosis, vascular invasion, and pancreatic duct dilatation | Multivariate Logistic Regression | AUC = 0.58, p = 0.006 |
| Attiyeh et al. (2019) [130] | 35 PDAC patients | Scout Liver Software, MATLAB | 255 features (GLCM, RLM, LBP, FD, IH, and ACM) | Fuzzy Minimum-Redundancy-Maximum-Relevance (fMRMR) | R2 = 0.731, RMSE = 19.5 |
| Lim et al. (2020) [131] | 48 PDAC patients | MIM v6.4, MATLAB (CGITA toolbox) | 35 PET-based radiomic features | Logistic Regression | AUC = 0.806 (KRAS), 0.727 (SMAD4) |
| Iwatate et al. (2020) [132] | 107 PDAC patients | PyRadiomics v2.2.0 | 2074 features (early- and late-phase CT) | XGBoost | AUC = 0.795 (p53), 0.683 (PD-L1) |
| Tang et al. (2024) [133] | 205 patients (151 internal, 54 CPTAC-PDAC) | ITK-SNAP, PyRadiomics | 1239 features | StepGBM + Elastic Net | AUC = 0.84 (train), 0.85 (val) |
| Hinzpeter et al. (2022) [134] | 47 PDAC patients | LIFEx v6.30 | Multiple HU and texture-based features | Logistic Regression | Youden Index: 0.67 (TP53), 0.56 (KRAS), and 0.50 (SMAD4, CDKN2A) |
| Iwatate et al. (2022) [135] | 107 PDAC patients (RNA-seq: 12) | ITK-SNAP | 3748 radiomic features | XGBoost | AUC = 0.697 (ITGAV), p = 0.048 (OS correlation) |
| Ref. | Dataset | Software/Tool/Prog. Lang. * | Features | ML/DL Model | Results |
|---|---|---|---|---|---|
| Dmitriev et al. (2017) [136] | 134 patients (4 pancreatic cyst types) | Scikit-learn, Keras (NVIDIA Titan X GPU) | 14 radiomic features + CNN deep features | Random Forest + CNN (Bayesian Fusion) | Acc = 83.6% |
| Ziegelmayer et al. (2020) [137] | 86 patients (44 AIP, 42 PDAC) | PyRadiomics, pretrained VGG19 | 1411 radiomic features + 256 deep features | Extremely Randomized Trees | AUC = 0.90, Sens = 89%, and Spec = 83% |
| Zhang et al. (2021) [138] | 98 PDAC patients (68 training, 30 validation) | PyRadiomics (v2.0.0) + 8-layer CNN (LungTrans) | 1428 radiomic features + 35 deep features | Risk Score-Based Fusion (Random Forest) | AUC = 0.84 |
| Wei et al. (2023) [139] | 112 patients (64 PDAC, 48 AIP) with 18F-FDG PET/CT | PyRadiomics + VGG11 CNN | Radiomics (texture, histogram) + CNN deep features | Multidomain Fusion Classifier | AUC = 96.4%, Acc = 90.1%, Sens = 87.5%, and Spec = 93.0% |
| Yao et al. (2023) [140] | 246 multi-center MRI scans (IPMN risk stratification) | nnUNet, multiple CNNs (DenseNet, ViT, etc.) | 107 radiomic features + deep CNN/ViT + clinical | Weighted Averaging-Based Fusion | Acc = 81.9% |
| Vétil et al. (2023) [141] | 2319 training + 1094 test CT scans (9 centers) | PyRadiomics + Variational Autoencoder (VAE) | Handcrafted radiomics + MI-minimized deep features | Logistic Regression (Fusion) | +1.13% AUC improvement over radiomics alone |
| Clinical Application | Public Dataset |
|---|---|
| Disease Detection | The Cancer Imaging Archive CPTAC-PDAC CT set |
| 4 × GEO expression profiles | |
| NIH Pancreas-CT dataset | |
| Public dataset—No name | |
| 16 × GEO expression profiles | |
| Longitudinal Cohort of Diabetes Patients (LHDB)–Taiwan NHI | |
| National Health Interview Survey (NHIS) | |
| The Health Improvement Network (THIN) primary-care database | |
| Medical Segmentation Decathlon (MSD)–pancreas task | |
| TCIA pancreas-CT collection (NTU study) | |
| Radiogenomics | cBioPortal PDAC sequencing cohorts |
| CPTAC-PDAC multi-omics set | |
| Disease Classification | N/A * |
| Survival Prediction | N/A |
| Treatment Response | N/A |
| Deep Radiomics Fusion | N/A |
| Clinical Application | Number of Features | Types of Features | Model Algorithm | Performance (Typical Metrics: AUC/Acc.) | Primary Purpose |
|---|---|---|---|---|---|
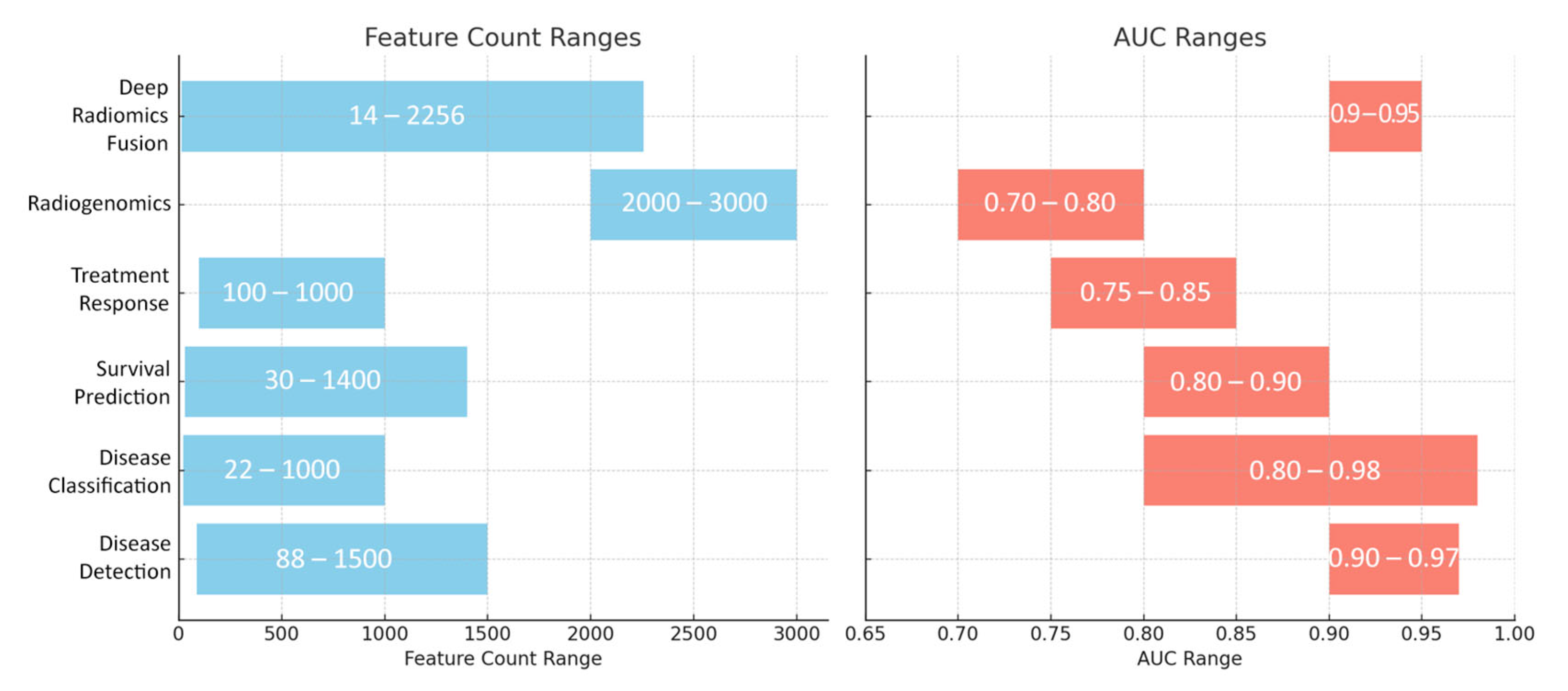
| Disease Detection | 88–1500+ (radiomics) or additional deep features | Handcrafted features such as shape, first-order intensity, GLCM texture, wavelet, and morphological descriptors; CNN embeddings capture abstract patterns. | 3D CNNs (ResNet variants, U-Net, and Faster R-CNN), patch-based CNNs, Random Forests, and SVM ensembles | AUCs: 0.90–0.97; Acc.: ~90–95% | Automatic detection of pancreatic cancer and differentiation from normal tissue or benign lesions. |
| Disease Classification | 22–1000+ (often reduced to 10–40 key features) | Texture descriptors (e.g., GLCM, GLRLM, fractal, and LBP), intensity histograms, and morphological indicators (volume, diameter, and shape factors). | SVMs, Random Forests, Logistic Regression, deep CNN classifiers, and ensemble methods (e.g., gradient boosting) | AUCs: 0.80–0.98; Acc.: ~85–95% | Differentiating among pancreatic pathologies such as PDAC, pancreatitis, PanNETs, and various cystic neoplasms. |
| Survival Prediction | 30–1400+ (often reduced to <10 or fused with clinical data) | Texture features (entropy, dissimilarity), first-order statistics, and morphological descriptors; often combined with clinical biomarkers (CA19-9, CEA). | Cox proportional hazards models, Random Forest Survival, Bayesian neural networks, and deep CNN survival models | C-index: ~0.65–0.75 (often >0.70); AUC: ~0.80–0.90 for early response | Predicting overall/disease-free survival and risk stratification in pancreatic cancer patients. |
| Treatment Response | 100–1000 (including delta-radiomics from daily scans) | Delta changes in texture (kurtosis, skewness, and entropy) and morphological features (tumor shrinkage, density changes) over time. | Logistic or Cox regression models, and deep CNN-based segmentation networks | AUC/Acc.: ~0.75–0.85 | Evaluating early treatment response and efficacy in neoadjuvant, chemoradiation, or SBRT settings. |
| Radiogenomics | 2000+–3000+ | Comprehensive radiomics encompassing texture, shape, and intensity measures correlated with genomic data (e.g., KRAS, TP53, SMAD4 mutations, and gene expression). | Random Forest, XGBoost, and SVM with recursive feature elimination and importance ranking | AUC: ~0.70–0.80 | Linking imaging phenotypes to molecular/genetic profiles to guide precision oncology. |
| Deep Radiomics Fusion | 14–2000 (handcrafted) plus ~256 deep features | Combination of interpretable handcrafted radiomics (GLCM, wavelet, etc.) and deep CNN embeddings capturing high-level image patterns. | Ensemble methods (Bayesian fusion, Random Forest, and Logistic Regression) with mutual information minimization techniques | Improvement of ~2–5% (often achieving AUC up to 0.90+) | Integrating complementary imaging biomarkers to boost performance in detection, classification, and survival prediction. |
| Category | Key Limitations/Gap | Possible Future Directions |
|---|---|---|
| Early Detection | Limited focus on early-stage PDAC; poor differentiation from benign/inflamed tissue | Develop models using pre-diagnostic data and biomarkers; enhance sensitivity to subtle features |
| Survival Prediction | Lack of multi-modal data; single time point analysis; and overfitting | Use longitudinal data; integrate clinical, genomic, proteomic, and metabolomic information |
| Treatment Response | Inability to predict individual therapy outcomes; limited data types used | Incorporate serial imaging, transcriptomics, and immune/metabolic markers |
| Radiogenomics | Narrow mutation scope; no temporal tracking; and limited datasets | Develop longitudinal radiogenomic models; include liquid biopsy and multi-omics data |
| Specificity and False Positives | Overlap with benign lesions; high false-positive rates | Combine imaging with histopathology/molecular profiling; design models with radiologist feedback |
| Data Availability | Predominant use of private data; poor reproducibility and benchmarking | Promote multi-institutional data sharing; build large, diverse, and publicly available datasets |
| Clinical Validation | Mostly retrospective studies; limited real-world testing | Conduct prospective trials; measure impact on diagnostic accuracy and patient outcomes |
| Workflow Integration | Models not designed for clinical systems; workflow disruptions | Develop plug-and-play AI tools integrated with PACS/RIS; utilize cloud-based real-time platforms |
| Bias and Ethics | Lack of fairness testing; biased datasets; and privacy issues | Ensure demographic diversity; apply fairness-aware training; and enforce strong data governance |
| Regulatory and Adoption Barriers | Lack of regulatory approvals (e.g., FDA, CE); unclear reimbursement; and clinician trust issues | Establish clear validation pathways and regulatory standards; include explainability mechanisms; align with reimbursement models; and promote clinician–AI co-pilot systems |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lekkas, G.; Vrochidou, E.; Papakostas, G.A. Advancements in Radiomics-Based AI for Pancreatic Ductal Adenocarcinoma. Bioengineering 2025, 12, 849. https://doi.org/10.3390/bioengineering12080849
Lekkas G, Vrochidou E, Papakostas GA. Advancements in Radiomics-Based AI for Pancreatic Ductal Adenocarcinoma. Bioengineering. 2025; 12(8):849. https://doi.org/10.3390/bioengineering12080849
Chicago/Turabian StyleLekkas, Georgios, Eleni Vrochidou, and George A. Papakostas. 2025. "Advancements in Radiomics-Based AI for Pancreatic Ductal Adenocarcinoma" Bioengineering 12, no. 8: 849. https://doi.org/10.3390/bioengineering12080849
APA StyleLekkas, G., Vrochidou, E., & Papakostas, G. A. (2025). Advancements in Radiomics-Based AI for Pancreatic Ductal Adenocarcinoma. Bioengineering, 12(8), 849. https://doi.org/10.3390/bioengineering12080849








