1.1. Research Background and Literature Review
Preterm infants, especially those classified as having an extremely low birth weight (ELBW) with a weight under 1000 g or gestational age less than 28 weeks, frequently encounter significant cardiac complications after birth [
1]. The abrupt transition from fetal to postnatal circulation imposes immense stress on their cardiovascular system, largely due to the sudden cessation of the low-resistance placental circulation. This interruption leads to a marked increase in systemic arterial resistance, thereby intensifying the afterload on the left ventricle [
2]. Such hemodynamic challenges are further amplified in the presence of a symptomatic patent ductus arteriosus (PDA), a condition that fails to resolve spontaneously in the majority of extremely preterm infants by day seven [
3]. As pulmonary vascular resistance decreases in the early days of life, a left-to-right shunt develops across the PDA, increasing pulmonary blood flow and potentially triggering complications like pulmonary edema, congestion, and, in severe cases, respiratory failure [
2]. These shifts also predispose infants to adverse outcomes including intraventricular hemorrhage (IVH), necrotizing enterocolitis (NEC), bronchopulmonary dysplasia (BPD), and increased mortality [
3].
The immature myocardium of preterm infants struggles to adapt to rapid changes in preload and afterload, elevating the risk of hemorrhagic events such as IVH and pulmonary hemorrhage. Understanding these hemodynamic alterations is therefore crucial for improving clinical outcomes. Echocardiographic techniques have been employed extensively to evaluate these changes. Takahashi et al. [
4] demonstrated that while very-low-birth-weight infants exhibit initially lower left ventricular contractility compared to term infants, contractility normalizes by day five. Toyoshima et al. [
2] proposed a tailored circulatory management approach based on the stress–velocity relationship to reduce IVH and mortality. Huang et al. [
5] highlighted the efficacy of customized circulatory strategies in mitigating hemorrhagic complications associated with PDA.
Echocardiography, a non-invasive imaging modality, remains the gold standard for assessing cardiac structure and function in preterm infants [
1,
6]. To enhance interpretation and reduce inter-observer variability, recent research has incorporated deep learning (DL) techniques. Early applications focused on image segmentation and classification tasks, particularly for left ventricle (LV) segmentation. Moradi et al. [
7] introduced MFP-U-Net, improving upon the U-Net architecture with extra convolutional layers to enhance LV segmentation accuracy. Kim et al. [
6] proposed convolutional neural networks (CNNs) for simultaneously segmenting the LV endocardium and myocardium, while Liu et al. [
8] employed deep pyramid local attention modules to refine structural delineation. Zhuang et al. [
9] leveraged YOLOv3 to identify critical points within the ventricular chamber, aiding in segmentation tasks. Similarly, Mortada et al. [
10] combined YOLOv7 with U-Net to segment the left ventricular endocardium, epicardium, and left atrium, demonstrating that integrating object detection with segmentation architectures can yield robust cardiac structural analyses.
While these studies mark significant progress, they predominantly address two-dimensional segmentation or classification tasks and rely heavily on static frames. In contrast, evaluating cardiac function in preterm infants often involves dynamic parameters extracted from motion-mode (M-mode) and pulsed-wave Doppler-mode echocardiograms. There remains a pronounced gap in the literature on applying DL-based object detection methodologies—originally advanced through large-scale benchmarks like the PASCAL VOC challenge [
11]—to these time-based echocardiographic modalities.
Object detection techniques have rapidly evolved, moving from region proposal-based frameworks like R-CNN [
12], Fast R-CNN [
13], and Faster R-CNN [
14] to single-stage detectors. Enhancements like Spatial Pyramid Pooling (SPP-net) [
15] and the adoption of powerful CNN architectures including VGG [
16], ResNet [
17], DenseNet [
18], and ResNeXt [
19] have improved accuracy and efficiency. Simultaneously, lightweight architectures such as MobileNets [
20], ShuffleNet [
21], and SqueezeNet [
22] have enabled deployment in resource-constrained environments.
On the optimization front, Adam [
23] and other gradient-based improvements [
24] accelerated convergence, while data pre-processing and augmentation techniques [
25] enhanced model generalization. Novel activation functions like Mish [
26] and specialized losses like focal loss [
27] further refined detection robustness. Single-stage detectors (e.g., SSD [
28], YOLO [
29], YOLO9000 [
30]) reframed detection as a direct regression problem, achieving near-real-time performance. YOLOV4 [
31] integrated CSPNet [
32], SPP modules, and Mish activation to balance speed and accuracy effectively—traits vital for medical applications that demand prompt, reliable assessments. Reviews by Zhao et al. [
33] and others underscore the need to weigh computational complexity against accuracy, especially in clinical contexts.
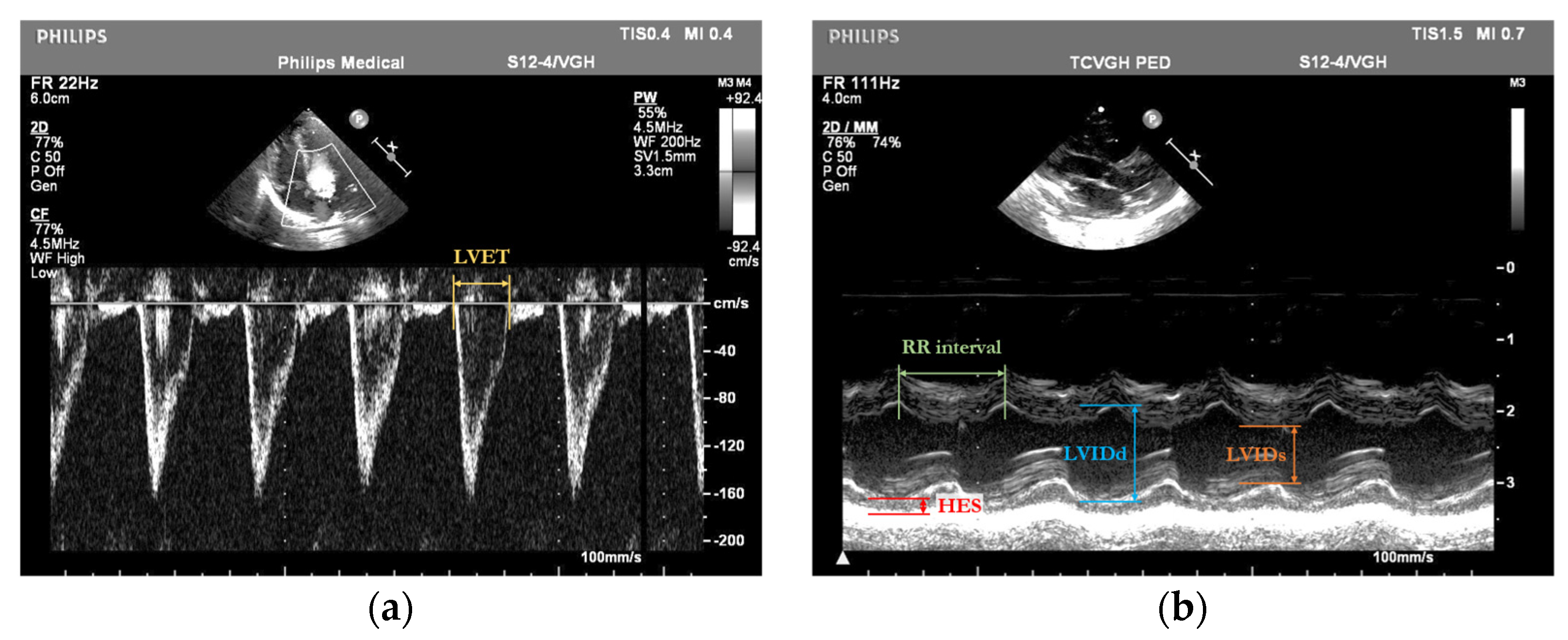
By integrating DL-based object detection, our approach aims to automate and enhance the precision of cardiac assessments in preterm infants, ultimately improving clinical outcomes for this high-risk population. We focus on several key cardiac parameters extracted from M-mode and pulsed-wave Doppler-mode echocardiograms of preterm infants. These include the left ventricular ejection time (LVET, measured in seconds), the left ventricular internal dimension at end-diastole (LVIDd, measured in centimeters), the left ventricular internal dimension at end-systole (LVIDs, measured in centimeters), the posterior wall thickness at end-systole (HES, measured in centimeters), and the RR interval (the time between two consecutive R-wave peaks on the ECG, measured in seconds). These fundamental variables are used to compute important derived indicators such as the rate-corrected mean velocity of circumferential fiber shortening (mVcfc, measured in circumferences per second, circ/s) and end-systolic wall stress (ESWS, measured in grams per square centimeter, g/cm
2). Automated measurement results from our system are directly compared with manual annotations by expert cardiologists, which serve as the clinical reference standard. The measurement errors observed in our deep learning approach are within the variability range previously reported for manual echocardiographic assessments [
34,
35], underscoring the reliability of our method as an alternative to traditional manual analysis.
In this study, we systematically compared a series of recent YOLO models, including YOLOV4, YOLOV8s [
36], YOLOv10s [
37], YOLOv11n/s [
38], and YOLOv12n/s [
39], on our clinical dataset of preterm infant echocardiograms. Meanwhile, YOLOV8’s anchor-free design and backbone enhancements achieved excellent precision in segmentation studies [
36], YOLOv10 introduced NMS-free dual-assignment training for efficiency gains [
37], and YOLOv11 improved depth with lightweight architectural refinements [
38]. Additionally, YOLOv12 introduces attention-centric modules and hierarchical architectures [
39], which, while effective in general scenes, did not translate into balanced performance on our clinical targets. To leverage these complementary strengths, we developed an ensemble framework that selects the best YOLO variant for each parameter and combines outputs via weighted fusion, resulting in the highest accuracy for derived clinical indices like the mVcfc and ESWS.
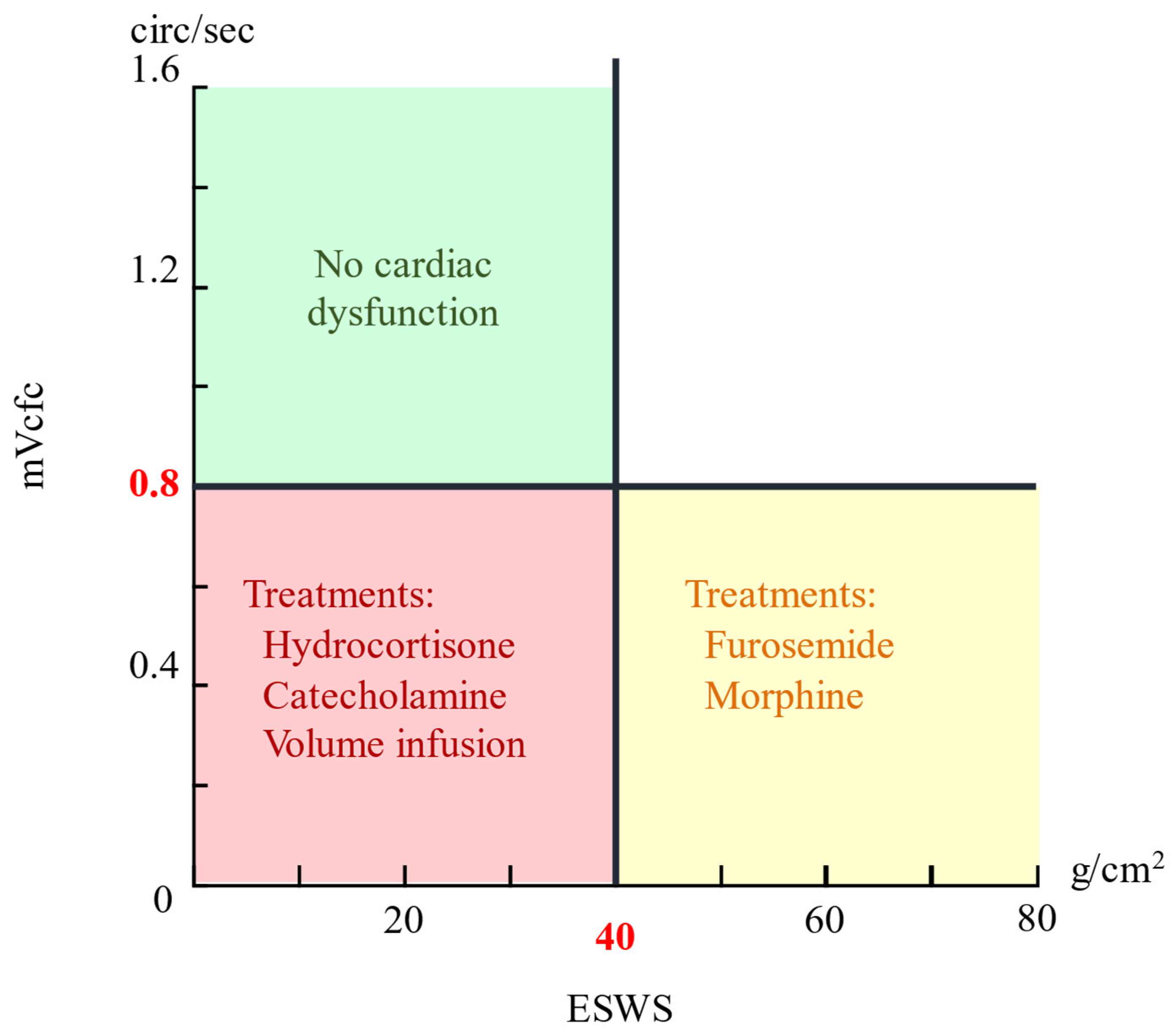
Building on these advances, we propose employing YOLO models to detect key cardiac parameters, the LVET, LVIDd, LVIDs, HES, and RR interval—from M-mode and pulsed-wave Doppler-mode echocardiograms of preterm infants. These parameters facilitate calculation of the stress–velocity relationship—quantified by the mean velocity of circumferential fiber shortening corrected for heart rate (mVcfc) and end-systolic wall stress (ESWS). By plotting the mVcfc against ESWS, clinicians can evaluate cardiac function and select treatments accordingly [
2].
The formulas for the
mVcfc and
ESWS are given by
where
MAP is the mean arterial pressure. If the
mVcfc > 0.8 circ/s and
ESWS < 40 g/cm
2, cardiac pump function is considered normal; deviations from these thresholds guide therapies ranging from hydrocortisone and volume expansion to furosemide or morphine [
2]. The Cartesian coordinate system representation in
Figure 1, with mVcfc and ESWS as axes, illustrates this relationship.
1.2. Echocardiogram Dataset of Preterm Infants
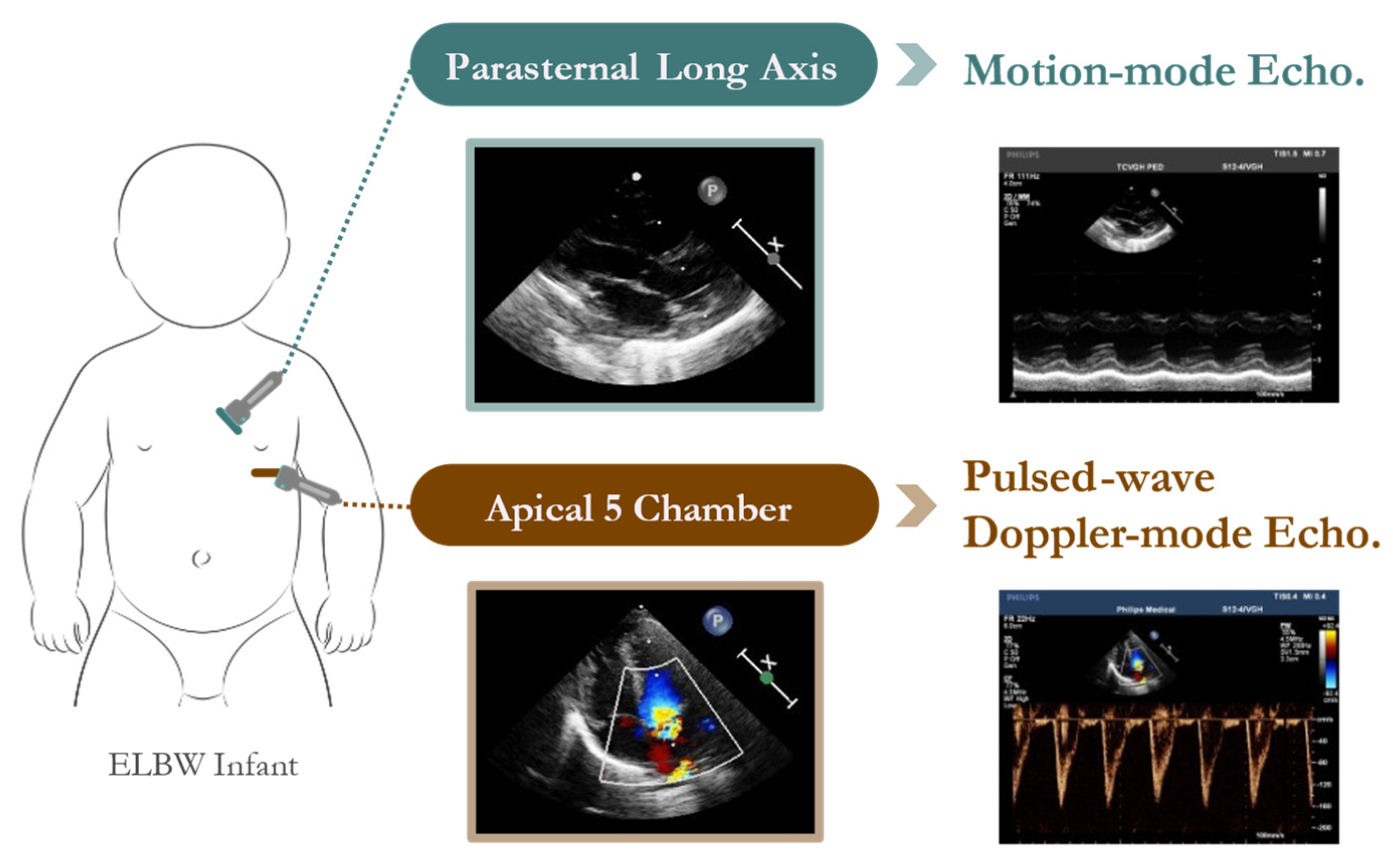
The dataset analyzed in this study was obtained from the Neonatal Intensive Care Unit (NICU) at Taichung Veterans General Hospital (TVGH) in Taiwan. The echocardiographic data were acquired using a high-resolution Philips IE33 cardiovascular ultrasound system equipped with a 4 MHz sector array transducer (Philips S12-4). Standardized probe placement ensured consistent acquisition of M-mode and pulsed-wave Doppler-mode echocardiograms (
Figure 2).
A total of 33 preterm infants were included, yielding 140 pulsed-wave Doppler-mode and 190 M-mode echocardiograms. Following best practices in DL-based medical image analysis, data pre-processing steps (e.g., grayscale conversion, normalization, resizing) and stochastic augmentations [
25] were applied to improve model robustness and generalization. This dataset provided the foundation for training and evaluating the YOLOV4-based detectors to measure the LVET, LVIDd, LVIDs, HES, and RR interval. Leveraging the optimized architectures [
17,
18,
22,
28,
29,
30,
32], advanced training strategies [
23,
24], and improved activation and loss functions [
26,
27], our integrated approach aims to streamline cardiac parameter detection.
By employing the latest advancements in object detection and CNN architectures [
12,
13,
14,
15,
16,
19,
20,
21,
31], as well as data augmentation [
25] and optimization techniques [
23,
24], this work bridges the gap in applying DL-based object detection to dynamic echocardiographic parameters in preterm infants. This synergy not only enhances quantitative cardiac assessments but also paves the way for more informed, timely therapeutic decisions and potentially better clinical outcomes in this fragile patient cohort.
For the LVET, our model achieves a mean percentage error (MPE) of 6.71%, corresponding to an average absolute error of approximately 2.8 ms (with a mean LVET of 41.8 ms). This level of error is within the range of inter-observer variability reported for echocardiographic timing measurements, typically 2–6% in the literature [
34,
35]. Previous studies have shown that manual LVET measurement by pulsed Doppler has a reproducibility error of about 4%, and most inter-observer differences are below 10% [
34,
35]. These results demonstrate that our automated measurement accuracy is comparable to traditional manual analysis.