Association Analysis Between Ischemic Stroke Risk Single Nucleotide Polymorphisms and Alzheimer’s Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Participates
2.2. Genotyping Data
2.3. CSF Biomarker Measurements
2.4. Brain Structures on MRI
2.5. Statistical Analysis
3. Results
3.1. Baseline Information of Participants
3.2. Characteristic of Enrolled SNPs
3.3. Association of IS-Risk SNPs with AD
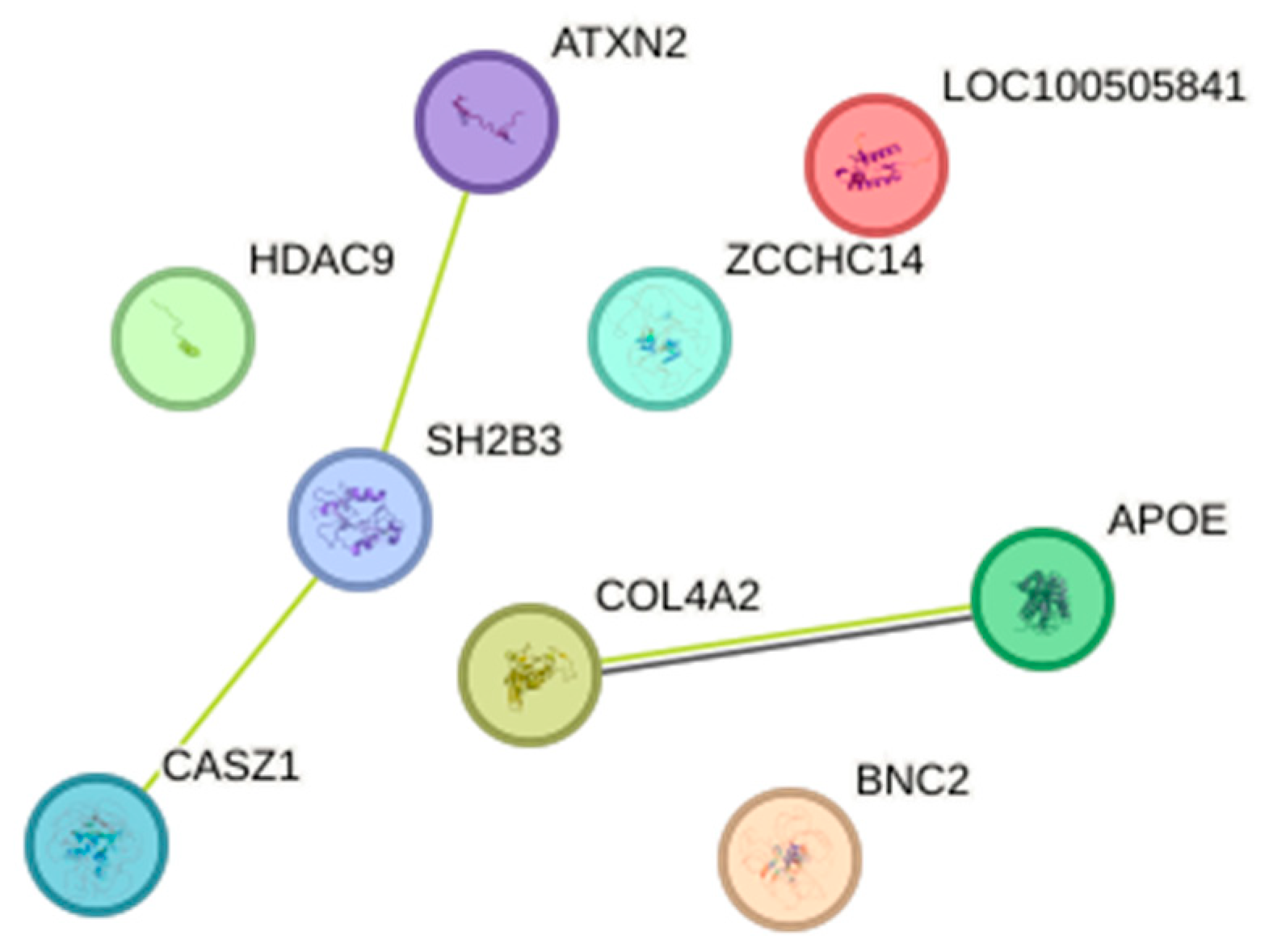
3.4. The Impact of SNP–SNP Interactions on AD Risk and PPI Analysis
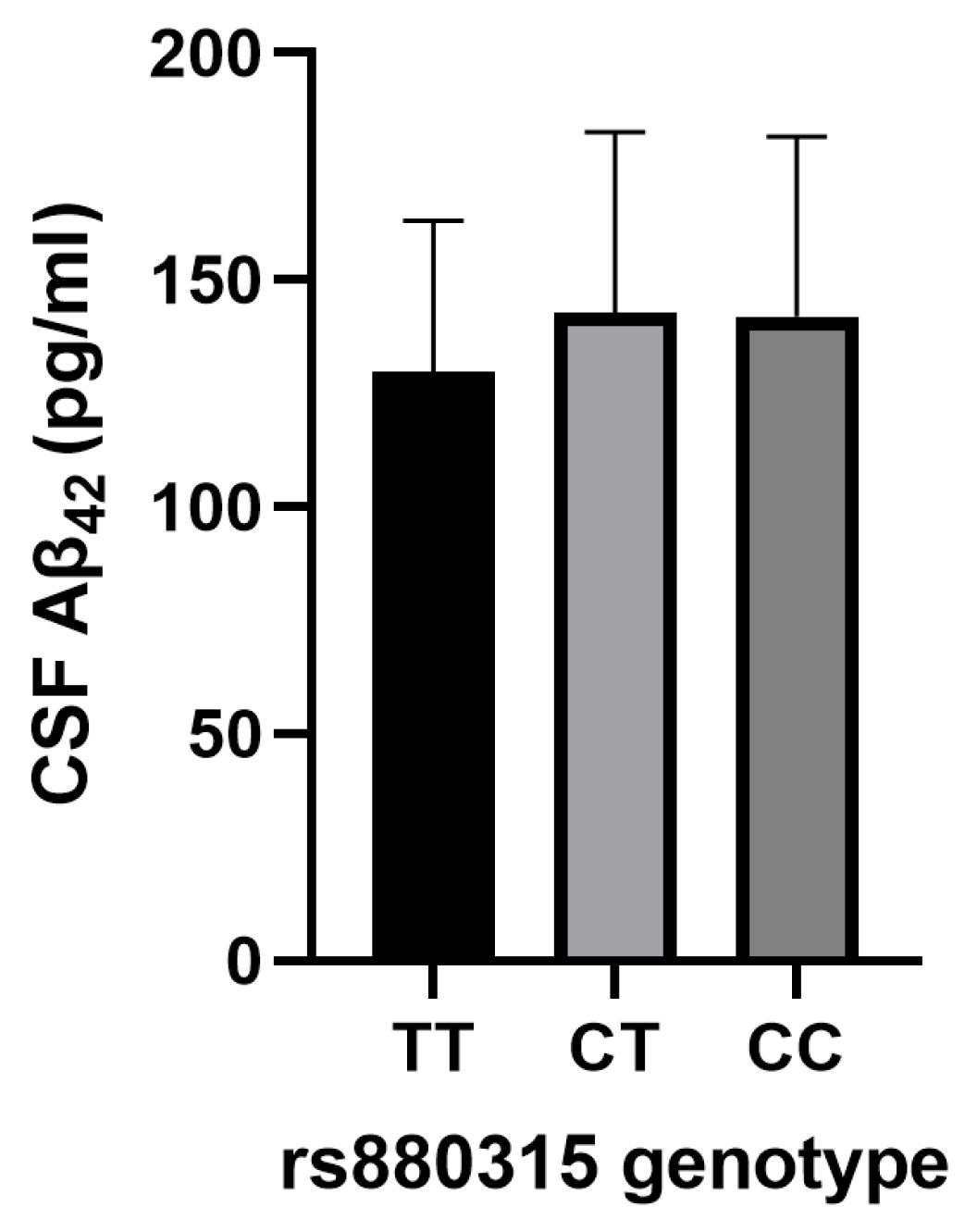
3.5. Association of IS Risk SNPs with AD CSF Biomarkers
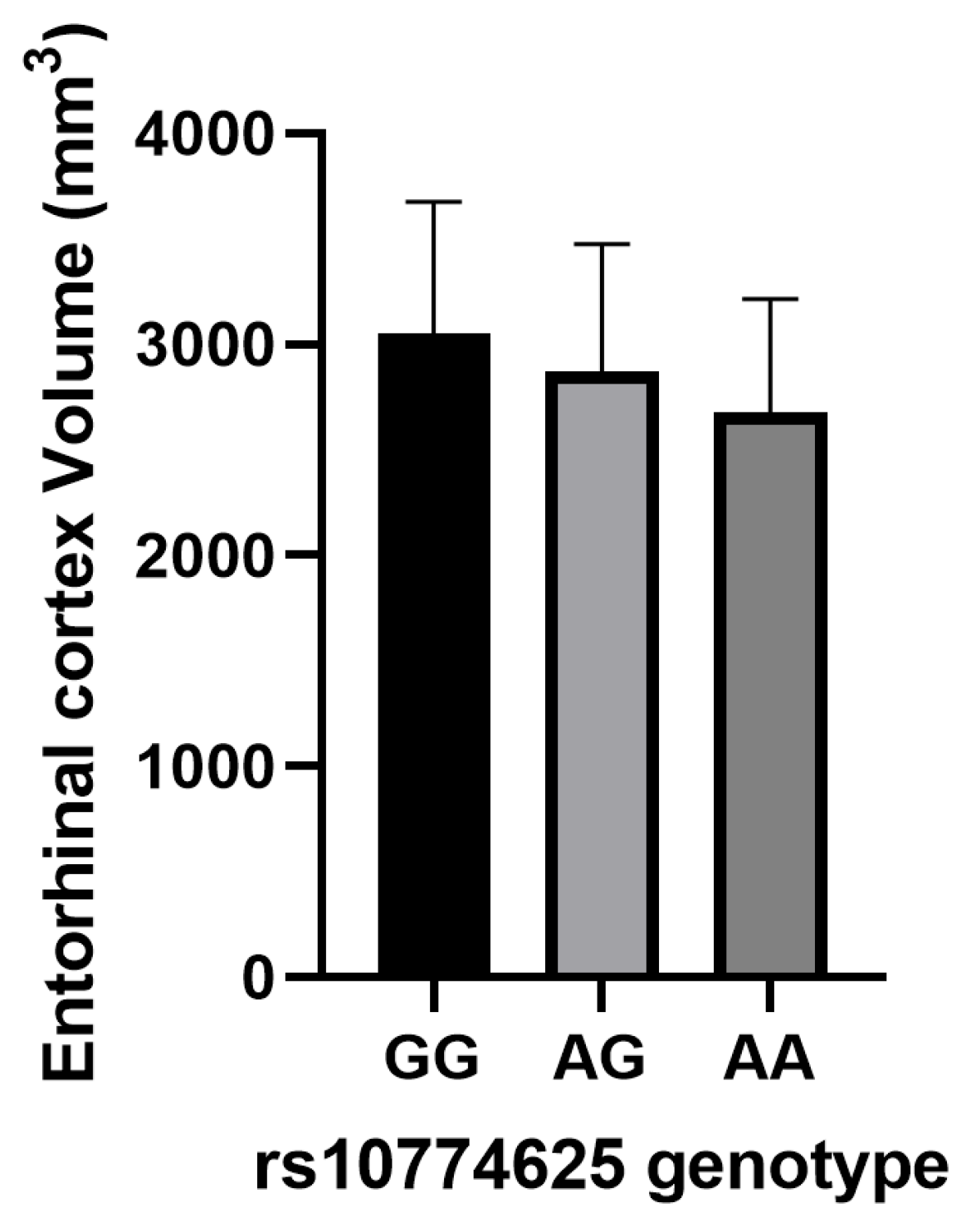
3.6. Association of IS-Risk SNPs with Neuroimaging Biomarkers in AD Patients
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Aβ42 | Amyloid-beta 1–42 |
| AD | Alzheimer’s disease |
| ADNI | Alzheimer’s Disease Neuroimaging Initiative |
| AIS | Any ischemic stroke |
| ApoE | Apolipoprotein E |
| ATP5H | Adenosine triphosphate synthase subunit d, mitochondrial |
| ATXN2 | Ataxin 2 |
| BBB | Blood–brain barrier |
| BETA | Regression coefficient |
| BNC2 | Basonuclin zinc finger protein 2 |
| CASZ1 | Castor zinc finger 1 |
| CBF | Cerebral blood flow |
| CI | Confidence interval |
| COL4A2 | Collagen type IV alpha 2 chain |
| CSF | Cerebrospinal fluid |
| EA | Effect allele |
| EAF | Effect allele frequency |
| ECM | Extracellular matrix |
| EPHA1 | EPH receptor A1 |
| eQTL | Expression quantitative trait loci |
| GMDR | Generalized multifactor dimensionality reduction |
| GWAS | Genome-wide association study |
| HDAC9 | Histone deacetylase 9 |
| HF | Heart failure |
| H-W | Hardy–Weinberg |
| ICH | Intracerebral hemorrhage |
| ICT1 | Lysophosphatidic acid acyltransferase ICT1 |
| IS | Ischemic stroke |
| KCTD2 | Potassium channel tetramerization domain containing 2 |
| LAS | Large artery stroke |
| LPS | Lipopolysaccharide |
| MCI | Mild cognitive impairment |
| MRI | Magnetic resonance imaging |
| MS4A4A | Membrane spanning 4-domains A4A |
| OA | Other allele |
| OR | Odds ratio |
| PET | Positron emission tomography |
| PPI | Protein–protein interactions |
| p-tau181 | Phosphorylated tau 181 |
| SH2B3 | SH2B adaptor protein 3 |
| SNP | Single-nucleotide polymorphism |
| SVS | Small vessel stroke |
| T1AM | Thyroid hormone derivative 3-iodothyronamine |
| Th | T helper |
| TH | Thyroid hormones |
| Treg | Regulatory T cell |
| TREM2 | Triggering receptor expressed on myeloid cells 2 |
| TSA | Trichostatin A |
| t-tau | Total tau |
| UBE2L3 | Ubiquitin-conjugating enzyme E2 L3 |
| WMH | White matter hyperintensities |
| ZCCHC14 | Zinc finger CCHC-type containing 14 |
References
- Srivastava, S.; Ahmad, R.; Khare, S.K. Alzheimer’s disease and its treatment by different approaches: A review. Eur. J. Med. Chem. 2021, 216, 113320. [Google Scholar] [CrossRef]
- O’Donnell, M.J.; Chin, S.L.; Rangarajan, S.; Xavier, D.; Liu, L.; Zhang, H.; Rao-Melacini, P.; Zhang, X.; Pais, P.; Agapay, S.; et al. Global and regional effects of potentially modifiable risk factors associated with acute stroke in 32 countries (INTERSTROKE): A case-control study. Lancet 2016, 388, 761–775. [Google Scholar] [CrossRef]
- Porsteinsson, A.P.; Isaacson, R.S.; Knox, S.; Sabbagh, M.N.; Rubino, I. Diagnosis of Early Alzheimer’s Disease: Clinical Practice in 2021. J. Prev. Alzheimers Dis. 2021, 8, 371–386. [Google Scholar] [CrossRef] [PubMed]
- Çakır, M.; Saçmacı, H. The relationship of salusins with Parkinson’s Disease, Alzheimer’s Disease, and acute ischemic stroke: A preliminary study. Neurosci. Lett. 2024, 824, 137683. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Wang, K. Childhood Secondhand Smoke Exposure and Risk of Dementia, Alzheimer’s Disease and Stroke in Adulthood: A Prospective Cohort Study. J. Prev. Alzheimers Dis. 2021, 8, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, H. Early-Onset Subgroup of Type 2 Diabetes and Risk of Dementia, Alzheimer’s Disease and Stroke: A Cohort Study. J. Prev. Alzheimers Dis. 2021, 8, 442–447. [Google Scholar] [CrossRef]
- Tang, C.; Ma, Y.; Lei, X.; Ding, Y.; Yang, S.; He, D. Hypertension linked to Alzheimer’s disease via stroke: Mendelian randomization. Sci. Rep. 2023, 13, 21606. [Google Scholar] [CrossRef]
- Wang, R.; Qiu, C.; Dintica, C.S.; Shang, Y.; Calderón Larrañaga, A.; Wang, H.X.; Xu, W. Shared risk and protective factors between Alzheimer’s disease and ischemic stroke: A population-based longitudinal study. Alzheimers Dement. 2021, 17, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Yin, L.; Zhang, L.; Zhang, Y.; Zha, T.; Zhang, W.; Gui, B. Diabetes accelerates Alzheimer’s disease progression in the first year post mild cognitive impairment diagnosis. Alzheimers Dement. 2024, 20, 4583–4593. [Google Scholar] [CrossRef]
- Durazzo, T.C.; Meyerhoff, D.J.; Yoder, K.K. Cigarette smoking is associated with cortical thinning in anterior frontal regions, insula and regions showing atrophy in early Alzheimer’s Disease. Drug Alcohol Depend. 2018, 192, 277–284. [Google Scholar] [CrossRef]
- Khan, A.D.; Elnagar, S.; Eltayeb, M.; Baluch, S.K.; Kumar, A.; Kumari, M.; Kumari, M.; Fareed, M.U.; Rehman, A.; Shehryar, A. The Impact of Hypertension on Cognitive Decline and Alzheimer’s Disease and Its Management: A Systematic Review. Cureus 2024, 16, e65194. [Google Scholar] [CrossRef]
- Gorelick, P.B.; Scuteri, A.; Black, S.E.; Decarli, C.; Greenberg, S.M.; Iadecola, C.; Launer, L.J.; Laurent, S.; Lopez, O.L.; Nyenhuis, D.; et al. Vascular contributions to cognitive impairment and dementia: A statement for healthcare professionals from the american heart association/american stroke association. Stroke 2011, 42, 2672–2713. [Google Scholar] [CrossRef]
- Zhou, J.; Yu, J.T.; Wang, H.F.; Meng, X.F.; Tan, C.C.; Wang, J.; Wang, C.; Tan, L. Association between stroke and Alzheimer’s disease: Systematic review and meta-analysis. J. Alzheimers Dis. 2015, 43, 479–489. [Google Scholar] [CrossRef]
- Song, I.U.; Kim, J.S.; Kim, Y.I.; Eah, K.Y.; Lee, K.S. Clinical significance of silent cerebral infarctions in patients with Alzheimer disease. Cogn. Behav. Neurol. 2007, 20, 93–98. [Google Scholar] [CrossRef]
- Suter, O.C.; Sunthorn, T.; Kraftsik, R.; Straubel, J.; Darekar, P.; Khalili, K.; Miklossy, J. Cerebral hypoperfusion generates cortical watershed microinfarcts in Alzheimer disease. Stroke 2002, 33, 1986–1992. [Google Scholar] [CrossRef] [PubMed]
- Gorenflo, M.P.; Davis, P.B.; Kendall, E.K.; Olaker, V.R.; Kaelber, D.C.; Xu, R. Association of Aspirin Use with Reduced Risk of Developing Alzheimer’s Disease in Elderly Ischemic Stroke Patients: A Retrospective Cohort Study. J. Alzheimers Dis. 2023, 91, 697–704. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Zhang, J.; Yang, K.; Xu, Z.; Zhang, H.; Chen, W.; Peng, T.; Wang, X.; Liu, Z.; Wei, P.; et al. Anti-Stroke Chinese Herbal Medicines Inhibit Abnormal Amyloid-β Protein Precursor Processing in Alzheimer’s Disease. J. Alzheimers Dis. 2022, 85, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Pinho, J.; Quintas-Neves, M.; Dogan, I.; Reetz, K.; Reich, A.; Costa, A.S. Incident stroke in patients with Alzheimer’s disease: Systematic review and meta-analysis. Sci. Rep. 2021, 11, 16385. [Google Scholar] [CrossRef]
- Waziry, R.; Chibnik, L.B.; Bos, D.; Ikram, M.K.; Hofman, A. Risk of hemorrhagic and ischemic stroke in patients with Alzheimer disease: A synthesis of the literature. Neurology 2020, 94, 265–272. [Google Scholar] [CrossRef]
- Belloy, M.E.; Andrews, S.J.; Le Guen, Y.; Cuccaro, M.; Farrer, L.A.; Napolioni, V.; Greicius, M.D. APOE Genotype and Alzheimer Disease Risk Across Age, Sex, and Population Ancestry. JAMA Neurol. 2023, 80, 1284–1294. [Google Scholar] [CrossRef]
- Schwartzentruber, J.; Cooper, S.; Liu, J.Z.; Barrio-Hernandez, I.; Bello, E.; Kumasaka, N.; Young, A.M.H.; Franklin, R.J.M.; Johnson, T.; Estrada, K.; et al. Genome-wide meta-analysis, fine-mapping and integrative prioritization implicate new Alzheimer’s disease risk genes. Nat. Genet. 2021, 53, 392–402. [Google Scholar] [CrossRef]
- Mishra, A.; Malik, R.; Hachiya, T.; Jürgenson, T.; Namba, S.; Posner, D.C.; Kamanu, F.K.; Koido, M.; Le Grand, Q.; Shi, M.; et al. Stroke genetics informs drug discovery and risk prediction across ancestries. Nature 2022, 611, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Huang, Y. Common Genetic Factors and Pathways in Alzheimer’s Disease and Ischemic Stroke: Evidences from GWAS. Genes 2023, 14, 353. [Google Scholar] [CrossRef]
- Traylor, M.; Adib-Samii, P.; Harold, D.; Dichgans, M.; Williams, J.; Lewis, C.M.; Markus, H.S. Shared genetic contribution to Ischaemic Stroke and Alzheimer’s Disease. Ann. Neurol. 2016, 79, 739–747. [Google Scholar] [CrossRef]
- Wei, C.J.; Cui, P.; Li, H.; Lang, W.J.; Liu, G.Y.; Ma, X.F. Shared genes between Alzheimer’s disease and ischemic stroke. CNS Neurosci. Ther. 2019, 25, 855–864. [Google Scholar] [CrossRef]
- Zhang, Z. Multiple imputation with multivariate imputation by chained equation (MICE) package. Ann. Transl. Med. 2016, 4, 30. [Google Scholar]
- Bobowski-Gerard, M.; Boulet, C.; Zummo, F.P.; Dubois-Chevalier, J.; Gheeraert, C.; Bou Saleh, M.; Strub, J.M.; Farce, A.; Ploton, M.; Guille, L.; et al. Functional genomics uncovers the transcription factor BNC2 as required for myofibroblastic activation in fibrosis. Nat. Commun. 2022, 13, 5324. [Google Scholar] [CrossRef]
- Hou, J.; Li, J. Identification of Anesthetic-Induced Cardiovascular Biomarkers in Off-Pump Coronary Artery Bypass Grafting Surgery Using Weighted Gene Co-Expression Network Analysis and Machine Learning. Heart. Surg. Forum 2023, 26, E740–E754. [Google Scholar] [CrossRef]
- Zhu, Y.; Chen, B.; Zu, Y. Identifying OGN as a Biomarker Covering Multiple Pathogenic Pathways for Diagnosing Heart Failure: From Machine Learning to Mechanism Interpretation. Biomolecules 2024, 14, 179. [Google Scholar] [CrossRef]
- Sun, W.; Zhuo, S.; Wu, H.; Cai, X. Association between Coronary Heart Disease, Heart Failure, and Risk of Alzheimer’s Disease: A Systematic Review and Meta-Analysis. Ann. Indian Acad. Neurol. 2023, 26, 958–965. [Google Scholar] [CrossRef] [PubMed]
- Qiu, C.; Winblad, B.; Marengoni, A.; Klarin, I.; Fastbom, J.; Fratiglioni, L. Heart failure and risk of dementia and Alzheimer disease: A population-based cohort study. Arch. Intern. Med. 2006, 166, 1003–1008. [Google Scholar] [CrossRef]
- Nakamura, A.; Rokosh, D.G.; Paccanaro, M.; Yee, R.R.; Simpson, P.C.; Grossman, W.; Foster, E. LV systolic performance improves with development of hypertrophy after transverse aortic constriction in mice. Am. J. Physiol. Heart Circ. Physiol. 2001, 281, H1104–H1112. [Google Scholar] [CrossRef] [PubMed]
- Kitaguchi, H.; Tomimoto, H.; Ihara, M.; Shibata, M.; Uemura, K.; Kalaria, R.N.; Kihara, T.; Asada-Utsugi, M.; Kinoshita, A.; Takahashi, R. Chronic cerebral hypoperfusion accelerates amyloid beta deposition in APPSwInd transgenic mice. Brain. Res. 2009, 1294, 202–210. [Google Scholar] [CrossRef]
- Pluta, R. The role of apolipoprotein E in the deposition of beta-amyloid peptide during ischemia-reperfusion brain injury. A model of early Alzheimer’s disease. Ann. N. Y. Acad. Sci. 2000, 903, 324–334. [Google Scholar] [CrossRef]
- de la Torre, J.C. Pathophysiology of neuronal energy crisis in Alzheimer’s disease. Neurodegener. Dis. 2008, 5, 126–132. [Google Scholar] [CrossRef]
- Kunkle, B.W.; Grenier-Boley, B.; Sims, R.; Bis, J.C.; Damotte, V.; Naj, A.C.; Boland, A.; Vronskaya, M.; van der Lee, S.J.; Amlie-Wolf, A.; et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 2019, 51, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Farrer, L.A.; Cupples, L.A.; Haines, J.L.; Hyman, B.; Kukull, W.A.; Mayeux, R.; Myers, R.H.; Pericak-Vance, M.A.; Risch, N.; van Duijn, C.M. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA 1997, 278, 1349–1356. [Google Scholar] [CrossRef]
- Wang, Y.; Li, F.; Qin, Q.; Li, T.; Wang, Q.; Li, Y.; Li, Y.; Jia, J. Influence of APOE ε4 on performance of CSF biomarkers in differentiating clinical Alzheimer’s disease. J. Prev. Alzheimers Dis. 2025, 12, 100065. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Xiao, D.; Su, B.B.; Saveron, J.M.; Gamez, D.; Navia, R.O.; Wang, N.; Roy, U.; Adjeroh, D.A.; Wang, K. Association of APOE gene with longitudinal changes of CSF amyloid beta and tau levels in Alzheimer’s disease: Racial differences. Neurol. Sci. 2024, 45, 1041–1050. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Cao, P. Circ-Bnc2 alleviates neuroinflammation in LPS-stimulated microglial cells to inhibit neuron cell apoptosis through regulating miR-497a-5p/HECTD1 axis. Brain Behav. 2023, 13, e2935. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, Y.; Pan, J.; Cao, J.; Sun, X.; Li, X.; Zhang, L.; Qin, C. Degradation of NLRP3 by p62-dependent-autophagy improves cognitive function in Alzheimer’s disease by maintaining the phagocytic function of microglia. CNS Neurosci. Ther. 2023, 29, 2826–2842. [Google Scholar] [CrossRef]
- Traylor, M.; Persyn, E.; Tomppo, L.; Klasson, S.; Abedi, V.; Bakker, M.K.; Torres, N.; Li, L.; Bell, S.; Rutten-Jacobs, L.; et al. Genetic basis of lacunar stroke: A pooled analysis of individual patient data and genome-wide association studies. Lancet Neurol. 2021, 20, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Graff-Radford, J.; Arenaza-Urquijo, E.M.; Knopman, D.S.; Schwarz, C.G.; Brown, R.D.; Rabinstein, A.A.; Gunter, J.L.; Senjem, M.L.; Przybelski, S.A.; Lesnick, T.; et al. White matter hyperintensities: Relationship to amyloid and tau burden. Brain 2019, 142, 2483–2491. [Google Scholar] [CrossRef]
- Bhaskaran, N.; Liu, Z.; Saravanamuthu, S.S.; Yan, C.; Hu, Y.; Dong, L.; Zelenka, P.; Zheng, L.; Bletsos, V.; Harris, R.; et al. Identification of Casz1 as a Regulatory Protein Controlling T Helper Cell Differentiation, Inflammation, and Immunity. Front. Immunol. 2018, 9, 184. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Park, S.Y.; Baek, H.; Lee, C.; Chung, G.; Liu, X.; Lee, J.H.; Kim, B.; Kwon, M.; Choi, H.; et al. Adoptive therapy with amyloid-β specific regulatory T cells alleviates Alzheimer’s disease. Theranostics 2022, 12, 7668–7680. [Google Scholar] [CrossRef] [PubMed]
- Wild, P.S.; Felix, J.F.; Schillert, A.; Teumer, A.; Chen, M.H.; Leening, M.J.G.; Völker, U.; Großmann, V.; Brody, J.A.; Irvin, M.R.; et al. Large-scale genome-wide analysis identifies genetic variants associated with cardiac structure and function. J. Clin. Investig. 2017, 127, 1798–1812. [Google Scholar] [CrossRef] [PubMed]
- Schaschl, H.; Göllner, T.; Morris, D.L. Positive selection acts on regulatory genetic variants in populations of European ancestry that affect ALDH2 gene expression. Sci. Rep. 2022, 12, 4563. [Google Scholar] [CrossRef] [PubMed]
- Brčić, L.; Barić, A.; Gračan, S.; Brdar, D.; Torlak Lovrić, V.; Vidan, N.; Zemunik, T.; Polašek, O.; Barbalić, M.; Punda, A.; et al. Association of established thyroid peroxidase autoantibody (TPOAb) genetic variants with Hashimoto’s thyroiditis. Autoimmunity 2016, 49, 480–485. [Google Scholar] [CrossRef] [PubMed]
- Sible, I.J.; Nation, D.A. Blood pressure variability and medial temporal atrophy in apolipoprotein ϵ4 carriers. Brain Imaging Behav. 2022, 16, 792–801. [Google Scholar] [CrossRef]
- Tozzi, F.; Rutigliano, G.; Borsò, M.; Falcicchia, C.; Zucchi, R.; Origlia, N. T(1)AM-TAAR1 signalling protects against OGD-induced synaptic dysfunction in the entorhinal cortex. Neurobiol. Dis. 2021, 151, 105271. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Li, H.; Li, K.; Fan, X.D. HDAC9 promotes brain ischemic injury by provoking IκBα/NF-κB and MAPKs signaling pathways. Biochem. Biophys. Res. Commun. 2018, 503, 1322–1329. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.L.; Hu, S.; Yang, P.; Long, H.C.; Ma, Q.H.; Yin, D.M.; Xu, G.Y. HDAC9-mediated calmodulin deacetylation induces memory impairment in Alzheimer’s disease. CNS Neurosci. Ther. 2024, 30, e14573. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Yin, L.; Kang, X.; Xue, W.; Wang, N.; Zhang, J.; Yuan, P.; Lin, L.; Li, Y. TFEB acetylation promotes lysosome biogenesis and ameliorates Alzheimer’s disease-relevant phenotypes in mice. J. Biol. Chem. 2022, 298, 102649. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.; Zhang, X.; Allen, M.; Wang, X.; Ma, Y.; Beecham, G.; Montine, T.J.; Younkin, S.G.; Dickson, D.W.; Golde, T.E.; et al. Genome-wide pleiotropy analysis of neuropathological traits related to Alzheimer’s disease. Alzheimers Res. Ther. 2018, 10, 22. [Google Scholar] [CrossRef]
| AD (N = 127) | NC (N = 155) | p Value | |
|---|---|---|---|
| Age (years) 1 2 | 74.513 (8.39) | 73.996 (6.019) | 0.549 |
| Gender (M/F) 3 | 75/52 | 80/75 | 0.212 |
| Education (years) 4 5 | 16.0 (4.0) | 16.0 (4.0) | 0.052 |
| ApoE ε4 (0/1/2) 3 | 23/59/45 | 116/35/4 | <0.001 |
| rs10774625 (AA/AG/GG) 3 | 21/70/36 | 36/72/47 | 0.573 |
| rs12445022 (AA/AG/GG) 3 | 13/52/62 | 22/60/73 | 0.492 |
| rs1487504 (AA/AG/GG) 3 | 2/28/97 | 2/21/132 | 0.085 |
| rs17148926 (AA/AC/CC) 3 | 87/35/5 | 113/39/3 | 0.308 |
| rs2107595 (AA/AG/GG) 3 | 3/49/75 | 4/43/108 | 0.105 |
| rs880315 (CC/CT/TT) 3 | 17/51/59 | 13/66/76 | 0.344 |
| rs9515201 (AA/AC/CC) 3 | 14/57/56 | 17/62/76 | 0.534 |
| CSF Aβ42 (pg/mL) 4 5 | 129.0 (35.0) | 205.0 (74.0) | <0.001 |
| CSF t-tau (pg/mL) 4 5 | 117.0 (73.3) | 57.1 (42.1) | <0.001 |
| CSF p-tau181 (pg/mL) 4 5 | 48.6 (32.4) | 28.3 (22.6) | <0.001 |
| Hippocampus (mm3) 1 2 | 5992.07 (928.034) | 7498.06 (860.923) | <0.001 |
| Whole brain (mm3) 1 2 | 1,004,979.63 (113,512.084) | 1,047,729.22 (103,976.995) | 0.001 |
| Entorhinal (mm3) 1 2 | 2890.39 (607.056) | 3845.45 (622.302) | <0.001 |
| Mid-temporal (mm3) 1 2 | 17,764.61 (3169.834) | 20,571.16 (2293.72) | <0.001 |
| SNP | Main Phenotype | Gene | EA/OA | Function | EAF | H-W (p) |
|---|---|---|---|---|---|---|
| rs10774625 | AIS | SH2B3/ATXN2 | A/G | intron variant | 0.454 | 0.792 |
| rs12445022 | SVS | ZCCHC14 | A/G | intergenic variant | 0.322 | 0.125 |
| rs1487504 | AIS | BNC2 | A/G | intergenic variant | 0.101 | 0.463 |
| rs17148926 | AIS | LOC100505841 | A/C | intron variant | 0.840 | 0.716 |
| rs2107595 | LAS | HDAC9 | A/G | intergenic variant | 0.188 | 0.248 |
| rs880315 | AIS | CASZ1 | C/T | intron variant | 0.314 | 0.538 |
| rs9515201 | SVS | COL4A2 | A/C | intron variant | 0.321 | 0.593 |
| SNP Allele | OR (95% CI) | p Value | |
|---|---|---|---|
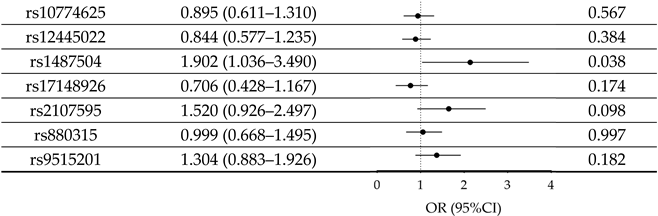 | |||
| Allele | ApoE ε4 Status | OR | 95% CI | p Value |
|---|---|---|---|---|
| rs1487504 | ApoE ε4+ | 1.038 | 0.406–2.651 | 0.938 |
| ApoE ε4− | 2.899 | 1.354–6.207 | 0.006 a |
| Model | Training Accuracy | Testing Accuracy | Sign Test (p) | Cross-Validation Consistency |
|---|---|---|---|---|
| ApoE ε4 | 0.697 | 0.6969 | 10 (0.0010) | 10/10 |
| rs1487504 ApoE ε4 | 0.7104 | 0.7043 | 10 (0.0010) | 10/10 |
| rs12445022 rs1487504 ApoE ε4 | 0.7265 | 0.6399 | 10 (0.0010) | 4/10 |
| rs1487504 rs17148926 rs9515201 ApoE ε4 | 0.758 | 0.651 | 9 (0.0107) | 4/10 |
| rs10774625 rs12445022 rs17148926 rs880315 ApoE ε4 | 0.802 | 0.6413 | 7 (0.1719) | 8/10 |
| rs10774625 rs12445022 rs17148926 rs2107595 rs880315 ApoE ε4 | 0.8543 | 0.5715 | 7 (0.1719) | 5/10 |
| rs10774625 rs12445022 rs17148926 rs2107595 rs880315 rs9515201 ApoE ε4 | 0.8977 | 0.5391 | 7 (0.1719) | 10/10 |
| AD Biomarker | SNP Allele | BETA | t | p Value |
|---|---|---|---|---|
| CSF Aβ42 (pg/mL) | rs10774625 | 0.136 | 1.633 | 0.105 |
| rs12445022 | 0.020 | 0.241 | 0.810 | |
| rs1487504 | −0.197 | −2.389 | 0.018 a | |
| rs17148926 | −0.161 | −1.967 | 0.051 | |
| rs2107595 | −0.016 | −0.191 | 0.849 | |
| rs880315 | 0.238 | 2.924 | 0.004 b | |
| rs9515201 | −0.002 | −0.029 | 0.977 | |
| CSF t-tau (pg/mL) | rs10774625 | −0.014 | −0.164 | 0.870 |
| rs12445022 | 0.129 | 1.541 | 0.126 | |
| rs1487504 | 0.177 | 2.094 | 0.038 a | |
| rs17148926 | 0.098 | 1.169 | 0.245 | |
| rs2107595 | 0.160 | 1.873 | 0.063 | |
| rs880315 | 0.079 | 0.924 | 0.358 | |
| rs9515201 | −0.143 | −1.680 | 0.095 | |
| CSF p-tau181 (pg/mL) | rs10774625 | −0.046 | −0.521 | 0.603 |
| rs12445022 | 0.083 | 0.948 | 0.345 | |
| rs1487504 | 0.038 | 0.432 | 0.666 | |
| rs17148926 | 0.18 | 2.088 | 0.039 a | |
| rs2107595 | −0.016 | −0.178 | 0.859 | |
| rs880315 | 0.006 | −0.066 | 0.947 | |
| rs9515201 | −0.059 | −0.665 | 0.507 |
| SNP Allele | BETA | t | p Value | |
|---|---|---|---|---|
| Hippocampus (mm3) | rs10774625 | −0.116 | −1.421 | 0.158 |
| rs12445022 | −0.041 | −0.500 | 0.618 | |
| rs1487504 | −0.070 | −0.859 | 0.392 | |
| rs17148926 | −0.074 | −0.918 | 0.360 | |
| rs2107595 | −0.178 | −2.180 | 0.031 a | |
| rs880315 | 0.020 | 0.240 | 0.811 | |
| rs9515201 | 0.072 | 0.882 | 0.379 | |
| Whole brain (mm3) | rs10774625 | −0.149 | −1.987 | 0.049 a |
| rs12445022 | −0.087 | −1.161 | 0.248 | |
| rs1487504 | 0.010 | 0.129 | 0.898 | |
| rs17148926 | −0.116 | −1.560 | 0.121 | |
| rs2107595 | −0.102 | −1.333 | 0.185 | |
| rs880315 | 0.093 | 1.224 | 0.223 | |
| rs9515201 | −0.045 | −0.594 | 0.554 | |
| Entorhinal cortex (mm3) | rs10774625 | −0.249 | −2.929 | 0.004 b |
| rs12445022 | −0.087 | −1.008 | 0.315 | |
| rs1487504 | −0.081 | −0.929 | 0.335 | |
| rs17148926 | −0.132 | −1.542 | 0.126 | |
| rs2107595 | −0.196 | −2.256 | 0.026 a | |
| rs880315 | 0.093 | 1.062 | 0.290 | |
| rs9515201 | 0.042 | 0.481 | 0.632 | |
| Mid-temporal lobe (mm3) | rs10774625 | −0.035 | −0.433 | 0.666 |
| rs12445022 | −0.025 | −0.317 | 0.752 | |
| rs1487504 | −0.076 | −0.929 | 0.355 | |
| rs17148926 | 0.016 | 0.197 | 0.844 | |
| rs2107595 | −0.001 | −0.017 | 0.987 | |
| rs880315 | 0.167 | 2.089 | 0.039 a | |
| rs9515201 | −0.155 | −1.931 | 0.056 |
| SNP | Gene | Potential Mechanisms That Connect IS and AD |
|---|---|---|
| rs1487504 | BNC2 | HF [29] and neuroinflammation [40] |
| rs10774625 | SH2B3/ATXN2 | Blood pressure variability [47] and decreased T1AM levels [48] |
| rs17148926 | LOC100505841 | White matter hyperintensities [42] |
| rs2107595 | HDAC9 | Neuroinflammation [51] and increased Aβ burden [53] |
| rs880315 | CASZ1 | T cell-associated inflammatory response [44] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, W.; Wang, W.; Li, M. Association Analysis Between Ischemic Stroke Risk Single Nucleotide Polymorphisms and Alzheimer’s Disease. Bioengineering 2025, 12, 804. https://doi.org/10.3390/bioengineering12080804
Dong W, Wang W, Li M. Association Analysis Between Ischemic Stroke Risk Single Nucleotide Polymorphisms and Alzheimer’s Disease. Bioengineering. 2025; 12(8):804. https://doi.org/10.3390/bioengineering12080804
Chicago/Turabian StyleDong, Wei, Wei Wang, and Mingxuan Li. 2025. "Association Analysis Between Ischemic Stroke Risk Single Nucleotide Polymorphisms and Alzheimer’s Disease" Bioengineering 12, no. 8: 804. https://doi.org/10.3390/bioengineering12080804
APA StyleDong, W., Wang, W., & Li, M. (2025). Association Analysis Between Ischemic Stroke Risk Single Nucleotide Polymorphisms and Alzheimer’s Disease. Bioengineering, 12(8), 804. https://doi.org/10.3390/bioengineering12080804





