Applications of Artificial Intelligence and Machine Learning in Spine MRI
Abstract
1. Introduction
2. Materials and Methods
2.1. Literature Search Strategy
2.2. Study Screening and Selection Criteria
2.3. Data Extraction and Reporting
- Study details: authorship, year of publication and journal name;
- Application and primary outcome measure;
- Study details: sample size, spine region studied, MRI sequences used;
- Artificial intelligence technique used;
- Key results and conclusion.
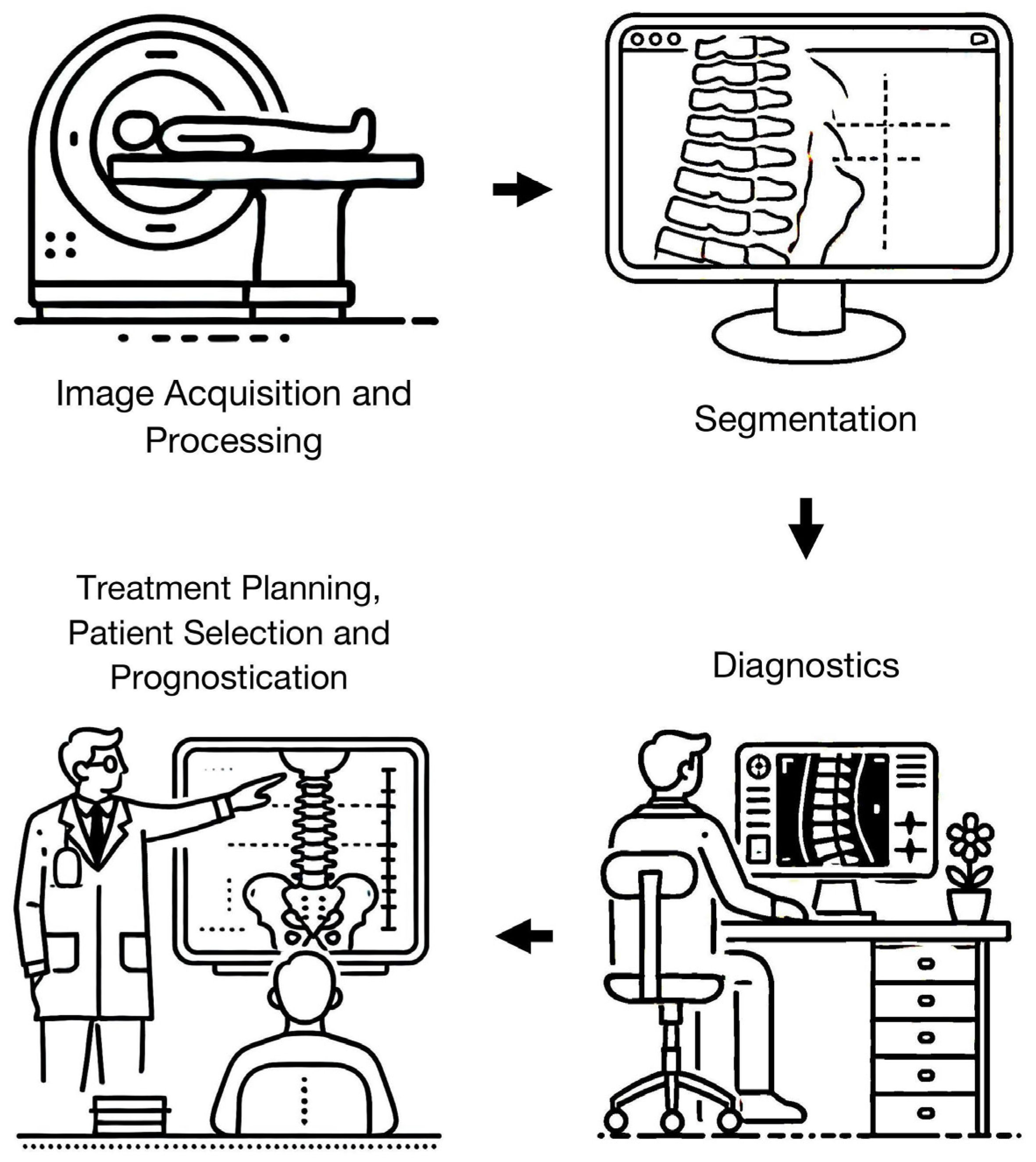
3. Results
3.1. Search Results
3.2. Image Acquisition and Processing
3.3. Segmentation
3.4. Diagnostics
3.4.1. Degenerative Disease
3.4.2. Neoplastic Diseases
3.4.3. Infection
3.4.4. Trauma
3.4.5. Spondyloarthropathy
3.5. Treatment Planning, Patient Selection, and Prognostication
3.6. Others
3.7. AI and ML Techniques
4. Discussion
4.1. Generalizability
4.2. Implementation
4.3. Study Limitations
4.4. Proposed Areas of Future Research
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, G.-U.; Chang, M.C.; Kim, T.U.; Lee, G.W. Diagnostic Modality in Spine Disease: A Review. Asian Spine J. 2020, 14, 910–920. [Google Scholar] [CrossRef] [PubMed]
- Leone, A.; Guglielmi, G.; Cassar-Pullicino, V.N.; Bonomo, L. Lumbar Intervertebral Instability: A Review. Radiology 2007, 245, 62–77. [Google Scholar] [CrossRef] [PubMed]
- Blackmore, C.C.; Mann, F.A.; Wilson, A.J. Helical CT in the Primary Trauma Evaluation of the Cervical Spine: An Evidence-Based Approach. Skelet. Radiol. 2000, 29, 632–639. [Google Scholar] [CrossRef]
- Selopranoto, U.S.; Soo, M.Y.; Fearnside, M.R.; Cummine, J.L. Ossification of the Posterior Longitudinal Ligament of the Cervical Spine. J. Clin. Neurosci. 1997, 4, 209–217. [Google Scholar] [CrossRef]
- Hartley, K.G.; Damon, B.M.; Patterson, G.T.; Long, J.H.; Holt, G.E. MRI Techniques: A Review and Update for the Orthopaedic Surgeon. J. Am. Acad. Orthop. Surg. 2012, 20, 775–787. [Google Scholar] [CrossRef]
- Alyas, F.; Saifuddin, A.; Connell, D. MR Imaging Evaluation of the Bone Marrow and Marrow Infiltrative Disorders of the Lumbar Spine. Magn. Reson. Imaging Clin. N. Am. 2007, 15, 199–219. [Google Scholar] [CrossRef] [PubMed]
- Henninger, B.; Kaser, V.; Ostermann, S.; Spicher, A.; Zegg, M.; Schmid, R.; Kremser, C.; Krappinger, D. Cervical Disc and Ligamentous Injury in Hyperextension Trauma: MRI and Intraoperative Correlation. J. Neuroimaging 2020, 30, 104–109. [Google Scholar] [CrossRef]
- Landman, J.A.; Hoffman, J.C., Jr.; Braun, I.F.; Barrow, D.L. Value of Computed Tomographic Myelography in the Recognition of Cervical Herniated Disk. AJNR Am. J. Neuroradiol. 1984, 5, 391–394. [Google Scholar]
- Runge, V.M.; Richter, J.K.; Heverhagen, J.T. Speed in Clinical Magnetic Resonance. Investig. Radiol. 2017, 52, 1–17. [Google Scholar] [CrossRef]
- Nölte, I.; Gerigk, L.; Brockmann, M.A.; Kemmling, A.; Groden, C. MRI of Degenerative Lumbar Spine Disease: Comparison of Non-Accelerated and Parallel Imaging. Neuroradiology 2008, 50, 403–409. [Google Scholar] [CrossRef]
- Gao, T.; Lu, Z.; Wang, F.; Zhao, H.; Wang, J.; Pan, S. Using the Compressed Sensing Technique for Lumbar Vertebrae Imaging: Comparison with Conventional Parallel Imaging. Curr. Med. Imaging Rev. 2021, 17, 1010–1017. [Google Scholar] [CrossRef] [PubMed]
- Hajiahmadi, S.; Shayganfar, A.; Askari, M.; Ebrahimian, S. Interobserver and Intraobserver Variability in Magnetic Resonance Imaging Evaluation of Patients with Suspected Disc Herniation. Heliyon 2020, 6, e05201. [Google Scholar] [CrossRef]
- SITNFlash. The History of Artificial Intelligence. Science in the News. Available online: https://sitn.hms.harvard.edu/flash/2017/history-artificial-intelligence/ (accessed on 16 June 2024).
- European Society of Radiology (ESR). What the Radiologist Should Know about Artificial Intelligence—An ESR White Paper. Insights Imaging 2019, 10, 44. [Google Scholar] [CrossRef] [PubMed]
- Noguerol, M.; Paulano-Godino, T.; Martín-Valdivia, F.; Menias, M.T.; Luna, C.O. Strengths, Weaknesses, Opportunities, and Threats Analysis of Artificial Intelligence and Machine Learning Applications in Radiology. J. Am. Coll. Radiol. 2019, 16 Pt B, 1239–1247. [Google Scholar] [CrossRef]
- Khan, S.A.; Hussain, S.; Yang, S. Contrast Enhancement of Low-Contrast Medical Images Using Modified Contrast Limited Adaptive Histogram Equalization. J. Med. Imaging Health Inform. 2020, 10, 1795–1803. [Google Scholar] [CrossRef]
- Khan, S.A.; Khan, M.A.; Song, O.-Y.; Nazir, M. Medical Imaging Fusion Techniques: A Survey Benchmark Analysis, Open Challenges and Recommendations. J. Med. Imaging Health Inform. 2020, 10, 2523–2531. [Google Scholar] [CrossRef]
- Nouman Noor, M.; Nazir, M.; Khan, S.A.; Song, O.-Y.; Ashraf, I. Efficient Gastrointestinal Disease Classification Using Pretrained Deep Convolutional Neural Network. Electronics 2023, 12, 1557. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, Y.; Wang, K.; Li, J.; Zhang, X.; Zhang, Y.; Li, J.; Wang, X. A Quantitative Evaluation of the Deep Learning Model of Segmentation and Measurement of Cervical Spine MRI in Healthy Adults. J. Appl. Clin. Med. Phys. 2024, 25, e14282. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Yang, Y.; Jiang, Z.; Zhang, K.; Zhang, X.; Lin, Y.; Shen, Y.; Jia, X.; Liu, H.; Yang, S.; et al. MRI Radiomics-Based Decision Support Tool for a Personalized Classification of Cervical Disc Degeneration: A Two-Center Study. Front. Physiol. 2023, 14, 1281506. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-N.; Liu, G.; Wang, L.; Chen, C.; Wang, Z.; Zhu, S.; Wan, W.-T.; Weng, Y.-Z.; Lu, W.W.; Li, Z.-Y.; et al. A Deep-Learning Model for Diagnosing Fresh Vertebral Fractures on Magnetic Resonance Images. World Neurosurg. 2024, 183, e818–e824. [Google Scholar] [CrossRef]
- Awan, K.M.; Goncalves Filho, A.L.M.; Tabari, A.; Applewhite, B.P.; Lang, M.; Lo, W.-C.; Sellers, R.; Kollasch, P.; Clifford, B.; Nickel, D.; et al. Diagnostic Evaluation of Deep Learning Accelerated Lumbar Spine MRI. Neuroradiol. J. 2024, 37, 323–331. [Google Scholar] [CrossRef]
- Lin, Y.; Chan, S.C.W.; Chung, H.Y.; Lee, K.H.; Cao, P. A Deep Neural Network for MRI Spinal Inflammation in Axial Spondyloarthritis. Eur. Spine J. 2024. ahead of print. [Google Scholar] [CrossRef]
- Kowlagi, N.; Kemppainen, A.; Panfilov, E.; McSweeney, T.; Saarakkala, S.; Nevalainen, M.; Niinimäki, J.; Karppinen, J.; Tiulpin, A. Semiautomatic Assessment of Facet Tropism from Lumbar Spine MRI Using Deep Learning: A Northern Finland Birth Cohort Study. Spine 2024, 49, 630–639. [Google Scholar] [CrossRef] [PubMed]
- Qu, Z.; Deng, B.; Sun, W.; Yang, R.; Feng, H. A Convolutional Neural Network for Automated Detection of Cervical Ossification of the Posterior Longitudinal Ligament Using Magnetic Resonance Imaging. Clin. Spine Surg. 2024, 37, E106–E112. [Google Scholar] [CrossRef]
- Kim, D.K.; Lee, S.-Y.; Lee, J.; Huh, Y.J.; Lee, S.; Lee, S.; Jung, J.-Y.; Lee, H.-S.; Benkert, T.; Park, S.-H. Deep Learning-Based k-Space-to-Image Reconstruction and Super Resolution for Diffusion-Weighted Imaging in Whole-Spine MRI. Magn. Reson. Imaging 2024, 105, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Wang, L.; You, S.-N.; Wang, Z.; Zhu, S.; Chen, C.; Ma, X.-L.; Yang, L.; Zhang, S.; Yang, Q. Automatic Detection and Classification of Modic Changes in MRI Images Using Deep Learning: Intelligent Assisted Diagnosis System. Orthop. Surg. 2024, 16, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.W.; Khil, E.K.; Lee, K.Y.; Choi, I.; Yoon, Y.S.; Cha, J.G.; Lee, J.H.; Kim, H.; Lee, S.Y. Deep Learning System for Automated Detection of Posterior Ligamentous Complex Injury in Patients with Thoracolumbar Fracture on MRI. Sci. Rep. 2023, 13, 19017. [Google Scholar] [CrossRef] [PubMed]
- Vitale, J.; Sconfienza, L.M.; Galbusera, F. Cross-Sectional Area and Fat Infiltration of the Lumbar Spine Muscles in Patients with Back Disorders: A Deep Learning-Based Big Data Analysis. Eur. Spine J. 2024, 33, 1–10. [Google Scholar] [CrossRef]
- Chen, Y.; Qin, S.; Zhao, W.; Wang, Q.; Liu, K.; Xin, P.; Yuan, H.; Zhuang, H.; Lang, N. MRI Feature-Based Radiomics Models to Predict Treatment Outcome after Stereotactic Body Radiotherapy for Spinal Metastases. Insights Imaging 2023, 14, 169. [Google Scholar] [CrossRef]
- Saravi, B.; Zink, A.; Ülkümen, S.; Couillard-Despres, S.; Wollborn, J.; Lang, G.; Hassel, F. Clinical and Radiomics Feature-Based Outcome Analysis in Lumbar Disc Herniation Surgery. BMC Musculoskelet. Disord. 2023, 24, 791. [Google Scholar] [CrossRef]
- Haim, O.; Agur, A.; Gabay, S.; Azolai, L.; Shutan, I.; Chitayat, M.; Katirai, M.; Sadon, S.; Artzi, M.; Lidar, Z. Differentiating Spinal Pathologies by Deep Learning Approach. Spine J. 2024, 24, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Chen, Z.; Su, Z.; Wang, Z.; Hai, J.; Huang, C.; Wang, Y.; Yan, B.; Lu, H. Deep Learning-Based Detection and Classification of Lumbar Disc Herniation on Magnetic Resonance Images. JOR Spine 2023, 6, e1276. [Google Scholar] [CrossRef] [PubMed]
- Tas, N.P.; Kaya, O.; Macin, G.; Tasci, B.; Dogan, S.; Tuncer, T. ASNET: A Novel AI Framework for Accurate Ankylosing Spondylitis Diagnosis from MRI. Biomedicines 2023, 11, 2441. [Google Scholar] [CrossRef]
- Masse-Gignac, N.; Flórez-Jiménez, S.; Mac-Thiong, J.-M.; Duong, L. Attention-Gated U-Net Networks for Simultaneous Axial/Sagittal Planes Segmentation of Injured Spinal Cords. J. Appl. Clin. Med. Phys. 2023, 24, e14123. [Google Scholar] [CrossRef] [PubMed]
- Yilizati-Yilihamu, E.E.; Yang, J.; Yang, Z.; Rong, F.; Feng, S. A Spine Segmentation Method Based on Scene Aware Fusion Network. BMC Neurosci. 2023, 24, 49. [Google Scholar] [CrossRef]
- Wang, W.; Fan, Z.; Zhen, J. MRI Radiomics-Based Evaluation of Tuberculous and Brucella Spondylitis. J. Int. Med. Res. 2023, 51, 3000605231195156. [Google Scholar] [CrossRef]
- Niemeyer, F.; Galbusera, F.; Tao, Y.; Phillips, F.M.; An, H.S.; Louie, P.K.; Samartzis, D.; Wilke, H.-J. Deep Phenotyping the Cervical Spine: Automatic Characterization of Cervical Degenerative Phenotypes Based on T2-Weighted MRI. Eur. Spine J. 2023, 32, 3846–3856. [Google Scholar] [CrossRef]
- Cai, J.; Shen, C.; Yang, T.; Jiang, Y.; Ye, H.; Ruan, Y.; Zhu, X.; Liu, Z.; Liu, Q. MRI-Based Radiomics Assessment of the Imminent New Vertebral Fracture after Vertebral Augmentation. Eur. Spine J. 2023, 32, 3892–3905. [Google Scholar] [CrossRef]
- Waldenberg, C.; Brisby, H.; Hebelka, H.; Lagerstrand, K.M. Associations between Vertebral Localized Contrast Changes and Adjacent Annular Fissures in Patients with Low Back Pain: A Radiomics Approach. J. Clin. Med. 2023, 12, 4891. [Google Scholar] [CrossRef]
- Roberts, M.; Hinton, G.; Wells, A.J.; Van Der Veken, J.; Bajger, M.; Lee, G.; Liu, Y.; Chong, C.; Poonnoose, S.; Agzarian, M.; et al. Imaging Evaluation of a Proposed 3D Generative Model for MRI to CT Translation in the Lumbar Spine. Spine J. 2023, 23, 1602–1612. [Google Scholar] [CrossRef]
- Tanenbaum, L.N.; Bash, S.C.; Zaharchuk, G.; Shankaranarayanan, A.; Chamberlain, R.; Wintermark, M.; Beaulieu, C.; Novick, M.; Wang, L. Deep Learning-Generated Synthetic MR Imaging STIR Spine Images Are Superior in Image Quality and Diagnostically Equivalent to Conventional STIR: A Multicenter, Multireader Trial. AJNR Am. J. Neuroradiol. 2023, 44, 987–993. [Google Scholar] [CrossRef] [PubMed]
- Küçükçiloğlu, Y.; Şekeroğlu, B.; Adalı, T.; Şentürk, N. Prediction of Osteoporosis Using MRI and CT Scans with Unimodal and Multimodal Deep-Learning Models. Diagn. Interv. Radiol. 2024, 30, 9–20. [Google Scholar] [CrossRef]
- Chiu, P.-F.; Chang, R.C.-H.; Lai, Y.-C.; Wu, K.-C.; Wang, K.-P.; Chiu, Y.-P.; Ji, H.-R.; Kao, C.-H.; Chiu, C.-D. Machine Learning Assisting the Prediction of Clinical Outcomes Following Nucleoplasty for Lumbar Degenerative Disc Disease. Diagnostics 2023, 13, 1863. [Google Scholar] [CrossRef]
- Mohanty, R.; Allabun, S.; Solanki, S.S.; Pani, S.K.; Alqahtani, M.S.; Abbas, M.; Soufiene, B.O. NAMSTCD: A Novel Augmented Model for Spinal Cord Segmentation and Tumor Classification Using Deep Nets. Diagnostics 2023, 13, 1417. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Jin, Y.; Feng, S.; Yu, H.; Zhang, Y.; Li, Y. Benign vs Malignant Vertebral Compression Fractures with MRI: A Comparison between Automatic Deep Learning Network and Radiologist’s Assessment. Eur. Radiol. 2023, 33, 5060–5068. [Google Scholar] [CrossRef] [PubMed]
- Liawrungrueang, W.; Kim, P.; Kotheeranurak, V.; Jitpakdee, K.; Sarasombath, P. Automatic Detection, Classification, and Grading of Lumbar Intervertebral Disc Degeneration Using an Artificial Neural Network Model. Diagnostics 2023, 13, 663. [Google Scholar] [CrossRef]
- Mukaihata, T.; Maki, S.; Eguchi, Y.; Geundong, K.; Shoda, J.; Yokota, H.; Orita, S.; Shiga, Y.; Inage, K.; Furuya, T.; et al. Differentiating Magnetic Resonance Images of Pyogenic Spondylitis and Spinal Modic Change Using a Convolutional Neural Network. Spine 2023, 48, 288–294. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, Z.; Zhang, J.; Duan, Y.; Qu, L.; Feng, C.; Huang, X.; Cheng, D.; Xu, X.; Sun, T.; Li, Z.; et al. Automated Classification of Intramedullary Spinal Cord Tumors and Inflammatory Demyelinating Lesions Using Deep Learning. Radiol. Artif. Intell. 2022, 4, e210292. [Google Scholar] [CrossRef] [PubMed]
- Kashiwagi, N.; Sakai, M.; Tsukabe, A.; Yamashita, Y.; Fujiwara, M.; Yamagata, K.; Nakamoto, A.; Nakanishi, K.; Tomiyama, N. Ultrafast Cervical Spine MRI Protocol Using Deep Learning-Based Reconstruction: Diagnostic Equivalence to a Conventional Protocol. Eur. J. Radiol. 2022, 156, 110531. [Google Scholar] [CrossRef]
- Chen, K.; Cao, J.; Zhang, X.; Wang, X.; Zhao, X.; Li, Q.; Chen, S.; Wang, P.; Liu, T.; Du, J.; et al. Differentiation between Spinal Multiple Myeloma and Metastases Originated from Lung Using Multi-View Attention-Guided Network. Front. Oncol. 2022, 12, 981769. [Google Scholar] [CrossRef]
- Alanazi, A.H.; Cradock, A.; Rainford, L. Development of Lumbar Spine MRI Referrals Vetting Models Using Machine Learning and Deep Learning Algorithms: Comparison Models vs. Healthcare Professionals. Radiography 2022, 28, 674–683. [Google Scholar] [CrossRef] [PubMed]
- Lim, D.S.W.; Makmur, A.; Zhu, L.; Zhang, W.; Cheng, A.J.L.; Sia, D.S.Y.; Eide, S.E.; Ong, H.Y.; Jagmohan, P.; Tan, W.C.; et al. Improved Productivity Using Deep Learning-Assisted Reporting for Lumbar Spine MRI. Radiology 2022, 305, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Hallinan, J.T.P.D.; Zhu, L.; Zhang, W.; Lim, D.S.W.; Baskar, S.; Low, X.Z.; Yeong, K.Y.; Teo, E.C.; Kumarakulasinghe, N.B.; Yap, Q.V.; et al. Deep Learning Model for Classifying Metastatic Epidural Spinal Cord Compression on MRI. Front. Oncol. 2022, 12, 849447. [Google Scholar] [CrossRef] [PubMed]
- Suri, A.; Jones, B.C.; Ng, G.; Anabaraonye, N.; Beyrer, P.; Domi, A.; Choi, G.; Tang, S.; Terry, A.; Leichner, T.; et al. Vertebral Deformity Measurements at MRI, CT, and Radiography Using Deep Learning. Radiol. Artif. Intell. 2022, 4, e210015. [Google Scholar] [CrossRef]
- Zhang, M.-Z.; Ou-Yang, H.-Q.; Liu, J.-F.; Jin, D.; Wang, C.-J.; Ni, M.; Liu, X.-G.; Lang, N.; Jiang, L.; Yuan, H.-S. Predicting Postoperative Recovery in Cervical Spondylotic Myelopathy: Construction and Interpretation of T2*-Weighted Radiomic-Based Extra Trees Models. Eur. Radiol. 2022, 32, 3565–3575. [Google Scholar] [CrossRef] [PubMed]
- Hwang, E.-J.; Kim, S.; Jung, J.-Y. Fully Automated Segmentation of Lumbar Bone Marrow in Sagittal, High-Resolution T1-Weighted Magnetic Resonance Images Using 2D U-NET. Comput. Biol. Med. 2022, 140, 105105. [Google Scholar] [CrossRef] [PubMed]
- Jujjavarapu, C.; Pejaver, V.; Cohen, T.A.; Mooney, S.D.; Heagerty, P.J.; Jarvik, J.G. A Comparison of Natural Language Processing Methods for the Classification of Lumbar Spine Imaging Findings Related to Lower Back Pain. Acad. Radiol. 2022, 29 (Suppl. S3), S188–S200. [Google Scholar] [CrossRef]
- Gotoh, M.; Nakaura, T.; Funama, Y.; Morita, K.; Sakabe, D.; Uetani, H.; Nagayama, Y.; Kidoh, M.; Hatemura, M.; Masuda, T.; et al. Virtual Magnetic Resonance Lumbar Spine Images Generated from Computed Tomography Images Using Conditional Generative Adversarial Networks. Radiography 2022, 28, 447–453. [Google Scholar] [CrossRef]
- Goedmakers, C.M.W.; Lak, A.M.; Duey, A.H.; Senko, A.W.; Arnaout, O.; Groff, M.W.; Smith, T.R.; Vleggeert-Lankamp, C.L.A.; Zaidi, H.A.; Rana, A.; et al. Deep Learning for Adjacent Segment Disease at Preoperative MRI for Cervical Radiculopathy. Radiology 2021, 301, E446. [Google Scholar] [CrossRef]
- Lemay, A.; Gros, C.; Zhuo, Z.; Zhang, J.; Duan, Y.; Cohen-Adad, J.; Liu, Y. Automatic Multiclass Intramedullary Spinal Cord Tumor Segmentation on MRI with Deep Learning. NeuroImage Clin. 2021, 31, 102766. [Google Scholar] [CrossRef]
- Liu, J.; Wang, C.; Guo, W.; Zeng, P.; Liu, Y.; Lang, N.; Yuan, H. A Preliminary Study Using Spinal MRI-Based Radiomics to Predict High-Risk Cytogenetic Abnormalities in Multiple Myeloma. Radiol. Med. 2021, 126, 1226–1235. [Google Scholar] [CrossRef] [PubMed]
- Merali, Z.; Wang, J.Z.; Badhiwala, J.H.; Witiw, C.D.; Wilson, J.R.; Fehlings, M.G. A Deep Learning Model for Detection of Cervical Spinal Cord Compression in MRI Scans. Sci. Rep. 2021, 11, 10473. [Google Scholar] [CrossRef] [PubMed]
- Hallinan, J.T.P.D.; Zhu, L.; Yang, K.; Makmur, A.; Algazwi, D.A.R.; Thian, Y.L.; Lau, S.; Choo, Y.S.; Eide, S.E.; Yap, Q.V.; et al. Deep Learning Model for Automated Detection and Classification of Central Canal, Lateral Recess, and Neural Foraminal Stenosis at Lumbar Spine MRI. Radiology 2021, 300, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Maki, S.; Furuya, T.; Horikoshi, T.; Yokota, H.; Mori, Y.; Ota, J.; Kawasaki, Y.; Miyamoto, T.; Norimoto, M.; Okimatsu, S.; et al. A Deep Convolutional Neural Network with Performance Comparable to Radiologists for Differentiating between Spinal Schwannoma and Meningioma. Spine 2020, 45, 694–700. [Google Scholar] [CrossRef]
- Gaonkar, B.; Beckett, J.; Villaroman, D.; Ahn, C.; Edwards, M.; Moran, S.; Attiah, M.; Babayan, D.; Ames, C.; Villablanca, J.P.; et al. Quantitative Analysis of Neural Foramina in the Lumbar Spine: An Imaging Informatics and Machine Learning Study. Radiol. Artif. Intell. 2019, 1, 180037. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Kim, S.; Lee, Y.H.; Lee, S.H.; Lee, H.S.; Kim, S. Performance of the Deep Convolutional Neural Network Based Magnetic Resonance Image Scoring Algorithm for Differentiating between Tuberculous and Pyogenic Spondylitis. Sci. Rep. 2018, 8, 13124. [Google Scholar] [CrossRef] [PubMed]
- Jamaludin, A.; Kadir, T.; Zisserman, A. SpineNet: Automated Classification and Evidence Visualization in Spinal MRIs. Med. Image Anal. 2017, 41, 63–73. [Google Scholar] [CrossRef]
- Pfirrmann, C.W.; Metzdorf, A.; Zanetti, M.; Hodler, J.; Boos, N. Magnetic Resonance Classification of Lumbar Intervertebral Disc Degeneration. Spine 2001, 26, 1873–1878. [Google Scholar] [CrossRef]
- Kumar, N.; Tan, W.L.B.; Wei, W.; Vellayappan, B.A. An Overview of the Tumors Affecting the Spine-inside to Out. Neuro-Oncol. Pract. 2020, 7 (Suppl. S1), i10–i17. [Google Scholar] [CrossRef]
- Hallinan, J.T.P.D.; Zhu, L.; Zhang, W.; Kuah, T.; Lim, D.S.W.; Low, X.Z.; Cheng, A.J.L.; Eide, S.E.; Ong, H.Y.; Muhamat Nor, F.E.; et al. Deep Learning Model for Grading Metastatic Epidural Spinal Cord Compression on Staging CT. Cancers 2022, 14, 3219. [Google Scholar] [CrossRef]
- Hallinan, J.T.P.D.; Zhu, L.; Zhang, W.; Ge, S.; Muhamat Nor, F.E.; Ong, H.Y.; Eide, S.E.; Cheng, A.J.L.; Kuah, T.; Lim, D.S.W.; et al. Deep Learning Assessment Compared to Radiologist Reporting for Metastatic Spinal Cord Compression on CT. Front. Oncol. 2023, 13, 1151073. [Google Scholar] [CrossRef] [PubMed]
- Hallinan, J.T.P.D.; Zhu, L.; Tan, H.W.N.; Hui, S.J.; Lim, X.; Ong, B.W.L.; Ong, H.Y.; Eide, S.E.; Cheng, A.J.L.; Ge, S.; et al. A Deep Learning-Based Technique for the Diagnosis of Epidural Spinal Cord Compression on Thoracolumbar CT. Eur. Spine J. 2023, 32, 3815–3824. [Google Scholar] [CrossRef]
- Kiryu, S.; Akai, H.; Yasaka, K.; Tajima, T.; Kunimatsu, A.; Yoshioka, N.; Akahane, M.; Abe, O.; Ohtomo, K. Clinical Impact of Deep Learning Reconstruction in MRI. Radiographics 2023, 43, e220133. [Google Scholar] [CrossRef]
- Antun, V.; Renna, F.; Poon, C.; Adcock, B.; Hansen, A.C. On Instabilities of Deep Learning in Image Reconstruction and the Potential Costs of AI. Proc. Natl. Acad. Sci. USA 2020, 117, 30088–30095. [Google Scholar] [CrossRef] [PubMed]
- Hsu, W.; Hoyt, A.C. Using Time as a Measure of Impact for AI Systems: Implications in Breast Screening. Radiol. Artif. Intell. 2019, 1, e190107. [Google Scholar] [CrossRef] [PubMed]
- Avanzo, M.; Wei, L.; Stancanello, J.; Vallières, M.; Rao, A.; Morin, O.; Mattonen, S.A.; El Naqa, I. Machine and Deep Learning Methods for Radiomics. Med. Phys. 2020, 47, e185–e202. [Google Scholar] [CrossRef]
- Willems, P.; de Bie, R.; Öner, C.; Castelein, R.; de Kleuver, M. Clinical Decision Making in Spinal Fusion for Chronic Low Back Pain. Results of a Nationwide Survey among Spine Surgeons. BMJ Open 2011, 1, e000391. [Google Scholar] [CrossRef]
- Fairbank, J.; Frost, H.; Wilson-MacDonald, J.; Yu, L.-M.; Barker, K.; Collins, R.; Spine Stabilisation Trial Group. Randomised Controlled Trial to Compare Surgical Stabilisation of the Lumbar Spine with an Intensive Rehabilitation Programme for Patients with Chronic Low Back Pain: The MRC Spine Stabilisation Trial. BMJ 2005, 330, 1233. [Google Scholar] [CrossRef]
- Azad, T.D.; Zhang, Y.; Weiss, H.; Alamin, T.; Cheng, I.; Huang, B.; Veeravagu, A.; Ratliff, J.; Malhotra, N.R. Fostering Reproducibility and Generalizability in Machine Learning for Clinical Prediction Modeling in Spine Surgery. Spine J. 2021, 21, 1610–1616. [Google Scholar] [CrossRef] [PubMed]
- Eche, T.; Schwartz, L.H.; Mokrane, F.-Z.; Dercle, L. Toward Generalizability in the Deployment of Artificial Intelligence in Radiology: Role of Computation Stress Testing to Overcome Underspecification. Radiol. Artif. Intell. 2021, 3, e210097. [Google Scholar] [CrossRef]
- Huisman, M.; Hannink, G. The AI Generalization Gap: One Size Does Not Fit All. Radiol. Artif. Intell. 2023, 5, e230246. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Jia, X.; Mei, Z.; Gu, X.; Lu, Y.; Fu, C.-C.; Zhang, R.; Gu, Y.; Chen, X.; Luo, X.; et al. Chinese Artificial Intelligence Alliance for Thyroid and Breast Ultrasound. Generalizability and Diagnostic Performance of AI Models for Thyroid US. Radiology 2023, 307, e221157. [Google Scholar] [CrossRef]
- RSNA Lumbar Spine Degenerative Classification AI Challenge. 2024. Rsna.org. Available online: https://www.rsna.org/rsnai/ai-image-challenge/lumbar-spine-degenerative-classification-ai-challenge (accessed on 12 July 2024).
- Kim, H.E.; Cosa-Linan, A.; Santhanam, N.; Jannesari, M.; Maros, M.E.; Ganslandt, T. Transfer Learning for Medical Image Classification: A Literature Review. BMC Med. Imaging 2022, 22, 69. [Google Scholar] [CrossRef]
- Xuan, J.; Ke, B.; Ma, W.; Liang, Y.; Hu, W. Spinal Disease Diagnosis Assistant Based on MRI Images Using Deep Transfer Learning Methods. Front. Public Health 2023, 11, 1044525. [Google Scholar] [CrossRef] [PubMed]
- Santomartino, S.M.; Putman, K.; Beheshtian, E.; Parekh, V.S.; Yi, P.H. Evaluating the Robustness of a Deep Learning Bone Age Algorithm to Clinical Image Variation Using Computational Stress Testing. Radiol. Artif. Intell. 2024, 6, e230240. [Google Scholar] [CrossRef] [PubMed]
- Brady, A.P.; Allen, B.; Chong, J.; Kotter, E.; Kottler, N.; Mongan, J.; Oakden-Rayner, L.; Pinto Dos Santos, D.; Tang, A.; Wald, C.; et al. Developing, Purchasing, Implementing and Monitoring AI Tools in Radiology: Practical Considerations. A Multi-Society Statement from the ACR, CAR, ESR, RANZCR and RSNA. J. Am. Coll. Radiol. 2021, 18, 710–717. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Romeijn, S.; van Buchem, M.; Mehrizi, M.H.R.; Grootjans, W. A Holistic Approach to Implementing Artificial Intelligence in Radiology. Insights Imaging 2024, 15, 22. [Google Scholar] [CrossRef]
- Suran, M.; Hswen, Y. How to Navigate the Pitfalls of AI Hype in Health Care. JAMA 2024, 331, 273–276. [Google Scholar] [CrossRef] [PubMed]
- Geis, J.R.; Brady, A.; Wu, C.C.; Spencer, J.; Ranschaert, E.; Jaremko, J.L.; Langer, S.G.; Kitts, A.B.; Birch, J.; Shields, W.F.; et al. Ethics of Artificial Intelligence in Radiology: Summary of the Joint European and North American Multisociety Statement. Insights Imaging 2019, 10, 101. [Google Scholar] [CrossRef]
- Jaremko, J.L.; Azar, M.; Bromwich, R.; Lum, A.; Alicia Cheong, L.H.; Gilbert, M.; Laviolette, F.; Gray, B.; Reinhold, C.; Cicero, M.; et al. Canadian Association of Radiologists White Paper on Ethical and Legal Issues Related to Artificial Intelligence in Radiology. Can. Assoc. Radiol. J. 2019, 70, 107–118. [Google Scholar] [CrossRef]
- Plackett, B. The Rural Areas Missing out on AI Opportunities. Nature 2022, 610, S17. [Google Scholar] [CrossRef]
- Celi, L.A.; Cellini, J.; Charpignon, M.-L.; Dee, E.C.; Dernoncourt, F.; Eber, R.; Mitchell, W.G.; Moukheiber, L.; Schirmer, J.; Situ, J.; et al. Sources of Bias in Artificial Intelligence That Perpetuate Healthcare Disparities-A Global Review. PLoS Digit. Health 2022, 1, e0000022. [Google Scholar] [CrossRef]
- Eltawil, F.A.; Atalla, M.; Boulos, E.; Amirabadi, A.; Tyrrell, P.N. Analyzing Barriers and Enablers for the Acceptance of Artificial Intelligence Innovations into Radiology Practice: A Scoping Review. Tomography 2023, 9, 1443–1455. [Google Scholar] [CrossRef]
- Borondy Kitts, A. Patient Perspectives on Artificial Intelligence in Radiology. J. Am. Coll. Radiol. 2023, 20, 243–250. [Google Scholar] [CrossRef]
- Brima, Y.; Atemkeng, M. Saliency-Driven Explainable Deep Learning in Medical Imaging: Bridging Visual Explainability and Statistical Quantitative Analysis. BioData Min. 2024, 17, 18. [Google Scholar] [CrossRef] [PubMed]
- Moor, M.; Banerjee, O.; Abad, Z.S.H.; Krumholz, H.M.; Leskovec, J.; Topol, E.J.; Rajpurkar, P. Foundation Models for Generalist Medical Artificial Intelligence. Nature 2023, 616, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Hafezi-Nejad, N.; Trivedi, P. Foundation AI Models and Data Extraction from Unlabeled Radiology Reports: Navigating Uncharted Territory. Radiology 2023, 308, e232308. [Google Scholar] [CrossRef] [PubMed]
- Seah, J.; Tang, C.; Buchlak, Q.D.; Holt, X.G.; Wardman, J.B.; Aimoldin, A. Effect of a Comprehensive Deep-Learning Model on the Accuracy of Chest X-ray Interpretation by Radiologists: A Retrospective, Multireader Multicase Study. Lancet Digit. Health 2021, 3, e496–e506. [Google Scholar] [CrossRef]
- van Beek, E.J.R.; Ahn, J.S.; Kim, M.J.; Murchison, J.T. Validation Study of Machine-Learning Chest Radiograph Software in Primary and Emergency Medicine. Clin. Radiol. 2023, 78, 1–7. [Google Scholar] [CrossRef]
- Niehoff, J.H.; Kalaitzidis, J.; Kroeger, J.R.; Schoenbeck, D.; Borggrefe, J.; Michael, A.E. Evaluation of the Clinical Performance of an AI-Based Application for the Automated Analysis of Chest X-rays. Sci. Rep. 2023, 13, 3680. [Google Scholar] [CrossRef]
- Hayashi, D. Deep Learning for Lumbar Spine MRI Reporting: A Welcome Tool for Radiologists. Radiology 2021, 300, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Gertz, R.J.; Bunck, A.C.; Lennartz, S. GPT-4 for Automated Determination of Radiological Study and Protocol Based on Radiology Request Forms: A Feasibility Study. Radiology 2023, 307, e230877. [Google Scholar] [CrossRef] [PubMed]
- Beddiar, D.-R.; Oussalah, M.; Seppänen, T. Automatic Captioning for Medical Imaging (MIC): A Rapid Review of Literature. Artif. Intell. Rev. 2023, 56, 4019–4076. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Ong, H.; Kennedy, P. Evaluating GPT4 on Impressions Generation in Radiology Reports. Radiology 2023, 307, e231259. [Google Scholar] [CrossRef]
- Ayers, J.W.; Poliak, A.; Dredze, M. Comparing Physician and Artificial Intelligence Chatbot Responses to Patient Questions Posted to a Public Social Media Forum. JAMA Intern. Med. 2023, 183, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Kuckelman, I.J.; Yi, P.H.; Bui, M.; Onuh, I.; Anderson, J.A.; Ross, A.B. Assessing AI-Powered Patient Education: A Case Study in Radiology. Acad. Radiol. 2024, 31, 338–342. [Google Scholar] [CrossRef]
- Wu, J.; Kim, Y.; Keller, E.C.; Chow, J.; Levine, A.P.; Pontikos, N.; Ibrahim, Z.; Taylor, P.; Williams, M.C.; Wu, H. Exploring Multimodal Large Language Models for Radiology Report Error-Checking. arXiv 2023, arXiv:2312.13103. [Google Scholar]
- Yu, F.; Moehring, A.; Banerjee, O.; Salz, T.; Agarwal, N.; Rajpurkar, P. Heterogeneity and Predictors of the Effects of AI Assistance on Radiologists. Nat. Med. 2024, 30, 837–849. [Google Scholar] [CrossRef]

| No | Study Title | Authorship | Year of Publication | Journal Name | Application and Primary Outcome Measure | Sample Size * | Spine Region Studied | MRI Sequences Used | Artificial Intelligence Technique Used | Key Results and Conclusion |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | A quantitative evaluation of the deep learning model of segmentation and measurement of cervical spine MRI in healthy adults | Y Zhu et al. [19] | 2024 | J Appl Clin Med Phys | Segmentation of cervical spine structures (subarachnoid space area/diameter, spinal cord area/diameter, anterior and posterior extra-spinal space) | 160 | Cervical | Sagittal T1w, T2w, axial T2w | 3D U-net | No comparative statistics |
| 2 | MRI radiomics-based decision support tool for a personalized classification of cervical disc degeneration: a two-center study | J Xie et al. [20] | 2024 | Front Physiol | Classification of cervical disc degeneration (Pfirrmann grading) | 435 | Cervical | Sagittal T1w, T2w | MedSAM | Disc segmentation Dice 0.93. Random forest overall performance AUC 0.95, accuracy 90%, precision 87% |
| 3 | A deep-learning model for diagnosing fresh vertebral fractures on magnetic resonance images | Y Wang et al. [21] | 2024 | World Neurosurg | Detection of fresh vertebral fractures | 716 | Whole spine | Midsagittal STIR | YoloV7, Resnet 50 | Accuracy 98%, sensitivity 98%, specificity 97%. External dataset accuracy 92% |
| 4 | Diagnostic evaluation of deep learning accelerated lumbar spine MRI | KM Awan et al. [22] | 2024 | Neuroradiol J | Comparison of deep learning accelerated protocol to conventional protocol for neural stenosis and facet arthropathy | 36 | Lumbar | Sagittal T1w, T2w, STIR, axial T2w | CNN | Non-inferior in all aspects however reduced signal-to-noise ratio and increased artifact perception. Interobserver variability κ = 0.50–0.76 |
| 5 | A deep neural network for MRI spinal inflammation in axial spondyloarthritis | Y Lin et al. [23] | 2024 | Eur Spine J | Detection of inflammatory lesions on STIR sequence for patients with axial spondyloarthritis | 330 | Whole spine | Sagittal STIR | U-net | AUC 0.87, sensitivity 80%, specificity 88%, comparable to a radiologist. True positive lesion Dice 0.55. |
| 6 | Semi-automatic assessment of facet tropism from lumbar spine MRI using deep learning: a Northern Finland birth cohort study | N Kowlagi et al. [24] | 2023 | Spine (Phila Pa 1976) | Measurement of facet joint angles | 490 | Lumbar (L3/4 to L5/S1) | Axial T2w | U-net | Dice 0.93, IOU 0.87 |
| 7 | A convolutional neural network for automated detection of cervical ossification of the posterior longitudinal ligament using magnetic resonance imaging | Z Qu et al. [25] | 2023 | Clin Spine Surg | Detection of ossification of posterior longitudinal ligament | 684 | Cervical | Sagittal MRI | ResNet | Accuracy 93–98%, AUC 0.91–0.97. ResNet50 and ResNet101 had higher accuracy and specificity than all human readers |
| 8 | Deep learning-based k-space-to-image reconstruction and super resolution for diffusion-weighted imaging in whole-spine MRI | DK Kim et al. [26] | 2024 | Magn Reson Imaging | K-space-to-image reconstruction for whole spine DWI in patients with hematologic and oncologic diseases | 67 | Whole spine | Axial single-shot echo-planar DWI | CNN | Higher diagnostic confidence scores and overall image quality |
| 9 | Automatic detection and classification of Modic changes in MRI images using deep learning: intelligent assisted diagnosis system | Gang L et al. [27] | 2024 | Orthop Surg | Detection and classification of Modic endplate changes | 168 | Lumbar | Median sagittal T1w and T2w | Single shot multibox detector, ResNet18 | Internal dataset: accuracy 86%, recall 88%, precision 85%, F1-score 86%, interobserver κ = 0.79 (95%CI 0.66–0.85). External dataset: accuracy 75%, recall 77%, precision 78%, F1-score 75%, interobserver κ = 0.68 (95%CI 0.51–0.68) |
| 10 | Deep learning system for automated detection of posterior ligamentous complex injury in patients with thoracolumbar fracture on MRI | SW Jo et al. [28] | 2023 | Sci Rep | Detection of posterior ligamentous complex injury in patients with acute thoracolumbar fractures | 500 | Thoracic and lumbar | Midline sagittal T2w | Attention U-net and Inception-ResNetv2 | AUC 0.92–0.93 (vs. 0.83–0.93 for radiologists) |
| 11 | Cross-sectional area and fat infiltration of the lumbar spine muscles in patients with back disorders: a deep learning-based big data analysis | J Vitale et al. [29] | 2023 | Eur Spine J | Segmentation of lumbar paravertebral muscles and correlation with age | 4434 | Lumbar | Axial T2w | U-net | Higher cross-sectional area in males (p < 0.001). Positive correlation between age and total fat infiltration (r = 0.73, p < 0.001), negligible negative correlation between cross-sectional area and age (r = −0.24, p < 0.001) |
| 12 | MRI feature-based radiomics models to predict treatment outcome after stereotactic body radiotherapy for spinal metastases | Y Chen et al. [30] | 2023 | Insights Imaging | Prediction of treatment outcome after stereotactic body radiotherapy for spine metastasis | 194 | Whole spine | Sagittal T1w, T2w, STIR, axial T2w | Multiple (including AdaBoost, XGBoost, RF, SVM) | Combined model AUC 0.83, clinical model AUC 0.73 |
| 13 | Clinical and radiomics feature-based outcome analysis in lumbar disc herniation surgery | B Saravi et al. [31] | 2023 | BMC Musculoskelet Disord | Combination of radiomics features and clinical features to predict lumbar disc herniation surgery outcomes | 172 | Lumbar | Sagittal T2w | Multiple (including XGBoost, Lagrangian SVM, RF radial basis function neural network) | Accuracy 88–93% (vs. 88–91% for clinical features alone) |
| 14 | Differentiating spinal pathologies by deep learning approach | O Haim et al. [32] | 2024 | Spine J | Differentiation of spinal lesions into infection, carcinoma, meningioma and schwannoma | 231 | Whole spine | Variable (T2w, T1w post-contrast) | Fast.ai | Accuracy 78% (validation), 93% (test) |
| 15 | Deep learning-based detection and classification of lumbar disc herniation on magnetic resonance images | W Zhang et al. [33] | 2023 | JOR Spine | Detection and classification of lumbar disc herniation according to the Michigan State University classification | 1115 | Lumbar | Axial T2w | Faster R-CNN, ResNeXt101 | Internal dataset: detection IOU 0.82, classification accuracy 88%, AUC 0.97, interclass correlation 0.87. External dataset: detection IOU 0.70, classification accuracy 74%, AUC 0.92, interclass correlation 0.79 |
| 16 | ASNET: a novel AI framework for accurate ankylosing spondylitis diagnosis from MRI | NP Tas et al. [34] | 2023 | Biomedicines | Prediction of ankylosing spondylitis diagnosis on MRI | 2036 | Sacroiliac joints | Axial, coronal STIR, coronal T1w post-contrast | DenseNet201, ResNet50, ShuffleNet | Accuracy 100%, recall 100%, precision 100%, F1-score 100% |
| 17 | Attention-gated U-Net networks for simultaneous axial/sagittal planes segmentation of injured spinal cords | N Masse-Gignac et al. [35] | 2023 | J Appl Clin Med Phys | Segmentation of the spinal cord in patients with traumatic injuries | 94 | All (mainly cervical) | Sagittal T2w | U-Net | Dice 0.95 |
| 18 | A spine segmentation method based on scene aware fusion network | EE Yilizati-Yilihamu et al. [36] | 2023 | BMC Neurosci | Segmentation of lumbar spine MRI into individual vertebrae and discs by level | 172 | Lumbar | Sagittal MRI | Scene-Aware Fusion Network (SAFNet) | Dice 0.79–0.81 (average 0.80) |
| 19 | MRI radiomics-based evaluation of Tuberculous and Brucella spondylitis | W Wang et al. [37] | 2023 | J Int Med Res | Differentiation of Tuberculous spondylitis from Brucella spondylitis, and culture positive from culture negative Tuberculous spondylitis | 190 | Whole spine | Sagittal T1w, T2w, fat suppressed | RF, SVM | SVM AUC 0.90–0.94, RF AUC 0.95 |
| 20 | Deep phenotyping the cervical spine: automatic characterization of cervical degenerative phenotypes based on T2-weighted MRI | F Niemeyer et al. [38] | 2023 | Eur Spine J | Classification of cervical spine into degenerative phenotypes based on disc and osteophyte configuration | 873 | Cervical | Sagittal MRI | 3D CNN | Disc κ = 0.55–0.68, disc displacement κ = 0.58–0.74, disc space narrowing κ = 0.65–0.72, osseous abnormalities κ = 0.18–0.49 |
| 21 | MRI-based radiomics assessment of the imminent new vertebral fracture after vertebral augmentation | J Cai et al. [39] | 2023 | Eur Spine J | Evaluation of risk of new vertebral fracture after vertebral augmentation | 168 | Lumbar | T2w | Multiple (logistic regression, RF, SVM, XGBoost) | AUC 0.90–0.93, superior to clinical features alone (p < 0.05) |
| 22 | Associations between vertebral localized contrast changes and adjacent annular fissures in patients with low back pain: a radiomics approach | C Waldenberg et al. [40] | 2023 | J Clin Med | Detection of adjacent level annular fissure based on vertebral changes on MRI | 61 | Lumbar | Sagittal T1w, T2w, discography, CT | Multilayer perceptron, RF, K-nearest neighbor | Accuracy 83%, sensitivity 97%, specificity 28%, AUC 0.76 |
| 23 | Imaging evaluation of a proposed 3D generative model for MRI to CT translation in the lumbar spine | M Roberts et al. [41] | 2023 | Spine J | Generation of 3D CT from sagittal MRI data | 420 | Lumbar | Sagittal T1w | 3D cycle-GAN | Measurements in sagittal plane <10% relative error, axial plane up to 34% relative error |
| 24 | Deep learning-generated synthetic MR imaging STIR spine images are superior in image quality and diagnostically equivalent to conventional STIR: a multicenter, multireader trial | LN Tanenbaum et al. [42] | 2023 | AJNR | Validation of synthetically created STIR images created from T1w and T2w | 93 | Whole spine | Sagittal T1w, T2w, STIR | CNN | No significant difference between synthetic and acquired STIR, higher image quality for synthetic STIR (p < 0.0001) |
| 25 | Prediction of osteoporosis using MRI and CT scans with unimodal and multimodal deep-learning models | Y Kucukciloglu et al. [43] | 2024 | Diagn Interv Radiol | Prediction of osteoporosis on lumbar spine MRI and CT against DEXA scans | 120 | Lumbar | Sagittal T1w, CT, DEXA | CNN | Accuracy 96–99% |
| 26 | Machine learning assisting the prediction of clinical outcomes following nucleoplasty for lumbar degenerative disc disease | PF Chiu et al. [44] | 2023 | Diagnostics (Basel) | Prediction of pain improvement after lumbar nucleoplasty for degenerative disc disease | 181 | Lumbar | Axial T2w | Multiple (SVM, light gradient boosting machine, XGBoost, XGBRF, CatBoost, iRF) | Improved RF: accuracy 76%, sensitivity 69%, specificity 83%, F1-score 0.73, AUC 0.77 |
| 27 | NAMSTCD: A novel augmented model for spinal cord segmentation and tumor classification using deep nets | R Mohanty et al. [45] | 2023 | Diagnostics (Basel) | Segmentation of spinal cord regions and tumour types | 5000 images | Whole spine | Not mentioned | Multiple (Multiple Mask Regional CNN (MRCNNs), VGGNet 19, YoLo V2, ResNet 101, GoogleNet | Classification accuracy 99% (versus 81–96% for other models) |
| 28 | Benign vs. malignant vertebral compression fractures with MRI: a comparison between automatic deep learning network and radiologist’s assessment | B Liu et al. [46] | 2023 | Eur Radiol | Differentiation of benign and malignant vertebral compression fractures | 209 | Whole spine | Median sagittal T1w, T2w fat suppressed | Two stream compare and contrast network (TSCCN) | AUC 92–99%, accuracy 90–96% (higher than radiologists), specificity 94–99% (higher than radiologists) |
| 29 | Automatic detection, classification, and grading of lumbar intervertebral disc degeneration using an artificial neural network model | W Liawrungrueang et al. [47] | 2023 | Diagnostics (Basel) | Classification of lumbar disc degeneration (Pfirrmann grading) | 515 | Lumbar | Sagittal T2w | Yolov5 | Accuracy > 95%, F1-score 0.98 |
| 30 | Differentiating magnetic resonance images of pyogenic spondylitis and spinal Modic change using a convolutional neural network | T Mukaihata et al. [48] | 2023 | Spine (Phila Pa 1976) | Differentiation of Modic changes from pyogenic spondylitis on MRI | 100 | Whole spine | Sagittal T1w, T2w, STIR | CNN | AUC 0.94–0.95, higher accuracy than clinicians (p < 0.05) |
| 31 | Automated classification of intramedullary spinal cord tumors and inflammatory demyelinating lesions using deep learning | Z Zhuo et al. [49] | 2022 | Radiol Artif Intell | Differentiation of cord tumors from demyelinating lesions | 647 | Whole spine | Sagittal T2w | MultiResU-net, DenseNet121 | Test cohort Dice 0.50–0.80, accuracy 79–96%, AUC 0.85–0.99 |
| 32 | Ultrafast cervical spine MRI protocol using deep learning-based reconstruction: diagnostic equivalence to a conventional protocol | N Kashiwagi et al. [50] | 2022 | Eur J Radiol | Validation of an ultrafast cervical spine MRI protocol | 50 | Cervical | Sagittal T1w, T2w, STIR, axial T2*w | CNN | κ = 0.60–0.98, individual equivalence index 95% CI < 5% |
| 33 | Differentiation between spinal multiple myeloma and metastases originated from lung using multi-view attention-guided network | K Chen et al. [51] | 2022 | Front Oncol | Differentiation of multiple myeloma lesions from metastasis on MRI | 217 | Whole spine | T2w, T1w post-contrast (3 planes) | Multi-view attention guided (MAGN), ResNet50, Class Activation Mapping | Accuracy 79–81%, AUC 0.77–0.78, F1-score 0.67–0.71 |
| 34 | Development of lumbar spine MRI referrals vetting models using machine learning and deep learning algorithms: Comparison models vs. healthcare professionals | AH Alanazi et al. [52] | 2022 | Radiography (Lond) | Vetting of MRI lumbar spine referrals for valid indications | 1020 | Lumbar | Nil | SVM, logistic regression, RF, CNN, bi-directional long-short term memory (Bi-LSTM) | RF AUC 0.99, CNN AUC 0.98 (outperforming radiographers) |
| 35 | Improved productivity using deep learning-assisted reporting for lumbar spine MRI | DSW Lim et al. [53] | 2022 | Radiology | Evaluation of time savings and accuracy for AI-assisted MRI lumbar spine reporting | 25 | Lumbar | Sagittal T1w, axial T2w | CNN, ResNet101 | Reduced interpretation time (p < 0.001), improved or equivalent interobserver agreement with DL assistance |
| 36 | Deep learning model for classifying metastatic epidural spinal cord compression on MRI | J Hallinan et al. [54] | 2022 | Front Oncol | Classification of metastatic vertebral and epidural disease (Bilsky classification) | 247 | Thoracic | Axial T2w | ResNet50 | Internal dataset: κ = 0.92–0.98, external dataset: κ = 0.94–0.95 |
| 37 | Vertebral deformity measurements at MRI, CT, and radiography using deep learning | A Suri et al. [55] | 2021 | Radiol Artif Intell | Measurement of vertebral deformity on MRI, CT and radiographs | 1744 | Whole spine | Sagittal T1w, T2w, CT, radiographs | Neural network | Vertebral measurement mean height percentage error 1.5–1.9% ± 0.2–0.4, lumbar lordosis angle mean absolute error 2.3–3.6° |
| 38 | Predicting postoperative recovery in cervical spondylotic myelopathy: construction and interpretation of T2*-weighted radiomic-based extra trees models | MZ Zhang et al. [56] | 2022 | Eur Radiol | Prediction of recovery rate after cervical spondylotic myelopathy surgery based on MRI and clinical features | 151 | Cervical | T2w, T2*w | Threshold selection, collinearity removal, tree-based feature selection | AUC 0.71–0.81 (vs. conventional clinical and radiologic models AUC 0.40–0.55) |
| 39 | Fully automated segmentation of lumbar bone marrow in sagittal, high-resolution T1-weighted magnetic resonance images using 2D U-NET | EJ Hwang et al. [57] | 2022 | Comput Biol Med | Segmentation of normal and pathological bone marrow on MRI lumbar spine | 100 | Lumbar | Sagittal T1w | U-net3D, Grow-cut | Healthy subjects Dice 0.91–0.96, diseased subjects Dice 0.83–0.95 |
| 40 | A comparison of natural language processing methods for the classification of lumbar spine imaging findings related to lower back pain | C Jujjavarapu et al. [58] | 2022 | Acad Radiol | Classification of spine MRI and radiograph reports into 26 findings | 871 | Lumbar | MRI, radiographs | Elastic-net logistic regression | AUC 0.96 for all findings (n-grams), AUC 0.95 for potentially clinically important findings |
| 41 | Virtual magnetic resonance lumbar spine images generated from computed tomography images using conditional generative adversarial networks | M Gotoh et al. [59] | 2022 | Radiography (Lond) | Generation of virtual MRI images from CT | 22 | Lumbar | MRI | Conditional GAN | No significant difference between virtual and conventional MRI, except in visualization of spinal canal structure. Peak signal-to-noise ratio 18.4 dB |
| 42 | Deep learning for adjacent segment disease at preoperative MRI for cervical radiculopathy | CMW Goedmakers et al. [60] | 2021 | Radiology | Prediction of adjacent segment disease after anterior cervical discectomy and fusion surgery on pre-operative MRI | 344 | Cervical | Sagittal T2w | CNN | Accuracy 95% (vs. 58% for clinicians), sensitivity 80% (vs. 60%), specificity 97% (vs. 58%) |
| 43 | Automatic multiclass intramedullary spinal cord tumor segmentation on MRI with deep learning | A Lemay et al. [61] | 2021 | Neuroimage Clin | Segmentation of three common spinal cord tumors | 343 | Whole spine | Sagittal T2w, T1w post-contrast | U-net | Dice 0.77 (all abnormal signal), 0.62 (tumour alone), true positive detection > 87% (all abnormal signal) |
| 44 | A preliminary study using spinal MRI-based radiomics to predict high-risk cytogenetic abnormalities in multiple myeloma | J Liu et al. [62] | 2021 | Radiol Med | Prediction of high-risk cytogenic abnormalities in multiple myeloma based on MRI | 248 lesions | Whole spine | Sagittal T1w, T2w, T2w fat suppressed | Logistic regression | AUC 0.86–0.87, sensitivity 79%, specificity 79%, PPV 75%, NPV 82%, accuracy 79% |
| 45 | A deep learning model for detection of cervical spinal cord compression in MRI scans | Z Merali et al. [63] | 2021 | Sci Rep | Dichotomous spinal cord compression for cervical spine | 289 | Cervical | Axial T2w | CNN | AUC 0.94, sensitivity 88%, specificity 89%, F1-score 0.82 |
| 46 | Deep learning model for automated detection and classification of central canal, lateral recess, and neural foraminal stenosis at lumbar spine MRI | J Hallinan et al. [64] | 2021 | Radiology | Grading of lumbar spinal canal, lateral recess and neural foraminal stenosis | 446 | Lumbar | Sagittal T1w, axial T2w | CNN | Recall 85–100%, dichotomous classification κ-range = 0.89–0.96 (vs. 0.92–0.98 for radiologists) |
| 47 | A deep convolutional neural network with performance comparable to radiologists for differentiating between spinal schwannoma and meningioma | S Maki et al. [65] | 2020 | Spine (Phila Pa 1976) | Differentiation of meningioma from schwannoma | 84 | Whole spine | Sagittal T2w, T1w post-contrast | CNN | AUC 0.87–0.88, sensitivity 78–85% (vs. 95–100% for radiologists), specificity 75–82% (vs. 26–58%), accuracy 80–81% (vs. 69–82%) |
| 48 | Quantitative analysis of neural foramina in the lumbar spine: an imaging informatics and machine learning study | B Gaonkar et al. [66] | 2019 | Radiol Artif Intell | Segmentation and statistical modelling of lumbar neural foraminal area | 1156 | Lumbar | Sagittal T2w | SVM, U-net | Dice 0.63–0.68 (neural foramen), 0.84–0.91 (disc) |
| 49 | Performance of the deep convolutional neural network based magnetic resonance image scoring algorithm for differentiating between Tuberculous and pyogenic spondylitis | K Kim et al. [67] | 2018 | Sci Rep | Differentiation of pyogenic from Tuberculous spondylitis | 161 | Whole spine | Axial T2w | Deep CNN | AUC 0.80 (vs. 0.73 for radiologists, p = 0.079) |
| 50 | SpineNet: automated classification and evidence visualization in spinal MRIs | A Jamaludin et al. [68] | 2017 | Med Image Anal | Detection and classification of multiple abnormalities (Pfirrmann grading, disc narrowing, endplate defects, marrow changes, spondylolisthesis, central canal stenosis) | 2009 | Lumbar spine | T2w sagittal | CNN | Pfirmann inter-rater κ = 0.69–0.81, overall accuracy 74% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, A.; Ong, W.; Makmur, A.; Ting, Y.H.; Tan, W.C.; Lim, S.W.D.; Low, X.Z.; Tan, J.J.H.; Kumar, N.; Hallinan, J.T.P.D. Applications of Artificial Intelligence and Machine Learning in Spine MRI. Bioengineering 2024, 11, 894. https://doi.org/10.3390/bioengineering11090894
Lee A, Ong W, Makmur A, Ting YH, Tan WC, Lim SWD, Low XZ, Tan JJH, Kumar N, Hallinan JTPD. Applications of Artificial Intelligence and Machine Learning in Spine MRI. Bioengineering. 2024; 11(9):894. https://doi.org/10.3390/bioengineering11090894
Chicago/Turabian StyleLee, Aric, Wilson Ong, Andrew Makmur, Yong Han Ting, Wei Chuan Tan, Shi Wei Desmond Lim, Xi Zhen Low, Jonathan Jiong Hao Tan, Naresh Kumar, and James T. P. D. Hallinan. 2024. "Applications of Artificial Intelligence and Machine Learning in Spine MRI" Bioengineering 11, no. 9: 894. https://doi.org/10.3390/bioengineering11090894
APA StyleLee, A., Ong, W., Makmur, A., Ting, Y. H., Tan, W. C., Lim, S. W. D., Low, X. Z., Tan, J. J. H., Kumar, N., & Hallinan, J. T. P. D. (2024). Applications of Artificial Intelligence and Machine Learning in Spine MRI. Bioengineering, 11(9), 894. https://doi.org/10.3390/bioengineering11090894







