Precision Medicine for Apical Lesions and Peri-Endo Combined Lesions Based on Transfer Learning Using Periapical Radiographs
Abstract
1. Introduction
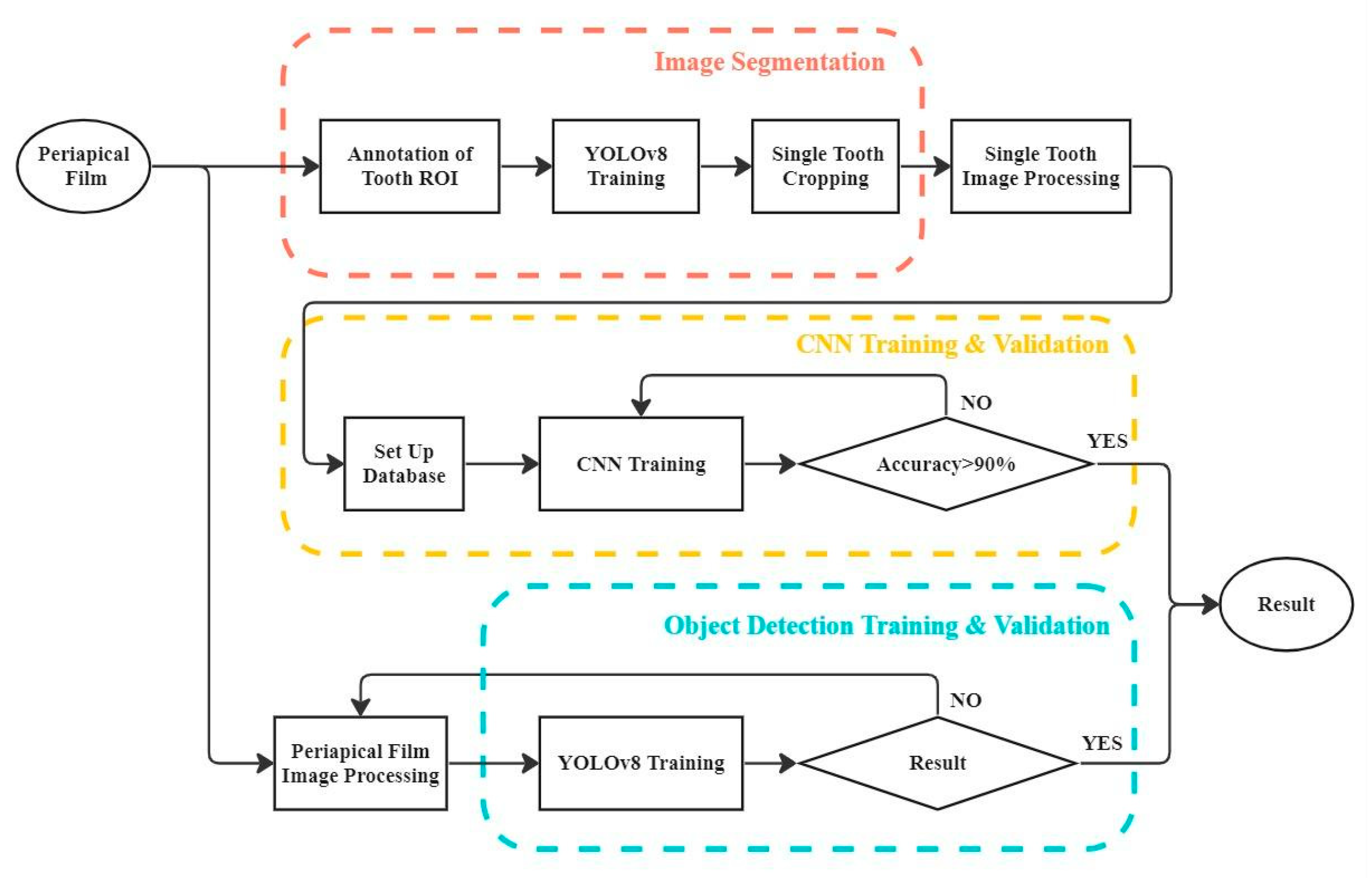
2. Materials and Methods
2.1. Image Segmentation
2.1.1. Tooth Annotation
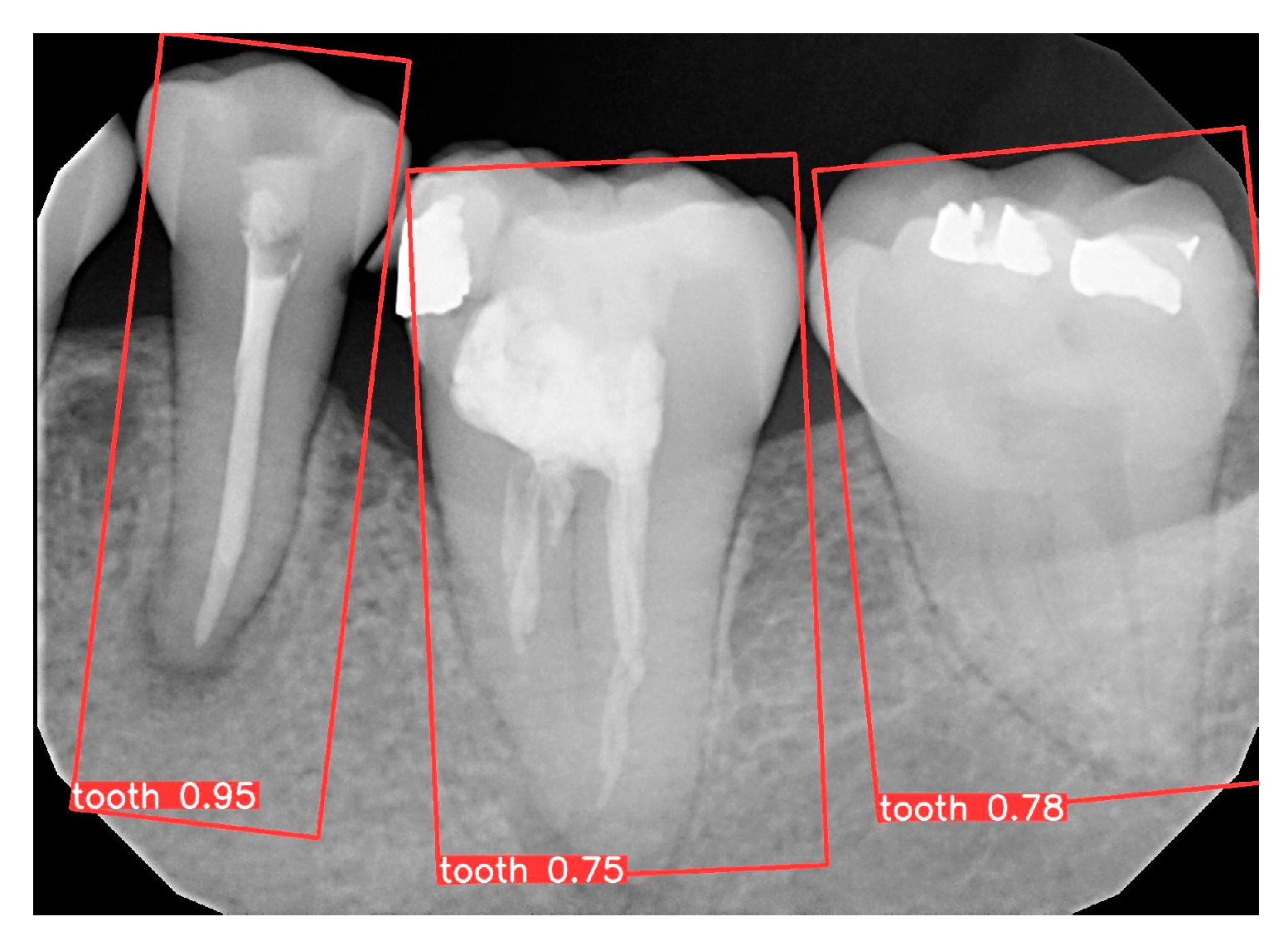
2.1.2. YOLOv8 OBB Model Training
2.1.3. Single-Tooth Cropping
- A.
- Image Rotation
- B.
- Coordinate Point Rotation
- C.
- Single-Tooth Cropping and Cropping Area Expansion
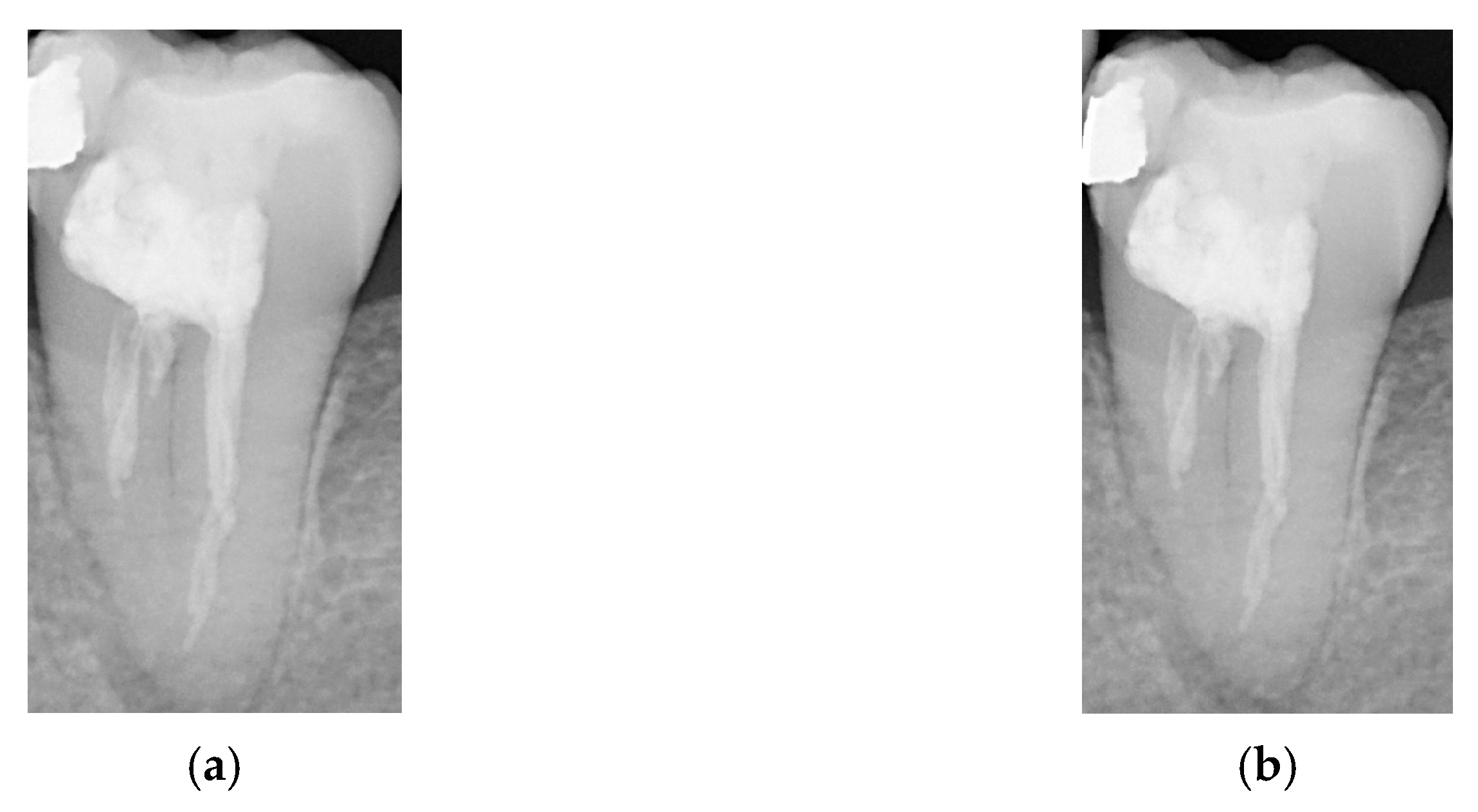
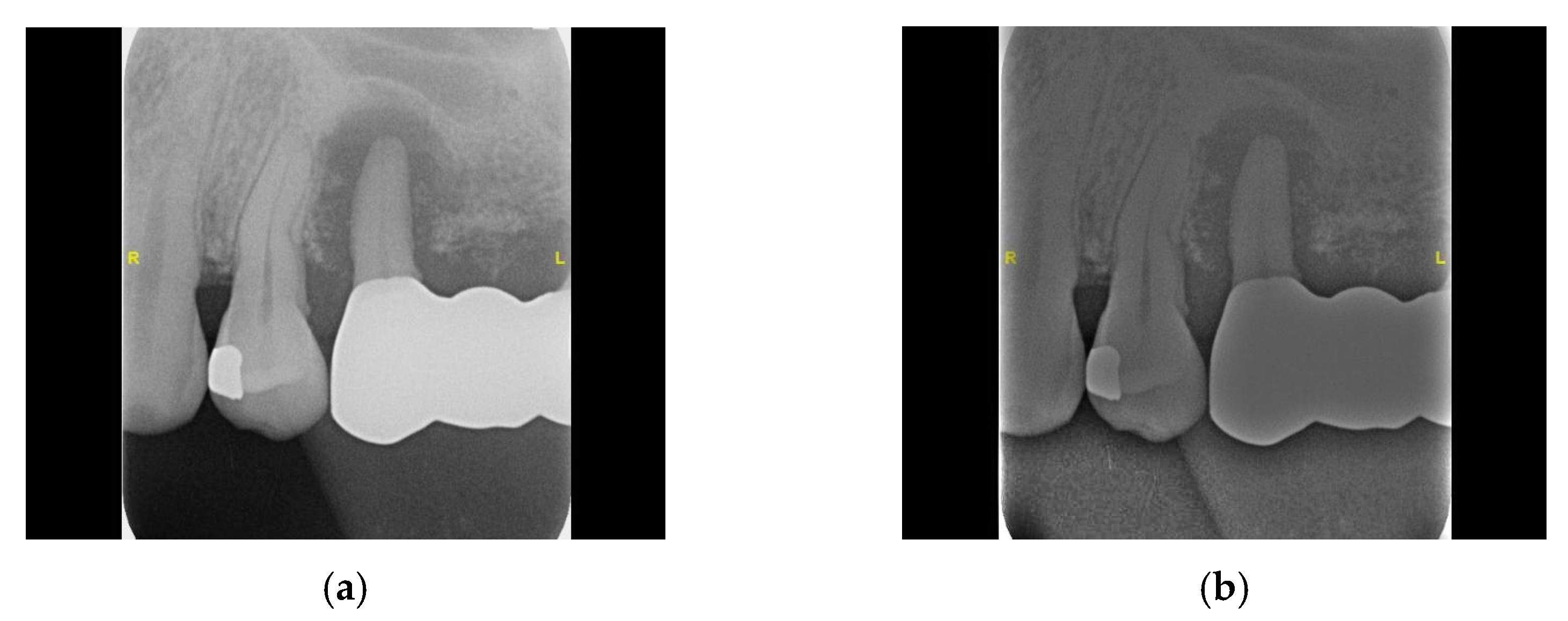
2.2. Image Processing
2.2.1. Grayscale
2.2.2. Gaussian High-Pass Filter
2.2.3. Adaptive Histogram Equalization
2.2.4. Flat-Field Correction
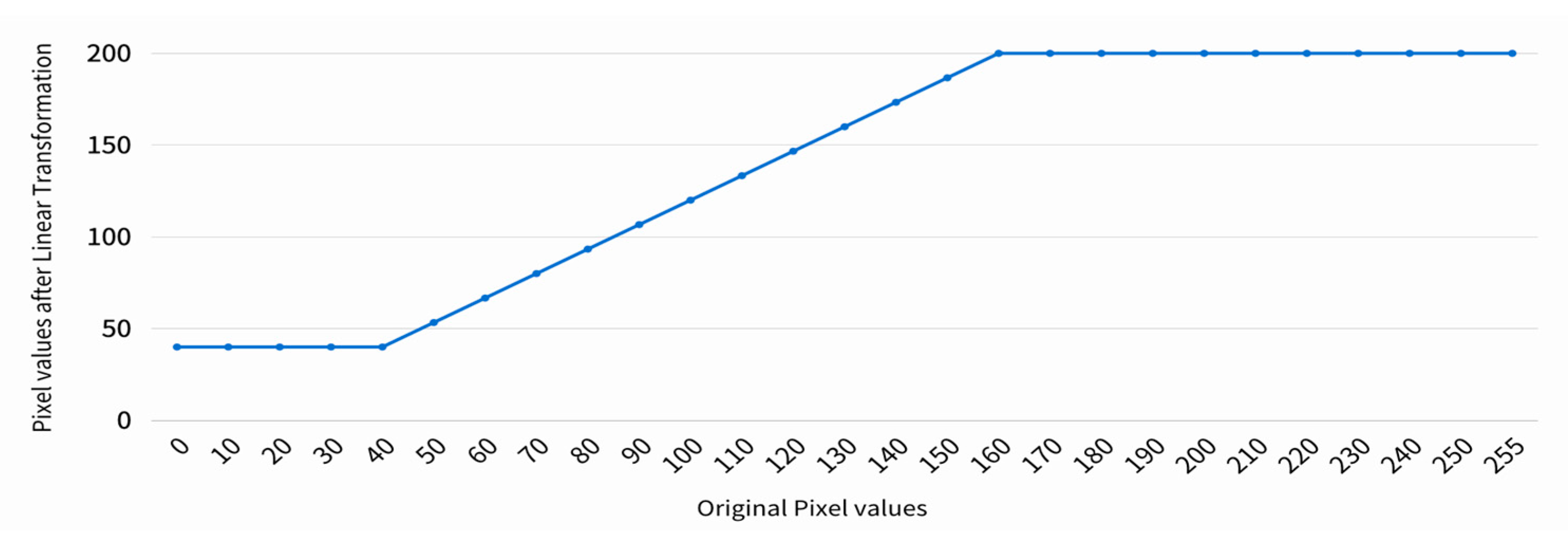
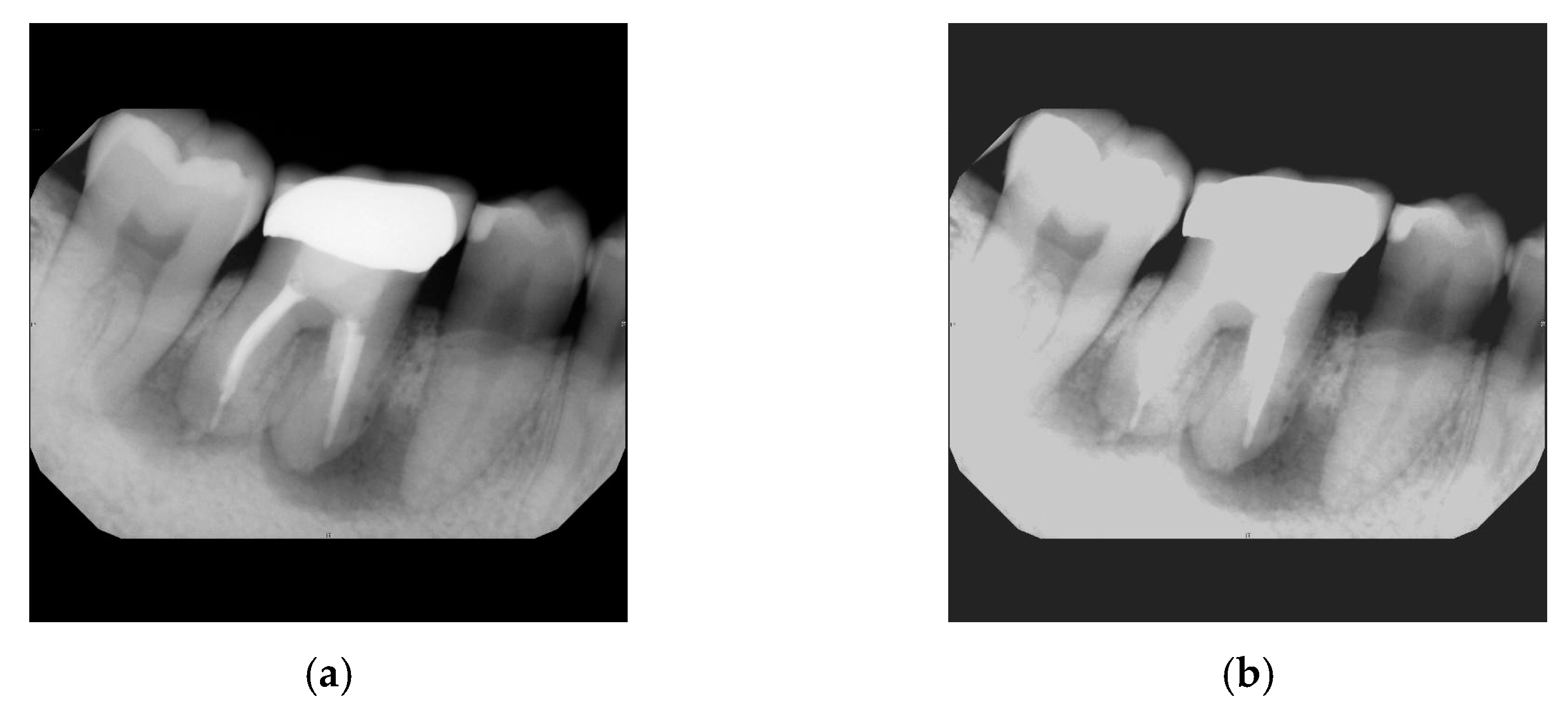
2.2.5. Linear Transformation
2.2.6. Negative Film Effect
2.3. CNN Training and Validation
2.3.1. CNN Architecture
2.3.2. Hyperparameter
2.3.3. Training and Validation
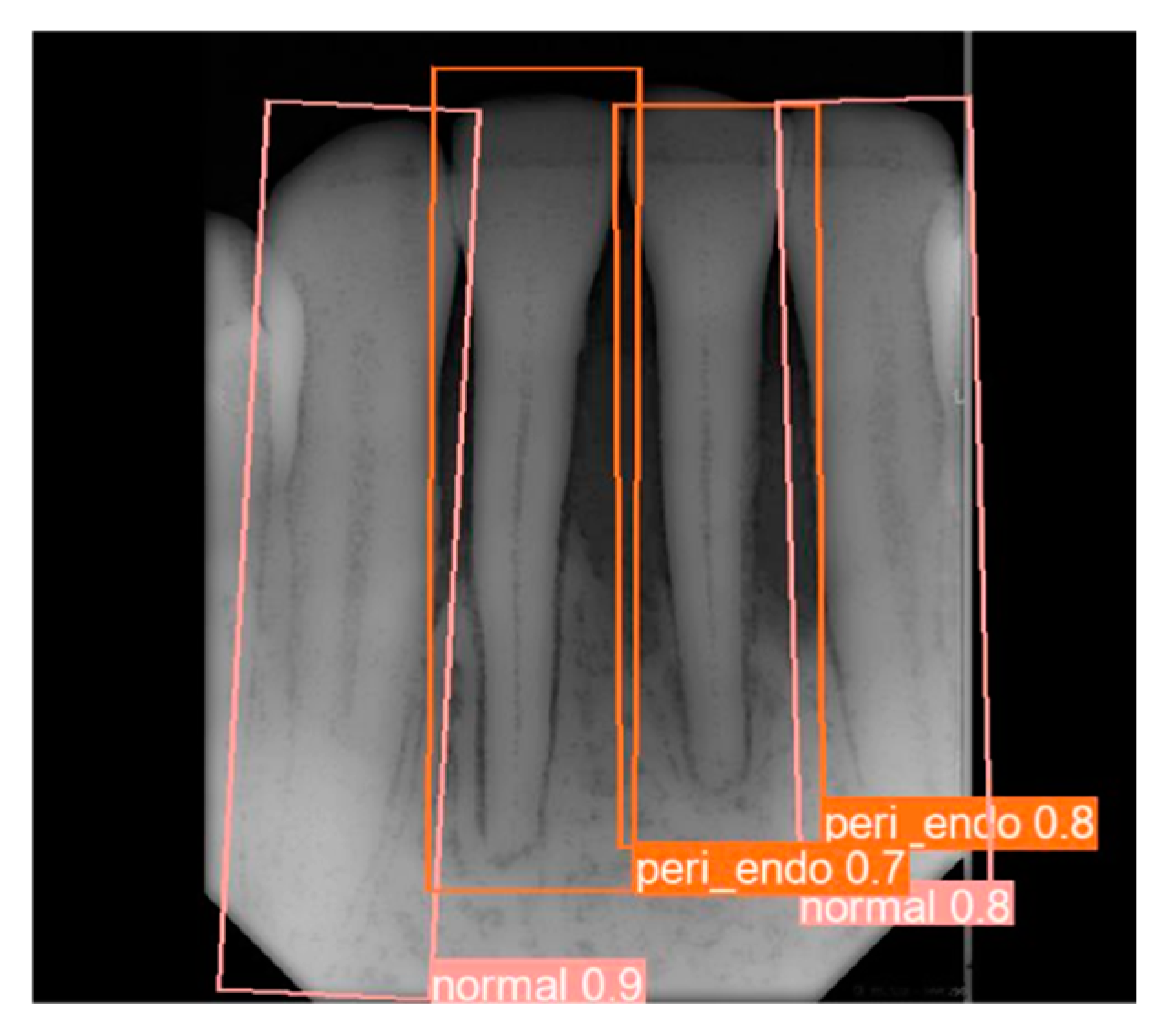
2.4. Object Detection Training and Validation
3. Results
3.1. YOLO Detection and Image Segmentation
3.2. CNN Training
- 1.
- Aspect 1: Black Padding
- 2.
- Aspect 2: Data Augmentation
- 3.
- Aspect 3: Expanding the Cropping Range
- 4.
- Aspect 4: Image Enhancement
3.3. YOLOv8
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Shenoy, N.; Shenoy, A. Endo-perio lesions: Diagnosis and clinical considerations. Indian J. Dent. Res. Off. Publ. Indian Soc. Dent. Res. 2010, 21, 579–585. [Google Scholar] [CrossRef] [PubMed]
- Northridge, M.E.; Kumar, A.; Kaur, R. Disparities in Access to Oral Health Care. Annu. Rev. Public Health 2020, 41, 513–535. [Google Scholar] [CrossRef] [PubMed]
- JOE Editorial Board. Endodontic-Periodontal Interrelationships: An Online Study Guide. J. Endod. 2008, 34, e71–e77. [Google Scholar] [CrossRef] [PubMed]
- Ehnevid, H.; Jansson, L.; Lindskog, S.; Weintraub, A.; Blomlöf, L. Endodontic Pathogens: Propagation of Infection through Patent Dentinal Tubules in Traumatized Monkey Teeth. Endod. Dent. Traumatol. 1995, 11, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-L.; Chou, H.-S.; Chuo, Y.; Lin, Y.-J.; Tsai, T.-H.; Peng, C.-H.; Tseng, A.-Y.; Li, K.-C.; Chen, C.-A.; Chen, T.-Y. Classification of the Relative Position between the Third Molar and the Inferior Alveolar Nerve Using a Convolutional Neural Network Based on Transfer Learning. Electronics 2024, 13, 702. [Google Scholar] [CrossRef]
- Chuo, Y.; Lin, W.-M.; Chen, T.-Y.; Chan, M.-L.; Chang, Y.-S.; Lin, Y.-R.; Lin, Y.-J.; Shao, Y.-H.; Chen, C.-A.; Chen, S.-L.; et al. A High-Accuracy Detection System: Based on Transfer Learning for Apical Lesions on Periapical Radiograph. Bioengineering 2022, 9, 777. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.-C.; Chen, T.-Y.; Chou, H.-S.; Lin, S.-Y.; Liu, S.-Y.; Chen, Y.-A.; Liu, Y.-L.; Chen, C.-A.; Huang, Y.-C.; Chen, S.-L.; et al. Caries and Restoration Detection Using Bitewing Film Based on Transfer Learning with CNNs. Sensors 2021, 21, 4613. [Google Scholar] [CrossRef] [PubMed]
- Song, I.-S.; Shin, H.-K.; Kang, J.-H.; Kim, J.-E.; Huh, K.-H.; Yi, W.-J.; Lee, S.-S.; Heo, M.-S. Deep Learning-Based Apical Lesion Segmentation from Panoramic Radiographs. Imaging Sci. Dent. 2022, 52, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-L.; Chen, T.-Y.; Mao, Y.-C.; Lin, S.-Y.; Huang, Y.-Y.; Chen, C.-A.; Lin, Y.-J.; Chuang, M.-H.; Abu, P.A.R. Detection of Various Dental Conditions on Dental Panoramic Radiography Using Faster R-CNN. IEEE Access 2023, 11, 127388–127401. [Google Scholar] [CrossRef]
- Fatima, A.; Shafi, I.; Afzal, H.; Mahmood, K.; Díez, I.D.; Lipari, V.; Ballester, J.B.; Ashraf, I. Deep Learning-Based Multiclass Instance Segmentation for Dental Lesion Detection. Healthcare 2023, 11, 347. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-L.; Chen, T.-Y.; Mao, Y.-C.; Lin, S.-Y.; Huang, Y.-Y.; Chen, C.-A.; Lin, Y.-J.; Hsu, Y.-M.; Li, C.-A.; Chiang, W.-Y.; et al. Automated Detection System Based on Convolution Neural Networks for Retained Root, Endodontic Treated Teeth, and Implant Recognition on Dental Panoramic Images. IEEE Sens. J. 2022, 22, 23293–23306. [Google Scholar] [CrossRef]
- Li, C.-W.; Lin, S.-Y.; Chou, H.-S.; Chen, T.-Y.; Chen, Y.-A.; Liu, S.-Y.; Liu, Y.-L.; Chen, C.-A.; Huang, Y.-C.; Chen, S.-L.; et al. Detection of Dental Apical Lesions Using CNNs on Periapical Radiograph. Sensors 2021, 21, 7049. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-L.; Chen, T.-Y.; Huang, Y.-C.; Chen, C.-A.; Chou, H.-S.; Huang, Y.-Y.; Lin, W.-C.; Li, T.-C.; Yuan, J.-J.; Abu, P.A.R.; et al. Missing Teeth and Restoration Detection Using Dental Panoramic Radiography Based on Transfer Learning with CNNs. IEEE Access 2022, 10, 118654–118664. [Google Scholar] [CrossRef]
- Chen, Y.-C.; Chen, M.-Y.; Chen, T.-Y.; Chan, M.-L.; Huang, Y.-Y.; Liu, Y.-L.; Lee, P.-T.; Lin, G.-J.; Li, T.-F.; Chen, C.-A.; et al. Improving Dental Implant Outcomes: CNN-Based System Accurately Measures Degree of Peri-Implantitis Damage on Periapical Film. Bioengineering 2023, 10, 640. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-C.; Chen, C.-A.; Chen, T.-Y.; Chou, H.-S.; Lin, W.-C.; Li, T.-C.; Yuan, J.-J.; Lin, S.-Y.; Li, C.-W.; Chen, S.-L.; et al. Tooth Position Determination by Automatic Cutting and Marking of Dental Panoramic X-ray Film in Medical Image Processing. Appl. Sci. 2021, 11, 11904. [Google Scholar] [CrossRef]
- Jiang, Y.; Liu, Z.; Li, Y.; Li, J.; Lian, Y.; Liao, N.; Li, Z.; Zhao, Z. A Digital Grayscale Generation Equipment for Image Display Standardization. Appl. Sci. 2020, 10, 2297. [Google Scholar] [CrossRef]
- Soora, N.R.; Vodithala, S.; Badam, J.S.H. Filtering Techniques to remove Noises from an Image. In Proceedings of the 2022 International Conference on Advances in Computing, Communication and Applied Informatics (ACCAI), Chennai, India, 28–29 January 2022; pp. 1–9. [Google Scholar] [CrossRef]
- Jiang, Y.; Ming, Y. Application of image sharpening based quality assessment model: Extraction of Traditional Chinese Embroidery Patterns as an Example. In Proceedings of the 2024 5th International Conference on Computer Engineering and Application (ICCEA), Hangzhou, China, 12–14 April 2024; pp. 994–1001. [Google Scholar] [CrossRef]
- Zhang, Y.; Zheng, X. Development of Image Processing Based on Deep Learning Algorithm. In Proceedings of the 2022 IEEE Asia-Pacific Conference on Image Processing, Electronics and Computers (IPEC), Dalian, China, 14–16 April 2022; pp. 1226–1228. [Google Scholar] [CrossRef]
- Mustafa, Z.; Nsour, H. Using Computer Vision Techniques to Automatically Detect Abnormalities in Chest X-rays. Diagnostics 2023, 13, 2979. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, X.; Li, P.; Wang, L.; Zhu, M.; Zhang, H.; Zeng, Z. An Improved YOLO Algorithm for Rotated Object Detection in Remote Sensing Images. In Proceedings of the 2021 IEEE 4th Advanced Information Management, Communicates, Electronic and Automation Control Conference (IMCEC), Chongqing, China, 18–20 June 2021; pp. 840–845. [Google Scholar] [CrossRef]
- Deng, L.Y.; Ho, S.S.; Lim, X.Y. Diseases Classification Utilizing Tooth X-ray Images Based on Convolutional Neural Network. In Proceedings of the 2020 International Symposium on Computer, Consumer and Control (IS3C), Taichung City, Taiwan, 13–16 November 2020; pp. 300–303. [Google Scholar] [CrossRef]
- Herbst, S.R.; Pitchika, V.; Krois, J.; Krasowski, A.; Schwendicke, F. Machine Learning to Predict Apical Lesions: A Cross-Sectional and Model Development Study. J. Clin. Med. 2023, 12, 5464. [Google Scholar] [CrossRef] [PubMed]
- Duman, Ş.B.; Çelik Özen, D.; Bayrakdar, I.Ş.; Baydar, O.; Alhaija, E.S.A.; Helvacioğlu Yiğit, D.; Çelik, Ö.; Jagtap, R.; Pileggi, R.; Orhan, K. Second Mesiobuccal Canal Segmentation with YOLOv5 Architecture Using Cone Beam Computed Tomography Images. Odontology 2024, 112, 552–561. [Google Scholar] [CrossRef] [PubMed]
- İçöz, D.; Terzioğlu, H.; Özel, M.A.; Karakurt, R. Evaluation of an Artificial Intelligence System for the Diagnosis of Apical Periodontitis on Digital Panoramic Images. Niger. J. Clin. Pract. 2023, 26, 1085. [Google Scholar] [CrossRef] [PubMed]






| Hardware Platform | Version | ||
| CPU | 11 Gen Intel(R) Core(TM) i9-11900@2.50GHz | ||
| GPU | NVIDIA GeForce RTX 3070 8G | ||
| DRAM | 32 GB | ||
| Software Platform | Version | Software Platform | Version |
| MATLAB | R2023b | Python | 3.11.8 |
| Deep Network designer | R2023b | PyTorch | 2.2.1 + cu121 |
| Deep Learning Toolbox | 23.2 | CUDA | 12.1 |
| Layer | Filters/Neuron | Filter Size | Stride | Padding | Size of Feature Map | Activation Functions |
|---|---|---|---|---|---|---|
| Input | 227 × 227 × 3 | |||||
| Conv 1 | 96 | 11 × 11 | 4 | 55 × 55 × 96 | ReLU | |
| MaxPool 1 | 3 × 3 | 2 | 27 × 27 × 96 | |||
| Conv 2 | 256 | 5 × 5 | 1 | 2 | 27 × 27 × 256 | ReLU |
| MaxPool 2 | 3 × 3 | 2 | 13 × 13 × 256 | |||
| Conv 3 | 384 | 3 × 3 | 1 | 1 | 13 × 13 × 384 | ReLU |
| Conv 4 | 384 | 3 × 3 | 1 | 1 | 13 × 13 × 384 | ReLU |
| Conv 5 | 356 | 3 × 3 | 1 | 1 | 13 × 13 × 256 | ReLU |
| MaxPool 3 | 3 × 3 | 2 | 6 × 6 × 256 | |||
| Dropout 1 | Rate = 0.7 | 6 × 6 × 256 | ||||
| Fc 1 | 4096 | ReLU | ||||
| Dropout 2 | Rate = 0.7 | 4096 | ||||
| Fc 2 | 4096 | ReLU | ||||
| Fc 3 | 3 | Softmax |
| Hyperparameter | Value | Hyper Parameter | Model | Value |
|---|---|---|---|---|
| Initial Learning Rate | 0.0001 | Max Epoch | AlexNet | 30 |
| Mini Batch Size | 16 | Places365-GoogLeNet | 20 | |
| Learning Rate Drop Factor | 0.1 | VGG-16 | 6 | |
| Learning Rate Drop Period | 10 | ResNet50 | 10 | |
| Shuffle | Every-epoch | GoogLeNet | 20 | |
| Validation Frequency | 100 | ConvNeXtv2_base | 10 |
| Disease | Training Set | Validation Set | Total |
| Normal | 53 | 14 | 67 |
| Apical Lesion | 53 | 14 | 67 |
| Peri-endo Combined Lesion | 53 | 14 | 67 |
| Total | 159 | 42 | 201 |
| Disease | Original | Augmentation |
| Normal | 67 | 268 |
| Apical Lesion | 67 | 268 |
| Peri-endo Combined Lesion | 67 | 268 |
| Hyperparameter | Value |
|---|---|
| Epoch | 100 |
| Batch | 8 |
| imgsize | 640 × 640 |
| lr0 | 0.01 |
| Number | No. 1 | No. 2 | No. 3 |
| Ground Truth | Peri-endo Combined Lesion | Normal | Apical Lesion |
| Validation | Peri-endo Combined Lesion | Normal | Apical Lesion |
| Accuracy | 99.94% | 60.47% | 85.02% |
| Method | Metrics | AlexNet | Places365-GoogLeNet | VGG16 | ResNet50 | GoogLeNet | ConvNeXtv2 |
| Original | Accuracy | 80.95% | 79.19% | 71.43% | 71.43% | 80.95% | 76.19% |
| Training time | 54 s | 1 m 20 s | 26 s | 1 m 19 s | 1 m 19 s | 6 m 4 s | |
| After padding | Accuracy | 83.33% | 80.95% | 80.95% | 73.81% | 85.71% | 85.71% |
| Training time | 48 s | 1 m 29 s | 24 s | 1 m 19 s | 1 m 17 s | 5 m 46 s |
| Method | Metrics | AlexNet | Places365-GoogLeNet | VGG16 | ResNet50 | GoogLeNet | ConvNeXtv2 |
| Padding | Accuracy | 83.33% | 80.95% | 80.95% | 73.81% | 85.71% | 85.71% |
| Training time | 48 s | 1 m 29 s | 24 s | 1 m 19 s | 1 m 17 s | 5 m 46 s | |
| Padding + Enhancement | Accuracy | 88.69% | 88.69% | 88.69% | 88.69% | 86.90% | 87.50% |
| Training time | 2 m 49 s | 5 m 17 s | 1 m 2 s | 4 m 33 s | 5 m 26 s | 21 m 58 s |
| Method | Metrics | AlexNet | Places365-GoogLeNet | VGG16 | ResNet50 | GoogLeNet | ConvNeXtv2 |
| Original YOLOv8 cropping | Accuracy | 88.69% | 88.69% | 88.69% | 88.69% | 86.90% | 87.50% |
| Training time | 2 m 49 s | 5 m 17 s | 1 m 21 s | 4 m 33 s | 5 m 26 s | 21 m 58 s | |
| Expand x = 20 pixels, y = 0 pixels | Accuracy | 91.67% | 89.29% | 90.48% | 85.71% | 89.29% | 89.28% |
| Training time | 1 m 30 s | 4 m 8 s | 1 m 37 s | 3 m 20 s | 4 m 39 s | 21 m 13 s | |
| Expand x = 0 pixels, y = 40 pixels | Accuracy | 88.10% | 91.67% | 90.48% | 88.10% | 88.69% | 88.09% |
| Training time | 2 m 38 s | 1 m 23 s | 1 m 16 s | 3 m 22 s | 5 m 31 s | 22 m 4 s | |
| Expand x = 20 pixels, y = 40 pixels | Accuracy | 89.29% | 89.88% | 88.10% | 89.88% | 88.10% | 91.07% |
| Training time | 2 m 1 s | 4 m 50 s | 1 m 29 s | 4 m 28 s | 4 m 37 s | 20 m 35 s |
| Method | Metrics | AlexNet | Places365- GoogLeNet | VGG16 | ResNet50 | GoogLeNet | Conv NeXtv2 |
| Original (padding, enhancement) | Accuracy | 88.69% | 88.69% | 88.69% | 88.69% | 86.90% | 87.50% |
| Training time | 2 m 49 s | 5 m 17 s | 1 m 21 s | 4 m 33 s | 5 m 26 s | 21 m 58 s | |
| Gaussian high-pass filter | Accuracy | 92.26% | 93..45% | 91.67% | 89.29% | 92.26% | 89.29% |
| Training time | 2 m 50 s | 6 m 3 s | 1 m 16 s | 4 m 3 s | 5 m 33 s | 20 m 53 s | |
| Adaptive histogram equalization | Accuracy | 88.10% | 92.86% | 92.26% | 91.07% | 89.88% | 95.23% |
| Training time | 50 s | 1 m 16 s | 1 m 21 s | 4 m 1 s | 5 m 14 s | 55 m 40 s | |
| Gaussian high-pass filter with adaptive histogram equalization | Accuracy | 93.45% | 91.07% | 92.86% | 90.48% | 88.10% | 93.45% |
| Training time | 2 m 24 s | 5 m 37 s | 1 m 30 s | 3 m 55 s | 5 m 48 s | 22 m 2 s |
| Disease | Actual | |||
| Normal | Apical Lesion | Peri-endo Combined Lesion | ||
| Predicted | Normal | 56 | 4 | 0 |
| Apical Lesion | 0 | 49 | 1 | |
| Peri-endo Combined Lesion | 0 | 3 | 55 | |
| Method | The Best Model in this Study | Method in [8] | Method in [23] | |||
| Model | ConvNeXtv2 | U-Net Model | Decision Tree | |||
| Disease | Normal | Apical Lesion | Peri-endo Combined Lesion | Total | Apical Lesion | Apical Lesion |
| Accuracy | 95.23% | No data | No data | |||
| Precision | 93.33% | 98.00% | 94.82% | 95.38% | No data | No data |
| Recall | 99.75% | 87.50% | 98.21% | 95.23% | No data | No data |
| F1-Score | 96.55% | 92.45% | 96.49% | 95.16% | 74.2% | 89% |
| Disease | Training Set | Validation Set | Total |
| Normal | 194 | 58 | 252 |
| Apical Lesion | 106 | 20 | 126 |
| Peri-endo Combined Lesion | 70 | 15 | 85 |
| Total | 370 | 93 | 463 |
| Method | Metrics | Normal | Apical Lesion | Peri-Endo Combined Lesion | Total |
| Original | mAP50 | 0.871 | 0.742 | 0.928 | 0.847 |
| Accuracy | 75.00% | ||||
| Original with data enhancement | mAP50 | 0.906 | 0.878 | 0.927 | 0.904 |
| Accuracy | 84.70% |
| Method | Metrics | Normal | Apical Lesion | Peri-Endo Combined Lesion | Total |
| Linear Transformation with Adaptive histogram equalization | mAP50 | 0.904 | 0.876 | 0.957 | 0.912 |
| Accuracy | 85.16% | ||||
| Flat-Field Correction with Adaptive histogram equalization | mAP50 | 0.888 | 0.857 | 0.971 | 0.905 |
| Accuracy | 87.79% | ||||
| Gaussian high-pass filter with Negative Film Effect | mAP50 | 0.918 | 0.913 | 0.923 | 0.918 |
| Accuracy | 92.13% |
| Disease | Actual | |||
| Normal | Apical Lesion | Peri-endo Combined Lesion | ||
| Predicted | Normal | 143 | 3 | 2 |
| Apical Lesion | 6 | 55 | 5 | |
| Peri-endo Combined Lesion | 2 | 2 | 36 | |
| Method | The Best Models of this Study Method | Method in [24] | Method in [25] | |||
| Model | YOLOv8 | YOLOv5x | YOLOv3 Darknet | |||
| Disease | Normal | Apical Lesion | Peri-endo Combined Lesion | Total | Apical Lesion | Apical Lesion |
| Accuracy | 92.13% | No data | No data | |||
| Precision | 69.3% | 91% | 86.4% | 82.2% | 83% | 56% |
| Recall | 84.2% | 95% | 91.7% | 90% | No data | 98% |
| mAP50 | 0.918 | 0.913 | 0.923 | 0.918 | 0.88 | No data |
| F1-Score | 87.46% | 80.13% | 88.49% | 85.92% | 87% | 71% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, P.-Y.; Mao, Y.-C.; Lin, Y.-J.; Li, X.-H.; Ku, L.-T.; Li, K.-C.; Chen, C.-A.; Chen, T.-Y.; Chen, S.-L.; Tu, W.-C.; et al. Precision Medicine for Apical Lesions and Peri-Endo Combined Lesions Based on Transfer Learning Using Periapical Radiographs. Bioengineering 2024, 11, 877. https://doi.org/10.3390/bioengineering11090877
Wu P-Y, Mao Y-C, Lin Y-J, Li X-H, Ku L-T, Li K-C, Chen C-A, Chen T-Y, Chen S-L, Tu W-C, et al. Precision Medicine for Apical Lesions and Peri-Endo Combined Lesions Based on Transfer Learning Using Periapical Radiographs. Bioengineering. 2024; 11(9):877. https://doi.org/10.3390/bioengineering11090877
Chicago/Turabian StyleWu, Pei-Yi, Yi-Cheng Mao, Yuan-Jin Lin, Xin-Hua Li, Li-Tzu Ku, Kuo-Chen Li, Chiung-An Chen, Tsung-Yi Chen, Shih-Lun Chen, Wei-Chen Tu, and et al. 2024. "Precision Medicine for Apical Lesions and Peri-Endo Combined Lesions Based on Transfer Learning Using Periapical Radiographs" Bioengineering 11, no. 9: 877. https://doi.org/10.3390/bioengineering11090877
APA StyleWu, P.-Y., Mao, Y.-C., Lin, Y.-J., Li, X.-H., Ku, L.-T., Li, K.-C., Chen, C.-A., Chen, T.-Y., Chen, S.-L., Tu, W.-C., & Abu, P. A. R. (2024). Precision Medicine for Apical Lesions and Peri-Endo Combined Lesions Based on Transfer Learning Using Periapical Radiographs. Bioengineering, 11(9), 877. https://doi.org/10.3390/bioengineering11090877









