Deep Attention Fusion Hashing (DAFH) Model for Medical Image Retrieval
Abstract
1. Introduction
2. Related Works
2.1. Conventional Medical Image Retrieval Algorithms
2.2. Deep Hashing Algorithms
3. Materials and Methods
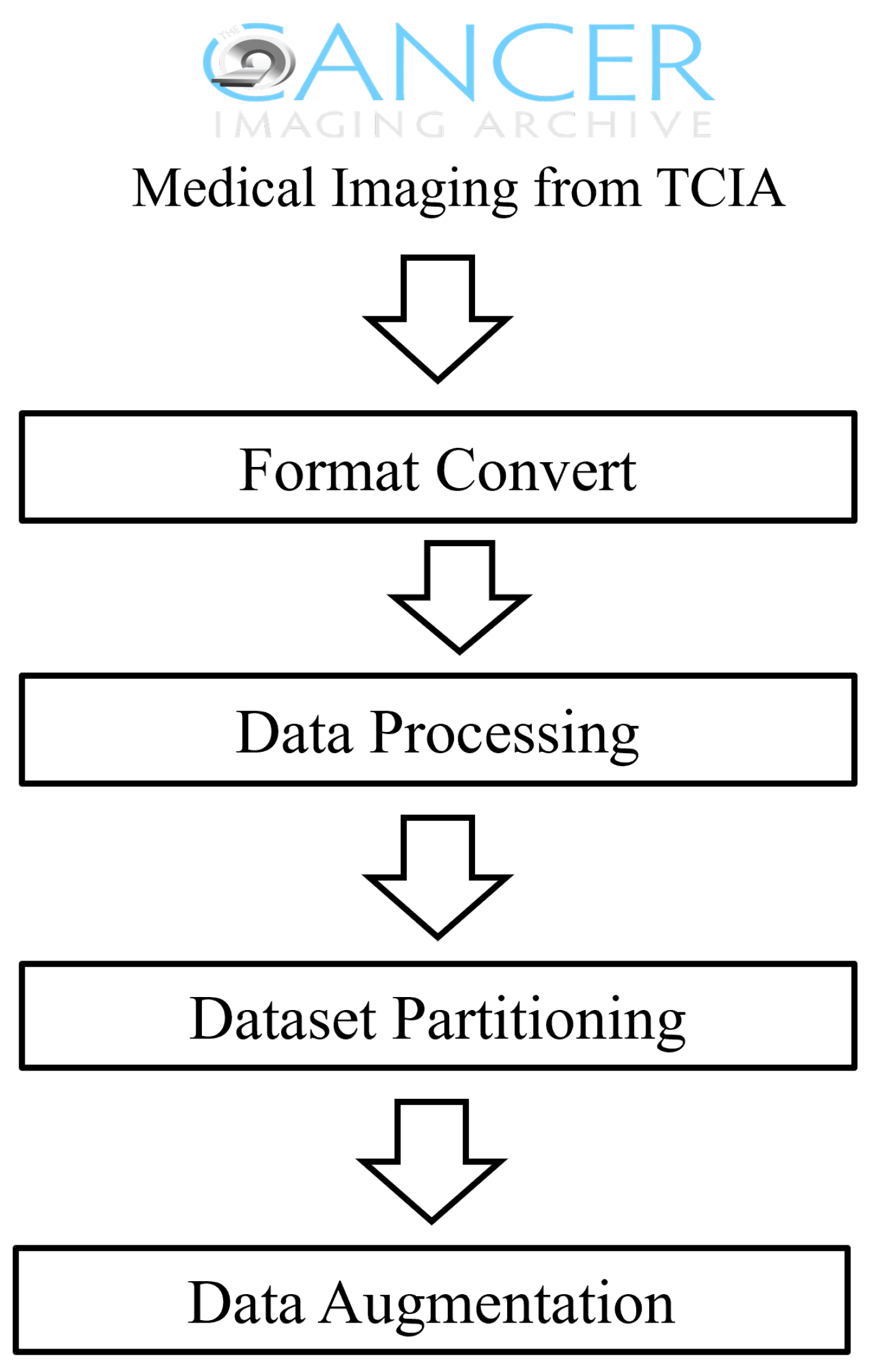
3.1. Data Acquisition and Preprocessing
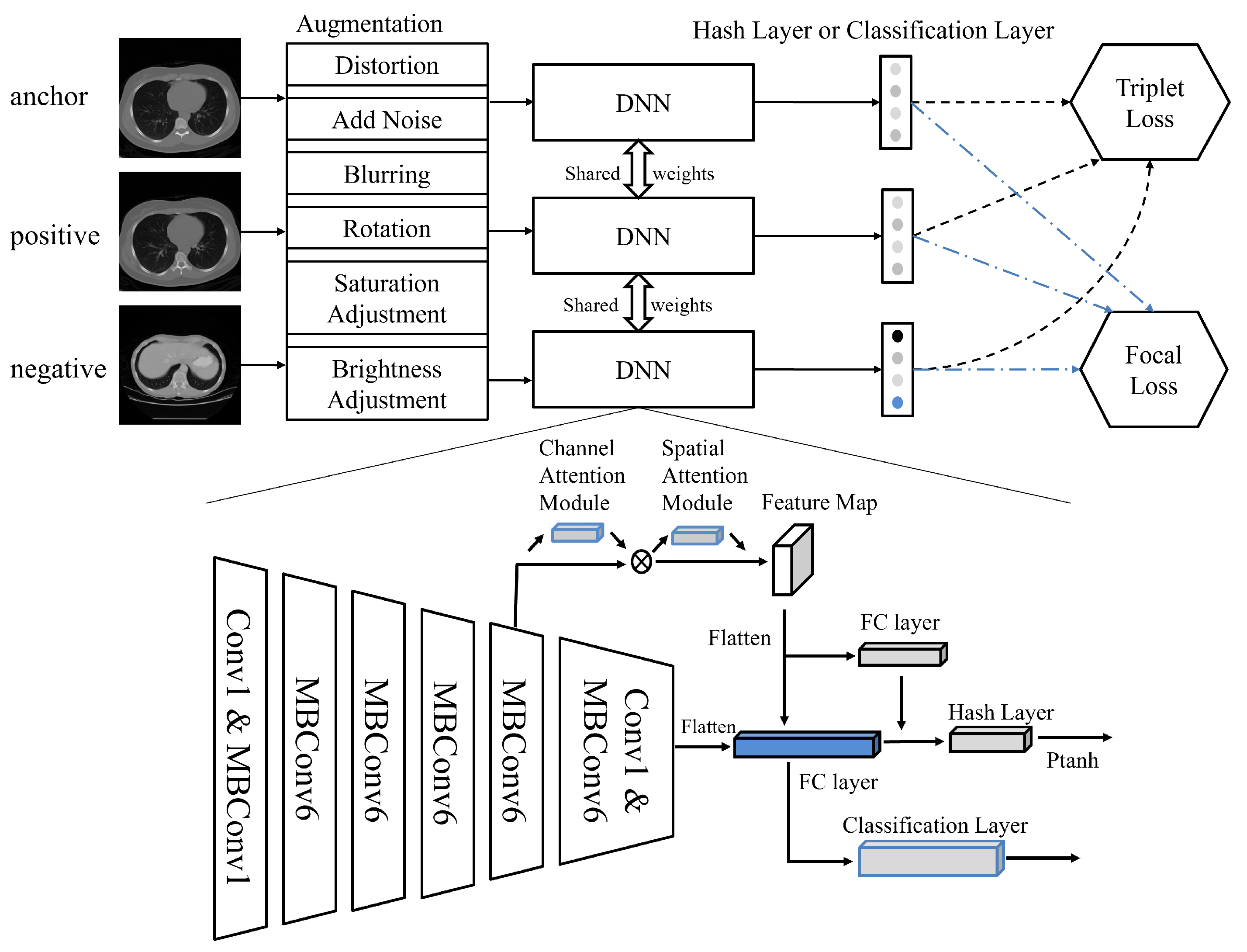
3.2. Medical Image Retrieval Model Based on Deep Triplet Hashing Network
- Data Augmentation: During the data preprocessing stage, we performed data augmentation on the collected medical image data, to enhance the model’s generalizability. This included operations such as rotation, scaling, and brightness adjustment to simulate different imaging conditions, thereby improving the model’s robustness against various transformations.
- Deep Feature Extraction: Utilizing EfficientNet from deep learning as the feature extractor, we independently extracted features from each medical image. EfficientNet is capable of capturing the deep features of images, providing rich information for subsequent image retrieval and classification tasks.
- Attention Mechanism and Inter-layer Feature Fusion: Building on the deep features, we introduced the Convolutional Block Attention Module (CBAM), to enhance the model’s focus on key areas within the images. Additionally, through inter-layer feature-fusion technology, we integrated deep features from different layers to obtain more comprehensive image representation.
- Binary Hash Code Retrieval: Finally, we mapped the continuous features to discrete binary hash codes, using a carefully designed quantization function. This process utilized a parameterized hyperbolic tangent function (Ptanh), which approximates the sign function, thus generating optimized hash codes for ease of similarity comparison during image retrieval.Through these four stages, the method proposed in this paper effectively extracts and integrates information from large-scale medical image data, achieving rapid and accurate medical image retrieval. The innovation of this method lies in its ability to handle multimodal data, as well as its adaptability to small samples and imbalanced datasets.
3.3. Architecture Details
3.3.1. Feature Extraction Based on EfficientNet
3.3.2. Attention and Multi-Layer Feature Fusion
3.3.3. Learnable Quantization Hashing Layer
3.3.4. Classification and Triplet Loss
3.3.5. Training and Implementation Details
| Algorithm 1 Deep Hashing Network Training |
|
3.3.6. Metrics
4. Results
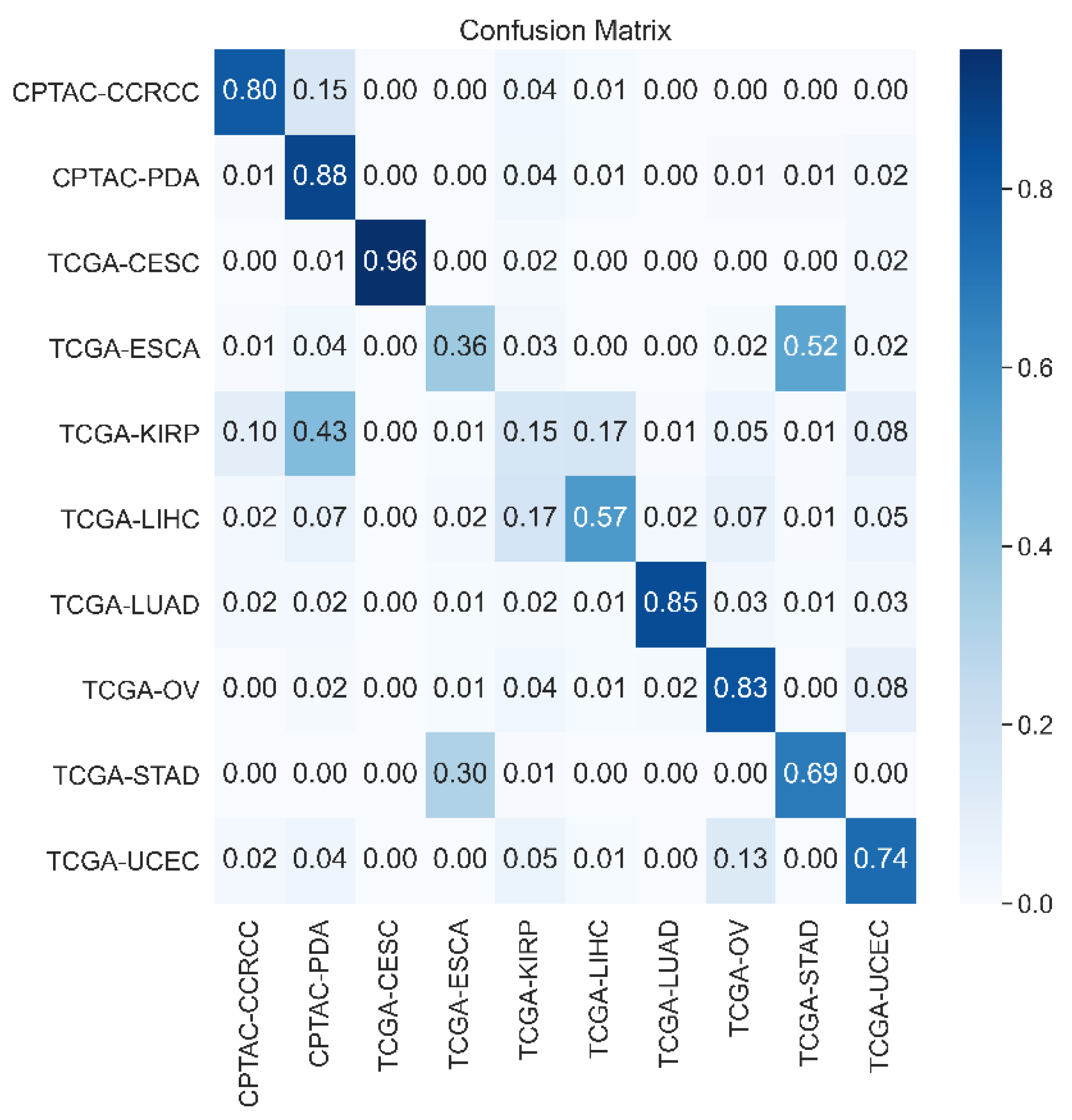
4.1. Comparative Analysis of Retrieval Performance
4.2. Impact of Backbone Network Architecture on Results
4.3. Ablation Analysis of Our Model
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alexander, A.; McGill, M.; Tarasova, A.; Ferreira, C.; Zurkiya, D. Scanning the future of medical imaging. J. Am. Coll. Radiol. 2019, 16, 501–507. [Google Scholar] [CrossRef]
- Shen, D.; Wu, G.; Suk, H.I. Deep learning in medical image analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef] [PubMed]
- Anwar, S.M.; Majid, M.; Qayyum, A.; Awais, M.; Alnowami, M.; Khan, M.K. Medical image analysis using convolutional neural networks: A review. J. Med. Syst. 2018, 42, 1–13. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Gao, K.; Liu, B.; Pan, C.; Liang, K.; Yan, L.; Ma, J.; He, F.; Zhang, S.; Pan, S.; et al. Advances in deep learning-based medical image analysis. Health Data Sci. 2021, 2021, 8786793. [Google Scholar] [CrossRef]
- Li, X.; Yang, J.; Ma, J. Recent developments of content-based image retrieval (CBIR). Neurocomputing 2021, 452, 675–689. [Google Scholar] [CrossRef]
- Shao, W.; Naghdy, G.; Phung, S.L. Automatic image annotation for semantic image retrieval. In Proceedings of the Advances in Visual Information Systems: 9th International Conference, VISUAL 2007, Shanghai, China, 28–29 June 2007; Revised Selected Papers 9. Springer: Berlin/Heidelberg, Germany, 2007; pp. 369–378. [Google Scholar]
- Smeulders, A.W.; Worring, M.; Santini, S.; Gupta, A.; Jain, R. Content-based image retrieval at the end of the early years. IEEE Trans. Pattern Anal. Mach. Intell. 2000, 22, 1349–1380. [Google Scholar] [CrossRef]
- Datta, R.; Joshi, D.; Li, J.; Wang, J.Z. Image retrieval: Ideas, influences, and trends of the new age. ACM Comput. Surv. CSUR 2008, 40, 1–60. [Google Scholar] [CrossRef]
- Rahman, M.M.; Bhattacharya, P.; Desai, B.C. A framework for medical image retrieval using machine learning and statistical similarity matching techniques with relevance feedback. IEEE Trans. Inf. Technol. Biomed. 2007, 11, 58–69. [Google Scholar] [CrossRef]
- Bhosle, N.; Kokare, M. Random forest-based active learning for content-based image retrieval. Int. J. Intell. Inf. Database Syst. 2020, 13, 72–88. [Google Scholar] [CrossRef]
- Lu, X.; Zheng, X.; Li, X. Latent semantic minimal hashing for image retrieval. IEEE Trans. Image Process. 2016, 26, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Jiang, Z.; Zhang, H.; Xie, F.; Zheng, Y.; Shi, H.; Zhao, Y. Breast histopathological image retrieval based on latent dirichlet allocation. IEEE J. Biomed. Health Inform. 2016, 21, 1114–1123. [Google Scholar] [CrossRef] [PubMed]
- Qayyum, A.; Anwar, S.M.; Awais, M.; Majid, M. Medical image retrieval using deep convolutional neural network. Neurocomputing 2017, 266, 8–20. [Google Scholar] [CrossRef]
- Qiu, C.; Cai, Y.; Gao, X.; Cui, Y. Medical image retrieval based on the deep convolution network and hash coding. In Proceedings of the 2017 10th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI), Shanghai, China, 14–16 October 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 1–6. [Google Scholar]
- Fang, J.; Fu, H.; Liu, J. Deep triplet hashing network for case-based medical image retrieval. Med. Image Anal. 2021, 69, 101981. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and operating a public information repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B. Computer-Aided Diagnosis in Mammography Using Content-Based Image Retrieval Approaches: Current Status and Future Perspectives. Algorithms 2009, 2, 828–849. [Google Scholar] [CrossRef] [PubMed]
- Lowe, D. Object recognition from local scale-invariant features. In Proceedings of the Seventh IEEE International Conference on Computer Vision, Kerkyra, Greece, 20–27 September 1999; Volume 2, pp. 1150–1157. [Google Scholar] [CrossRef]
- Cao, Y.; Steffey, S.; He, J.; Xiao, D.; Tao, C.; Chen, P.; Müller, H. Medical image retrieval: A multimodal approach. Cancer Inform. 2014, 13, CIN-S14053. [Google Scholar] [CrossRef] [PubMed]
- Jagtap, J.; Bhosle, N. A comprehensive survey on the reduction of the semantic gap in content-based image retrieval. Int. J. Appl. Pattern Recognit. 2021, 6, 254–271. [Google Scholar] [CrossRef]
- Hwang, K.H.; Lee, H.; Choi, D. Medical image retrieval: Past and present. Healthc. Inform. Res. 2012, 18, 3–9. [Google Scholar] [CrossRef]
- Sharma, A.K.; Nandal, A.; Dhaka, A.; Dixit, R. A survey on machine learning based brain retrieval algorithms in medical image analysis. Health Technol. 2020, 10, 1359–1373. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1–9. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- The Cancer Imaging Archive. TCGA-LUAD | The Cancer Genome Atlas Lung Adenocarcinoma Collection. 2016. Available online: https://www.cancerimagingarchive.net/collection/tcga-luad/ (accessed on 10 June 2024).
- The Cancer Imaging Archive. TCGA-CESC | The Cancer Genome Atlas Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma Collection. 2016. Available online: https://www.cancerimagingarchive.net/collection/tcga-cesc/ (accessed on 10 June 2024).
- The Cancer Imaging Archive. TCGA-STAD | The Cancer Genome Atlas Stomach Adenocarcinoma Collection. 2016. Available online: https://www.cancerimagingarchive.net/collection/tcga-stad/ (accessed on 10 June 2024).
- The Cancer Imaging Archive. CPTAC-PDA | The Clinical Proteomic Tumor Analysis Consortium Pancreatic Ductal Adenocarcinoma Collection. 2016. Available online: https://www.cancerimagingarchive.net/collection/cptac-pda/ (accessed on 10 June 2024).
- The Cancer Imaging Archive. TCGA-ESCA | The Cancer Genome Atlas Esophageal Carcinoma Collection. 2016. Available online: https://www.cancerimagingarchive.net/collection/tcga-esca/ (accessed on 10 June 2024).
- The Cancer Imaging Archive. TCGA-OV | The Cancer Genome Atlas Ovarian Cancer Collection. 2016. Available online: https://www.cancerimagingarchive.net/collection/tcga-ov/ (accessed on 10 June 2024).
- The Cancer Imaging Archive. TCGA-UCEC | The Cancer Genome Atlas Uterine Corpus Endometrial Carcinoma Collection. 2016. Available online: https://www.cancerimagingarchive.net/collection/TCGA-UCEC/ (accessed on 10 June 2024).
- The Cancer Imaging Archive. CPTAC-CCRCC | The Clinical Proteomic Tumor Analysis Consortium Clear Cell Renal Cell Carcinoma Collection. 2016. Available online: https://www.cancerimagingarchive.net/collection/cptac-ccrcc/ (accessed on 10 June 2024).
- The Cancer Imaging Archive. TCGA-LIHC | The Cancer Genome Atlas Liver Hepatocellular Carcinoma Collection. 2016. Available online: https://www.cancerimagingarchive.net/collection/tcga-lihc/ (accessed on 10 June 2024).
- The Cancer Imaging Archive. TCGA-KIRP | The Cancer Genome Atlas Cervical Kidney Renal Papillary Cell Carcinoma Collection. 2016. Available online: https://www.cancerimagingarchive.net/collection/tcga-kirp/ (accessed on 10 June 2024).
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, PMLR, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. Cbam: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal loss for dense object detection. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2980–2988. [Google Scholar]
- Balntas, V.; Riba, E.; Ponsa, D.; Mikolajczyk, K. Learning local feature descriptors with triplets and shallow convolutional neural networks. In Proceedings of the BMVC, York, UK, 19–22 September 2016; Volume 1, p. 3. [Google Scholar]
- Cao, Z.; Long, M.; Wang, J.; Yu, P.S. Hashnet: Deep learning to hash by continuation. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 5608–5617. [Google Scholar]
- Li, W.J.; Wang, S.; Kang, W.C. Feature learning based deep supervised hashing with pairwise labels. arXiv 2015, arXiv:1511.03855. [Google Scholar]
- Gao, J.; Jagadish, H.V.; Lu, W.; Ooi, B.C. DSH: Data sensitive hashing for high-dimensional k-nnsearch. In Proceedings of the 2014 ACM SIGMOD International Conference on Management of Data, Snowbird, UT, USA, 22–27 June 2014; pp. 1127–1138. [Google Scholar]
- Fan, L.; Ng, K.W.; Ju, C.; Zhang, T.; Chan, C.S. Deep Polarized Network for Supervised Learning of Accurate Binary Hashing Codes. In Proceedings of the IJCAI, Online, 7–15 January 2020; pp. 825–831. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2010, arXiv:2010.11929. [Google Scholar]
- Jin, E.; Zhao, D.; Wu, G.; Zhu, J.; Wang, Z.; Wei, Z.; Zhang, S.; Wang, A.; Tang, B.; Chen, X.; et al. OBIA: An open biomedical imaging archive. Genom. Proteom. Bioinform. 2023, 21, 1059–1065. [Google Scholar] [CrossRef]
| Collection | Cancer Type | Location | Subjects | Data Type |
|---|---|---|---|---|
| TCGA-KIRP | Kidney Renal Papillary Cell Carcinoma | Kidney | 33 | CT, MR, PT, Pathology |
| TCGA-LIHC | Liver Hepatocellular Carcinoma | Liver | 97 | MR, CT, PT, Pathology |
| CPTAC-CCRCC | Clear Cell Carcinoma | Kidney | 222 | Clinical, Genomics, Proteomics |
| TCGA-UCEC | Uterine Corpus Endometrial Carcinoma | Uterus | 65 | CT, CR, MR, PT, Pathology |
| TCGA-OV | Ovarian Serous Cystadenocarcinoma | Ovary | 143 | CT, MR, Pathology |
| TCGA-ESCA | Esophageal Carcinoma | Esophagus | 16 | CT, Pathology |
| CPTAC-PDA | Ductal Adenocarcinoma | Pancreas | 168 | CT, MR, PT, US, Pathology |
| TCGA-STAD | Stomach Adenocarcinoma | Stomach | 46 | CT, Pathology |
| TCGA-CESC | Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma | Cervix | 54 | MR, Pathology |
| TCGA-LUAD | Lung Adenocarcinoma | Lung | 69 | CT, PT, NM, Pathology |
| Parameters | Value |
|---|---|
| Epochs | 50 |
| Learning Rates | 1 × |
| Optimizer | Adam |
| Regularization Rate | 1 × |
| Triplet Loss Margin | 0.5 |
| Focal Loss Gamma | 1.5 |
| Model | 16-bit | 32-bit | 48-bit |
|---|---|---|---|
| HashNet | 0.550 | 0.727 | 0.736 |
| DSH | 0.578 | 0.581 | 0.596 |
| DPSH | 0.540 | 0.717 | 0.740 |
| DPN | 0.679 | 0.670 | 0.681 |
| Ours (DAFH) | 0.711 | 0.754 | 0.762 |
| Model | 16-bit | 32-bit | 48-bit |
|---|---|---|---|
| ResNet-50 | - | 0.658 | - |
| ResNet-50+Attention | 0.584 | 0.685 | 0.705 |
| ViT+ResNet-50 | 0.670 | 0.685 | 0.691 |
| EfficientNet | 0.711 | 0.754 | 0.762 |
| Model | MAP@1 | MAP@10 |
|---|---|---|
| EfficientNet | 0.370 | 0.498 |
| EfficientNet+Ptanh | 0.405 | 0.532 |
| EfficientNet+AT | 0.718 | 0.734 |
| EfficientNet+LF | 0.716 | 0.742 |
| EfficientNet+Ptanh+AT | 0.653 | 0.695 |
| EfficientNet+Ptanh+LF | 0.704 | 0.705 |
| EfficientNet+AT+LF | 0.691 | 0.728 |
| Proposed | 0.745 | 0.754 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, G.; Jin, E.; Sun, Y.; Tang, B.; Zhao, W. Deep Attention Fusion Hashing (DAFH) Model for Medical Image Retrieval. Bioengineering 2024, 11, 673. https://doi.org/10.3390/bioengineering11070673
Wu G, Jin E, Sun Y, Tang B, Zhao W. Deep Attention Fusion Hashing (DAFH) Model for Medical Image Retrieval. Bioengineering. 2024; 11(7):673. https://doi.org/10.3390/bioengineering11070673
Chicago/Turabian StyleWu, Gangao, Enhui Jin, Yanling Sun, Bixia Tang, and Wenming Zhao. 2024. "Deep Attention Fusion Hashing (DAFH) Model for Medical Image Retrieval" Bioengineering 11, no. 7: 673. https://doi.org/10.3390/bioengineering11070673
APA StyleWu, G., Jin, E., Sun, Y., Tang, B., & Zhao, W. (2024). Deep Attention Fusion Hashing (DAFH) Model for Medical Image Retrieval. Bioengineering, 11(7), 673. https://doi.org/10.3390/bioengineering11070673








