Comparative Analysis of Large Language Models in Chinese Medical Named Entity Recognition
Abstract
1. Introduction
2. Related Work
2.1. Biomedical Named Entity Recognition
2.2. Large Language Models and In-Context Learning
3. Methodology
3.1. Zero-Shot Prompting
3.2. Few-Shot Prompting
3.3. Instruction Fine-Tuning
4. Experiments Set
4.1. Dataset
4.2. Evaluation Metrics
4.3. Models
- -
- Source code: https://github.com/ymcui/Chinese-BERT-wwm, accessed on 2 November 2021.
- -
- Source code: https://github.com/thudm/chatglm2-6b, accessed on 25 June 2023.
- -
- Unofficial demo: https://huggingface.co/spaces/mikeee/chatglm2-6b-4bit, accessed on 25 June 2023.
- -
- Source code: https://github.com/THUDM/GLM-130B, accessed on 31 July 2023.
- -
- Official online demo: https://chatglm.cn/detail, accessed on 31 July 2023.
- -
- Web application: https://chat.openai.com/, accessed on 30 November 2022.
- -
- Web application: https://chat.openai.com/, accessed on 14 March 2023.
4.4. Experimental Environment
5. Experiment Results
5.1. Zero-Shot Prompting
5.2. Few-Shot Prompting
5.3. Instruction Fine-Tuning
5.3.1. The Comparative Results with Existing State-of-the-Art Methods
5.3.2. The Effects of Data Content
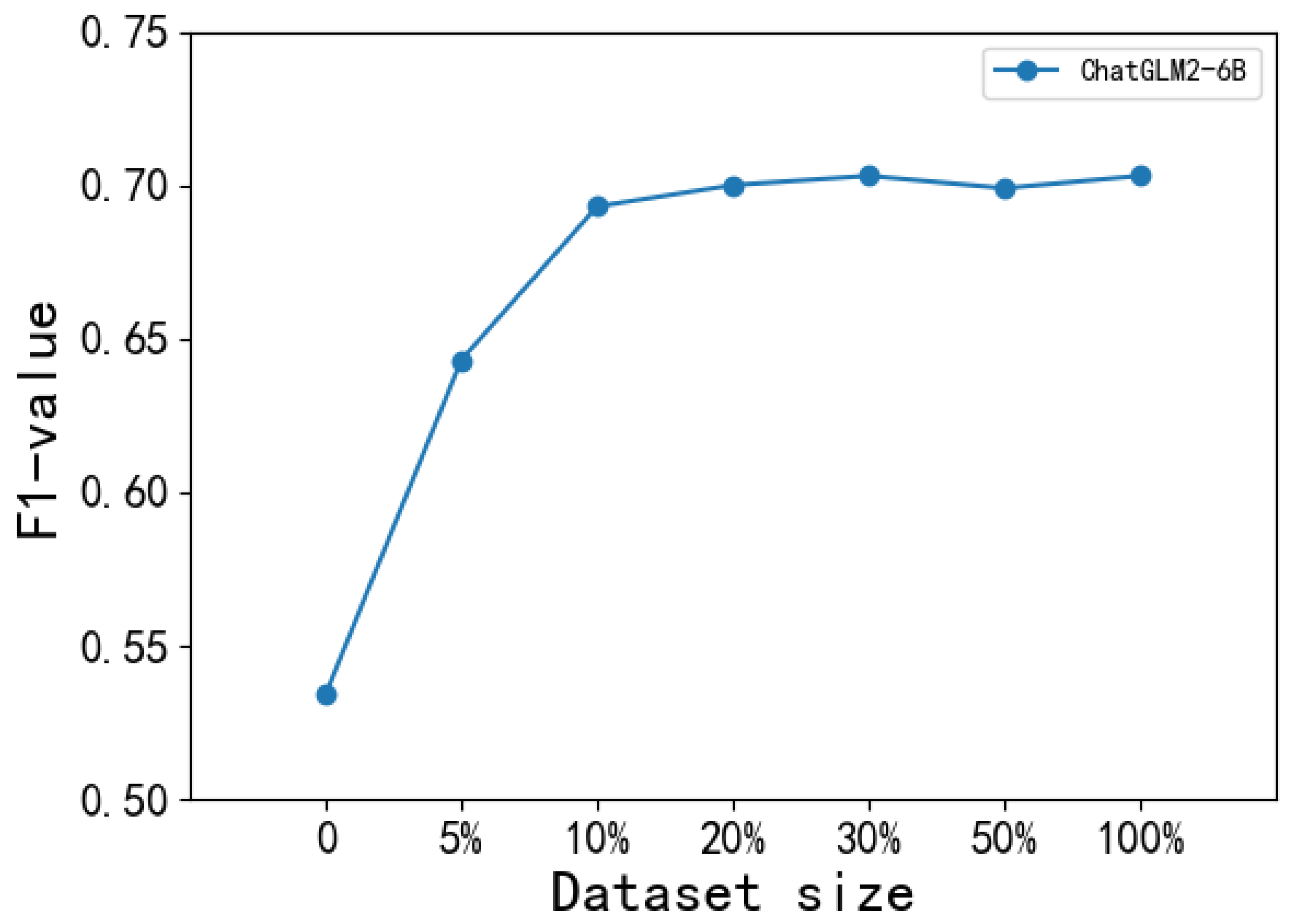
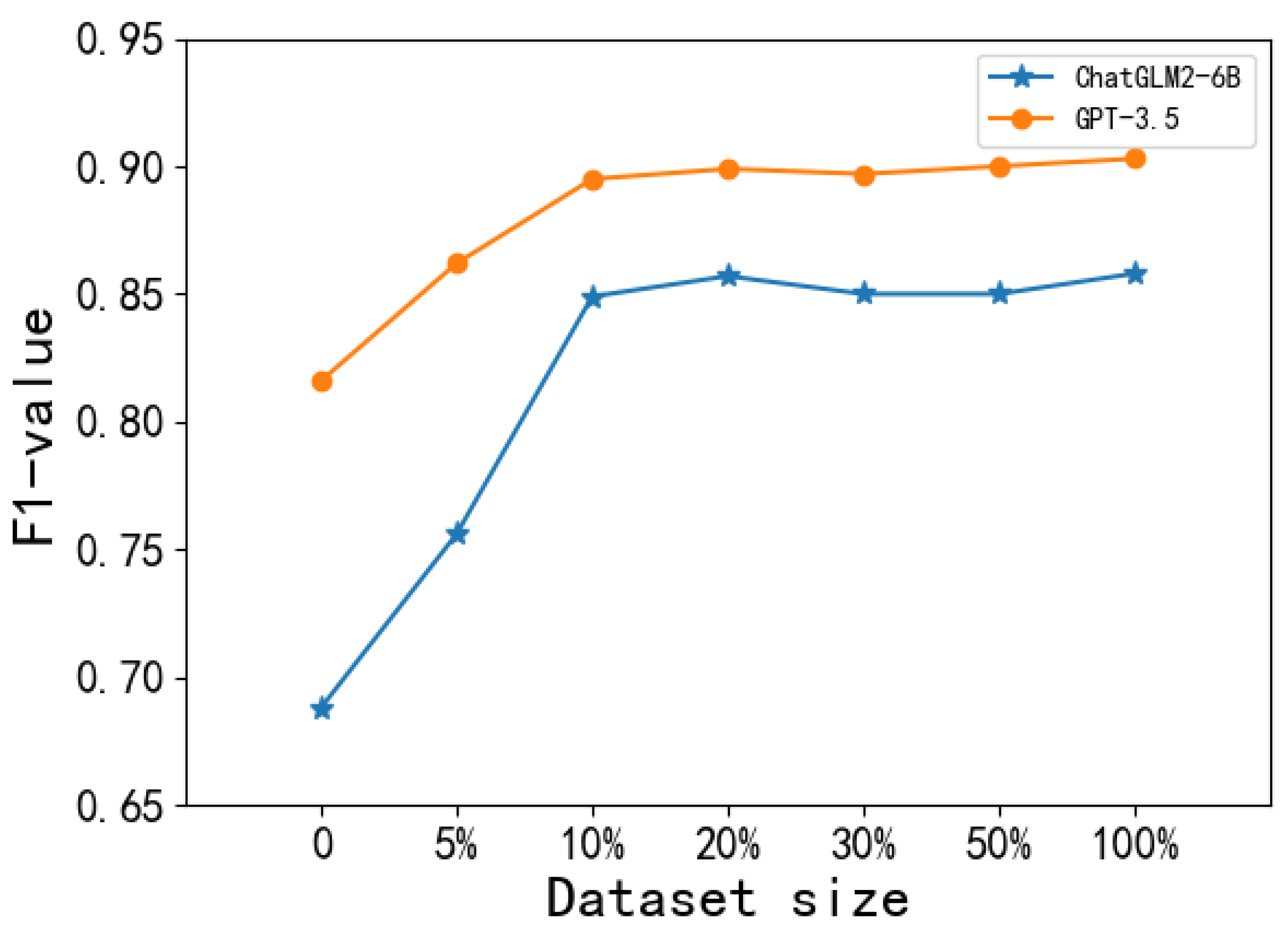
5.3.3. The Effects of Data Scale
5.4. Resource Cost
6. Discussion
6.1. Guidelines for Empowering LLMs for Chinese BNER Tasks
6.2. Bias and Privacy
6.3. Limitations
6.4. Future Work Directions
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhao, Q.; Li, J.; Zhao, L.; Zhu, Z. Knowledge guided feature aggregation for the prediction of chronic obstructive pulmonary disease with Chinese EMRs. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022, 20, 3343–3352. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Xu, D.; Li, J.; Zhao, L.; Rajput, F.A. Knowledge guided distance supervision for biomedical relation extraction in Chinese electronic medical records. Expert Syst. Appl. 2022, 204, 117606. [Google Scholar] [CrossRef]
- Zhao, Q.; Xu, H.; Li, J.; Rajput, F.A.; Qiao, L. The Application of Artificial Intelligence in Alzheimer’s Research. Tsinghua Sci. Technol. 2023, 29, 13–33. [Google Scholar] [CrossRef]
- Chen, X.; Cheng, Q. Acute Complication Prediction and Diagnosis Model CLSTM-BPR: A Fusion Method of Time Series Deep Learning and Bayesian Personalized Ranking. Tsinghua Sci. Technol. 2024, 29, 1509–1523. [Google Scholar] [CrossRef]
- Tan, L.; Liang, Y.; Xia, J.; Wu, H.; Zhu, J. Detection and Diagnosis of Small Target Breast Masses Based on Convolutional Neural Networks. Tsinghua Sci. Technol. 2024, 29, 1524–1539. [Google Scholar] [CrossRef]
- Lin, X.; Lei, Y.; Chen, J.; Xing, Z.; Yang, T.; Wang, Q.; Wang, C. A Case-Finding Clinical Decision Support System to Identify Subjects with Chronic Obstructive Pulmonary Disease Based on Public Health Data. Tsinghua Sci. Technol. 2023, 28, 525–540. [Google Scholar] [CrossRef]
- Li, D.; Ma, H.; Li, W.; Zhao, B.; Zhao, J.; Liu, Y.; Fu, J. KTI-RNN: Recognition of Heart Failure from Clinical Notes. Tsinghua Sci. Technol. 2023, 28, 117–130. [Google Scholar] [CrossRef]
- Yu, Y.; Duan, J.; Li, M. Fusion Model for Tentative Diagnosis Inference Based on Clinical Narratives. Tsinghua Sci. Technol. 2023, 28, 686–695. [Google Scholar] [CrossRef]
- Li, J.; Sun, A.; Han, J.; Li, C. A Survey on Deep Learning for Named Entity Recognition. IEEE Trans. Knowl. Data Eng. 2018, 34, 50–70. [Google Scholar] [CrossRef]
- Gokgol, M.K.; Orhan, Z. OP41 Intercultural Medical Decision Support System Using Natural Language Processing (NLP). Int. J. Technol. Assess. Health Care 2019, 35, 10. [Google Scholar] [CrossRef]
- Trujillo, A.; Orellana, M.; Acosta, M.I. Design of emergency call record support system applying natural language processing techniques. In Proceedings of the Conference on Information Technologies and Communication of Ecuador(TIC.EC), Cuenca City, Ecuador, 27–29 November 2019; pp. 53–65. [Google Scholar]
- Redjdal, A.; Novikava, N.; Kempf, E.; Bouaud, J.; Seroussi, B. Leveraging Rule-Based NLP to Translate Textual Reports as Structured Inputs Automatically Processed by a Clinical Decision Support System. Stud. Health Technol. Inform. 2024, 316, 1861–1865. [Google Scholar] [PubMed]
- Parmar, J.; Koehler, W.C.; Bringmann, M.; Volz, K.S.; Kapicioglu, B. Biomedical Information Extraction for Disease Gene Prioritization. arXiv 2020, arXiv:2011.05188. [Google Scholar]
- Zhu, Z.; Li, J.; Zhao, Q.; Wei, Y.; Jia, Y. Medical named entity recognition of Chinese electronic medical records based on stacked Bidirectional Long Short-Term Memory. In Proceedings of the 2021 IEEE 45th Annual Computers 2021, Software, and Applications Conference (COMPSAC), Madrid, Spain, 16 July 2021; pp. 1930–1935. [Google Scholar]
- Ma, Z.; Zhao, L.; Li, J.; Xu, X.; Li, J. SiBERT: A Siamese-based BERT Network for Chinese Medical Entities Alignment. Methods 2022, 205, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Bommasani, R.; Hudson, D.A.; Adeli, E.; Altman, R.; Arora, S.; von Arx, S.; Bernstein, M.S.; Bohg, J.; Bosselut, A.; Brunskill, E.; et al. On the Opportunities and Risks of Foundation Models. arXiv 2021, arXiv:2108.07258. [Google Scholar]
- Zhou, C.; Li, Q.; Li, C.; Yu, J.; Liu, Y.; Wang, G.; Zhang, K.; Ji, C.; Yan, Q.; He, L.; et al. A Comprehensive Survey on Pretrained Foundation Models: A History from BERT to ChatGPT. arXiv 2023, arXiv:2302.09419. [Google Scholar]
- Wei, J.; Tay, Y.; Bommasani, R.; Raffel, C.; Zoph, B.; Borgeaud, S.; Bosma, M.; Zhou, D.; Metzler, D.; Chi, E.H.; et al. Emergent Abilities of Large Language Models. arXiv 2022, arXiv:2206.07682. [Google Scholar]
- Brown, T.; Mann, B.; Ryder, N.; Subbiah, M.; Kaplan, J.D.; Dhariwal, P.; Neelakantan, A.; Shyam, P.; Sastry, G.; Askell, A.; et al. Language Models are Few-Shot Learners. arXiv 2020, arXiv:2005.14165. [Google Scholar]
- Ouyang, L.; Wu, J.; Jiang, X.; Almeida, D.; Wainwright, C.; Mishkin, P.; Zhang, C.; Agarwal, S.; Slama, K.; Ray, A.; et al. Training language models to follow instructions with human feedback. NeurIPS 2022, 35, 730–744. [Google Scholar]
- OpenAI. GPT-4 Technical Report. arXiv 2023, arXiv:2303.08774. [Google Scholar]
- Li, J.; Li, H.; Pan, Z.; Pan, G. Prompt ChatGPT In MNER: Improved multimodal named entity recognition method based on auxiliary refining knowledge from ChatGPT. arXiv 2023, arXiv:2305.12212. [Google Scholar]
- Chokwijitkul, T.; Nguyen, A.; Hassanzadeh, H.; Perez, S. Identifying risk factors for heart disease in electronic medical records: A deep learning approach. In Proceedings of the BioNLP 2018 workshop, Melbourne, Australia, 19 July 2018; pp. 18–27. [Google Scholar]
- Xu, K.; Zhou, Z.; Hao, T.; Liu, W. A bidirectional LSTM and conditional random fields approach to medical named entity recognition. In Proceedings of the International Conference on Advanced Intelligent Systems and Informatics 2017; Springer: Berlin/Heidelberg, Germany, 2017; pp. 355–365. [Google Scholar] [CrossRef]
- Unanue, I.J.; Borzeshi, E.Z.; Piccardi, M. Recurrent neural networks with specialized word embeddings for health-domain named-entity recognition. J. Biomed. Inform. 2017, 76, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhao, J.; Hou, L.; Zhai, Y.; Shi, J.; Cui, F. An attention-based deep learning model for clinical named entity recognition of Chinese electronic medical records. BMC Med. Inform. Decis. Mak. 2019, 19, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Yang, Z.; Kang, P.; Wang, Q.; Liu, W. Document-level attention-based BiLSTM-CRF incorporating disease dictionary for disease named entity recognition. Comput. Biol. Med. 2019, 108, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Li, J.; Zhao, Q.; Akhtar, F. A dictionary-guided attention network for biomedical named entity recognition in Chinese electronic medical records. Expert Syst. Appl. 2023, 231, 120709. [Google Scholar] [CrossRef]
- Rae, J.W.; Borgeaud, S.; Cai, T.; Millican, K.; Hoffmann, J.; Song, F.; Aslanides, J.; Henderson, S.; Ring, R.; Young, S.Y.; et al. Scaling language models: Methods, analysis & insights from training gopher. arXiv 2021, arXiv:2112.11446. [Google Scholar]
- Smith, S.; Patwary, M.; Norick, B.; LeGresley, P.; Rajbhandari, S.; Casper, J.; Liu, Z.; Prabhumoye, S.; Zerveas, G.; Korthikanti, V.; et al. Using deepspeed and megatron to train megatron-turing nlg 530b, a large-scale generative language model. arXiv 2022, arXiv:2201.11990. [Google Scholar]
- Hoffmann, J.; Borgeaud, S.; Mensch, A.; Buchatskaya, E.; Cai, T.; Rutherford, E.; Casas, D.d.; Hendricks, L.A.; Welbl, J.; Clark, A.; et al. Training compute-optimal large language models. arXiv 2022, arXiv:2203.15556. [Google Scholar]
- Chowdhery, A.; Narang, S.; Devlin, J.; Bosma, M.; Mishra, G.; Roberts, A.; Barham, P.; Chung, H.W.; Sutton, C.; Gehrmann, S.; et al. Palm: Scaling language modeling with pathways. arXiv 2022, arXiv:2204.02311. [Google Scholar]
- Hegselmann, S.; Buendia, A.; Lang, H.; Agrawal, M.; Jiang, X.; Sontag, D. Tabllm: Few-shot classification of tabular data with large language models. arXiv 2022, arXiv:2210.10723. [Google Scholar]
- Vilar, D.; Freitag, M.; Cherry, C.; Luo, J.; Ratnakar, V.; Foster, G. Prompting palm for translation: Assessing strategies and performance. arXiv 2022, arXiv:2211.09102. [Google Scholar]
- Perez, E.; Kiela, D.; Cho, K. True few-shot learning with language models. Adv. Neural Inf. Process. Syst. 2021, 34, 11054–11070. [Google Scholar]
- Pietrzak, B.; Swanson, B.; Mathewson, K.; Dinculescu, M.; Chen, S. Story Centaur: Large Language Model Few Shot Learning as a Creative Writing Tool; Association for Computational Linguistics: Toronto, QC, Canada, 2021. [Google Scholar]
- Wei, J.; Bosma, M.; Zhao, V.Y.; Guu, K.; Yu, A.W.; Lester, B.; Du, N.; Dai, A.M.; Le, Q.V. Finetuned language models are zero-shot learners. arXiv 2021, arXiv:2109.01652. [Google Scholar]
- Raffel, C.; Shazeer, N.; Roberts, A.; Lee, K.; Narang, S.; Matena, M.; Zhou, Y.A.; Li, W.; Liu, P.J. Exploring the limits of transfer learning with a unified text-to-text transformer. J. Mach. Learn. Res. 2020, 21, 5485–5551. [Google Scholar]
- Gururangan, S.; Swayamdipta, S.; Levy, O.; Schwartz, R.; Bowman, S.R.; Smith, N.A. Annotation artifacts in natural language inference data. arXiv 2018, arXiv:1803.02324. [Google Scholar]
- Roberts, A.; Raffel, C.; Shazeer, N. How much knowledge can you pack into the parameters of a language model? arXiv 2020, arXiv:2002.08910. [Google Scholar]
- Guu, K.; Lee, K.; Tung, Z.; Pasupat, P.; Chang, M. Retrieval augmented language model pre-training. arXiv 2020, arXiv:2002.08909. [Google Scholar]
- Liu, J.; Shen, D.; Zhang, Y.; Dolan, B.; Carin, L.; Chen, W. What makes good in-context examples for gpt-3? arXiv 2021, arXiv:2101.06804. [Google Scholar]
- Rubin, O.; Herzig, J.; Berant, J. Learning to retrieve prompts for in-context learning. arXiv 2021, arXiv:2112.08633. [Google Scholar]
- Lee, D.; Kadakia, A.; Tan, K.; Agarwal, M.; Feng, X.; Shibuya, T.; Mitani, R.; Sekiya, T.; Pujara, J.; Ren, X. Good examples make a faster learner: Simple demonstration-based learning for low-resource ner. arXiv 2021, arXiv:2110.08454. [Google Scholar]
- Wang, X.; Zhu, W.; Wang, W.Y.A. Large language models are implicitly topic models: Explaining and finding good demonstrations for in-context learning. arXiv 2023, arXiv:2301.11916. [Google Scholar]
- Kojima, T.; Gu, S.S.; Reid, M.; Matsuo, Y.; Iwasawa, Y. Large language models are zero-shot reasoners. Adv. Neural Inf. Process. Syst. 2022, 35, 22199–22213. [Google Scholar]
- Radford, A.; Wu, J.; Child, R.; Luan, D.; Amodei, D.; Sutskever, I. Language Models are Unsupervised Multitask Learners. OpenAI Blog 2019, 1, 9. [Google Scholar]
- Dong, Y.; Jiang, X.; Jin, Z.; Li, G. Self-collaboration Code Generation via ChatGPT. arXiv 2023, arXiv:2304.07590. [Google Scholar] [CrossRef]
- Devlin, J.; Chang, M.; Lee, K.; Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Dai, Z.; Wang, X.; Ni, P.; Li, Y.; Li, G.; Bai, X. Named entity recognition using BERT BiLSTM CRF (BBC) for Chinese electronic health records. In Proceedings of the 2019 12th International Congress on Image and Signal Processing, Biomedical Engineering and Informatics (CISP-BMEI), Suzhou, China, 21 October 2019; pp. 1–5. [Google Scholar]
- Yang, Z.; Salakhutdinov, R.; Cohen, W. Multi-task cross-lingual sequence tagging from scratch. arXiv 2016, arXiv:1603.06270. [Google Scholar]
- Li, X.; Zhang, H.; Zhou, X.H. Chinese clinical named entity recognition with variant neural structures based on BERT methods. J. Biomed. Inform. 2020, 107, 103422. [Google Scholar] [CrossRef]
- Maslej, N.; Fattorini, L.; Brynjolfsson, E.; Etchemendy, J.; Ligett, K.; Lyons, T.; Manyika, J.; Ngo, H.; Niebles, J.; Parli, V.; et al. Artificial Intelligence Index Report 2023. arXiv 2023, arXiv:2310.03715. [Google Scholar]
- Hoover, A. An Eating Disorder Chatbot is Suspended for Giving Harmful Advice. 2023. Available online: https://www.wired.com/story/tessachatbot-suspended/ (accessed on 14 March 2023).
- Ghosh, S.; Caliskan, A. ChatGPT Perpetuates Gender Bias in Machine Translation and Ignores Non-Gendered Pronouns: Findings across Bengali and Five other Low-Resource. arXiv 2023, arXiv:2305.10510. [Google Scholar]
- Abid, A.; Farooqi, M.; Zou, J. Persistent anti-muslim bias in large language models. In Proceedings of the 2021 AAAI/ACM Conference on AI, Ethics, and Society, New York, NY, USA, 19–21 May 2021; pp. 298–306. [Google Scholar]
- Pessach, D.; Shmueli, E. A review on fairness in machine learning. ACM Comput. Surv. (CSUR) 2022, 55, 1–44. [Google Scholar] [CrossRef]
- Gemalmaz, M.A.; Yin, M. Accounting for Confirmation Bias in Crowdsourced Label Aggregation. In Proceedings of the International Joint Conference on Artificial Intelligence (IJCAI), Montreal, QC, Canada, 19–27 August 2021; pp. 1729–1735. [Google Scholar]
- Touvron, H.; Lavril, T.; Izacard, G.; Martinet, X.; Lachaux, M.; Lacroix, T.; Roziere, B.; Goyal, N.; Hambro, E.; Azhar, F.; et al. LLaMA: Open and Efficient Foundation Language Models. arXiv 2023, arXiv:2302.13971. [Google Scholar]
- Touvron, H.; Martin, L.; Stone, K.R.; Albert, P.; Almahairi, A.; Babaei, Y.; Bashlykov, N.; Batra, S.; Bhargava, P.; Bhosale, S.; et al. Llama 2: Open Foundation and Fine-Tuned Chat Models. arXiv 2023, arXiv:2307.09288. [Google Scholar]
- Wei, J.; Wang, X.; Schuurmans, D.; Bosma, M.; Chi, E.H.; Xia, F.; Le, Q.; Zhou, D. Chain of Thought Prompting Elicits Reasoning in Large Language Models. arXiv 2022, arXiv:2201.11903. [Google Scholar]
- Miao, N.; Teh, Y.W.; Rainforth, T. SelfCheck: Using LLMs to Zero-Shot Check Their Own Step-by-Step Reasoning. arXiv 2023, arXiv:2308.00436. [Google Scholar]
- Kairouz, P.; McMahan, H.B.; Avent, B.; Bellet, A.; Bennis, M.; Bhagoji, A.N.; Bonawitz, K.; Charles, Z.; Cormode, G.; Cummings, R. Advances and Open Problems in Federated Learning. Found. Trends Mach. Learn. 2019, 14, 1–210. [Google Scholar] [CrossRef]
- Warnat-Herresthal, S.; Schultze, H.; Shastry, K.L.; Manamohan, S.; Mukherjee, S.; Garg, V.; Sarveswara, R.; Handler, K.; Pickkers, P.; Aziz, N.A.; et al. Swarm Learning for decentralized and confidential clinical machine learning. Nature 2021, 594, 265–270. [Google Scholar] [CrossRef] [PubMed]
| S_text | |
|---|---|
| Basic | {} |
| Context enhancement | (1) 这是一个命名实体识别任务, 注意实体的类别只有“症状、检查、检查结果、疾病、治疗”五种。(This is a named entity recognition task, note that there are only five categories of entities: ‘symptom’, ‘test’, ‘test result’, ‘disease’, and ‘treatment’.) (2) 这是一个命名实体识别任务, 注意只考虑“症状、检查、检查结果、疾病、治疗”这五种实体类别。(This is a named entity recognition task, note that only considering five entity categories: ‘symptom’, ‘test’, ‘test result’, ‘disease’, and ‘treatment’.) |
| Role-playing | (1) 作为一名医学专家, 请阅读这条电子病历文本并回答这个问题。(As a medical expert, please read this electronic medical record text and answer this question.) (2) 如果你是一名医学专家, 请阅读这条电子病历文本并回答这个问题。(If you are a medical expert, please read this electronic medical record text and answer this question.) |
| Context and Role-playing | (1) 这是一个命名实体识别任务, 注意实体类别只有“症状、检查、检查结果、疾病、治疗”五种。作为一名医学专家, 请阅读这条电子病历文本并回答这个问题。(This is a named entity recognition task, note that there are only five categories of entities: ‘symptom’, ‘test’, ‘test result’, ‘disease’, and ‘treatment’. As a medical expert, please read this electronic medical record text and answer this question.) (2) 这是一个命名实体识别任务, 注意只考虑“症状、检查、检查结果、疾病、治疗”这五种实体类别。作为一名医学专家, 请阅读这条电子病历文本并回答这个问题。(This is a named entity recognition task, note that only considering five entity categories: ‘symptom’, ‘test’, ‘test result’, ‘disease’, and ‘treatment’. As a medical expert, please read this electronic medical record text and answer this question.) (3) 这是一个命名实体识别任务, 注意实体类别只有“症状、检查、检查结果、疾病、治疗”五种。如果你是一名医学专家, 请阅读这条电子病历文本并回答这个问题。(This is a named entity recognition task, note that there are only five categories of entities: ‘symptom’, ‘test’, ‘test result’, ‘disease’, and ‘treatment’. If you are a medical expert, please read this electronic medical record text and answer this question.) (4) 这是一个命名实体识别任务, 注意只考虑“症状、检查、检查结果、疾病、治疗”这五种实体类别。如果你是一名医学专家, 请阅读这条电子病历文本并回答这个问题。(This is a named entity recognition task, note that only considering five entity categories: ‘symptom’, ‘test’, ‘test result’, ‘disease’, and ‘treatment’. If you are a medical expert, please read this electronic medical record text and answer this question.) |
| Q_text | |
| Entity recognition and type prediction | (1) 请标记出这条文本中的医学实体, 并相应的给出这些实体所属的类型。(Please label the biomedical entities in this text and provide their corresponding types.) (2) 这条文本中的医学实体有哪些, 请识别出这些实体并给出其所属的类型。(What are the biomedical entities in this text? Please recognize these entities and provide their corresponding types.) |
| Processor | Cache | GPU | Hard Disk |
|---|---|---|---|
| 13th Gen Intel Core i9-13900KF × 32 | 128 GB | NVIDIA Corporation 4090 24 GB | 3.0 TB |
| Type | Category | Models | P | R | F1 |
|---|---|---|---|---|---|
| Supervised | Baseline | BC [24] | 0.681 ± 0.001 | 0.670 ± 0.002 | 0.675 ± 0.001 |
| BERT [49] | 0.744 ± 0.017 | 0.721 ± 0.010 | 0.732 ± 0.015 | ||
| BBC [50] | 0.748 ± 0.008 | 0.732 ± 0.015 | 0.740 ± 0.012 | ||
| RSBGC [51] | 0.756 ± 0.006 | 0.763± 0.010 | 0.760 ± 0.008 | ||
| FBBCE [52] | 0.802 ± 0.005 | 0.803 ± 0.010 | 0.802 ± 0.007 | ||
| DABLC [27] | 0.815 ± 0.004 | 0.812 ± 0.004 | 0.813 ± 0.003 | ||
| DGAN [28] | 0.832 ± 0.000 | 0.820 ± 0.000 | 0.826 ± 0.000 | ||
| LLMs | Zero-shot | 0.388 ± 0.023 | 0.402 ± 0.015 | 0.395 ± 0.020 | |
| 0.500 ± 0.031 | 0.592 ± 0.041 | 0.542 ± 0.036 | |||
| 0.482 ± 0.010 | 0.501 ± 0.039 | 0.491 ± 0.022 | |||
| 0.551 ± 0.031 | 0.567 ± 0.020 | 0.559 ± 0.025 | |||
| LLMs | Few-shot | 0.600 ± 0.014 | 0.588 ± 0.019 | 0.594 ± 0.018 | |
| 0.613 ± 0.017 | 0.609 ± 0.010 | 0.611 ± 0.012 | |||
| LLMs | Fine-tuning | 0.694 ± 0.032 | 0.730 ± 0.023 | 0.711 ± 0.025 | |
| 0.576 ± 0.021 | 0.590 ± 0.018 | 0.583 ± 0.018 | |||
| 0.720 ± 0.010 | 0.721 ± 0.017 | 0.720 ± 0.012 |
| Type | Category | Models | P | R | F1 |
|---|---|---|---|---|---|
| Supervised | Baseline | BC [24] | 0.836 ± 0.001 | 0.849 ± 0.001 | 0.842 ± 0.001 |
| BERT [49] | 0.879 ± 0.003 | 0.858 ± 0.007 | 0.868 ± 0.004 | ||
| BBC [50] | 0.877 ± 0.009 | 0.873 ± 0.010 | 0.875 ± 0.007 | ||
| RSBGC [51] | 0.891 ± 0.004 | 0.884 ± 0.003 | 0.887 ± 0.004 | ||
| FBBCE [52] | 0.920 ± 0.003 | 0.913 ± 0.002 | 0.916 ± 0.003 | ||
| DABLC [27] | 0.925 ± 0.002 | 0.919 ± 0.001 | 0.922 ± 0.001 | ||
| DGAN [28] | 0.950 ± 0.001 | 0.954 ± 0.001 | 0.952 ± 0.001 | ||
| LLMs | Zero-shot | 0.546 ± 0.024 | 0.600 ± 0.037 | 0.572 ± 0.030 | |
| 0.615 ± 0.041 | 0.652 ± 0.038 | 0.633 ± 0.040 | |||
| 0.600 ± 0.031 | 0.567 ± 0.043 | 0.583 ± 0.037 | |||
| 0.672 ± 0.056 | 0.641 ± 0.022 | 0.648 ± 0.039 | |||
| 0.642 ± 0.004 | 0.599 ± 0.019 | 0.620 ± 0.013 | |||
| 0.771 ± 0.030 | 0.723 ± 0.011 | 0.746 ± 0.020 | |||
| 0.648 ± 0.022 | 0.627 ± 0.039 | 0.637 ± 0.033 | |||
| 0.748 ± 0.005 | 0.804 ± 0.044 | 0.775 ± 0.039 | |||
| 0.663 ± 0.041 | 0.621 ± 0.044 | 0.641 ± 0.045 | |||
| 0.788 ± 0.009 | 0.740 ± 0.014 | 0.763 ± 0.011 | |||
| 0.732 ± 0.066 | 0.677 ± 0.017 | 0.703 ± 0.049 | |||
| 0.802 ± 0.022 | 0.776 ± 0.030 | 0.789 ± 0.025 | |||
| 0.704 ± 0.077 | 0.660 ± 0.064 | 0.681 ± 0.065 | |||
| 0.732 ± 0.036 | 0.819 ± 0.038 | 0.773 ± 0.038 | |||
| 0.750 ± 0.017 | 0.711 ± 0.015 | 0.730 ± 0.016 | |||
| 0.784 ± 0.031 | 0.801 ± 0.009 | 0.792 ± 0.019 | |||
| LLMs | Few-shot | 0.660 ± 0.035 | 0.710 ± 0.024 | 0.684 ± 0.030 | |
| 0.799 ± 0.010 | 0.773 ± 0.017 | 0.786 ± 0.015 | |||
| 0.820 ± 0.027 | 0.802 ± 0.014 | 0.811 ± 0.021 | |||
| 0.813 ± 0.011 | 0.832 ± 0.005 | 0.822 ± 0.007 | |||
| 0.675 ± 0.047 | 0.702 ± 0.056 | 0.688 ± 0.060 | |||
| 0.818 ± 0.009 | 0.800 ± 0.032 | 0.809 ± 0.020 | |||
| 0.837 ± 0.016 | 0.796 ± 0.020 | 0.816 ± 0.018 | |||
| 0.822 ± 0.006 | 0.838 ± 0.008 | 0.830 ± 0.006 | |||
| LLMs | Fine-tuning | 0.759 ± 0.023 | 0.702 ± 0.018 | 0.729 ± 0.021 | |
| 0.830 ± 0.006 | 0.861 ± 0.004 | 0.845 ± 0.005 | |||
| 0.867 ± 0.012 | 0.860 ± 0.009 | 0.863 ± 0.010 | |||
| 0.897 ± 0.009 | 0.915 ± 0.003 | 0.906 ± 0.005 |
| Models | PCHD | CCKS2017 | ||||
|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | |
| ↑0.112 | ↑0.190 | ↑0.147 | ↑0.069 | ↑0.052 | ↑0.061 | |
| ↑0.094 | ↑0.099 | ↑0.096 | ↑0.054 | ↓0.033 | ↑0.011 | |
| ↑0.163 | ↑0.165 | ↑0.164 | ↑0.126 | ↑0.041 | ↑0.076 | |
| - | - | - | ↑0.129 | ↑0.124 | ↑0.126 | |
| - | - | - | ↑0.006 | ↑0.028 | ↑0.017 | |
| - | - | - | ↑0.105 | ↑0.205 | ↑0.155 | |
| - | - | - | ↑0.125 | ↑0.119 | ↑0.122 | |
| - | - | - | ↑0.069 | ↑0.056 | ↑0.062 | |
| - | - | - | ↑0.139 | ↑0.155 | ↑0.148 | |
| - | - | - | ↑0.028 | ↑0.159 | ↑0.092 | |
| - | - | - | ↑0.046 | ↑0.051 | ↑0.049 | |
| - | - | - | ↑0.080 | ↑0.141 | ↑0.111 | |
| Models | PCHD | CCKS2017 | ||||
|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | |
| ↑0.049 | ↓0.021 | ↑0.035 | ↓0.012 | ↑0.069 | ↑0.036 | |
| - | - | - | ↑0.051 | ↓0.031 | ↑0.011 | |
| - | - | - | ↑0.018 | ↑0.026 | ↑0.021 | |
| - | - | - | ↑0.029 | ↑0.031 | ↑0.030 | |
| ↑0.062 | ↑0.042 | ↑0.052 | ↑0.003 | ↑0.061 | ↑0.040 | |
| - | - | - | ↑0.070 | ↓0.004 | ↑0.034 | |
| - | - | - | ↑0.035 | ↑0.020 | ↑0.027 | |
| - | - | - | ↑0.038 | ↑0.037 | ↑0.038 | |
| Models | PCHD | CCKS2017 | ||||
|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | |
| ↑0.143 | ↑0.163 | ↑0.152 | ↑0.087 | ↑0.061 | ↑0.081 | |
| ↑0.025 | ↓0.023 | ↑0.024 | ↑0.158 | ↑0.220 | ↑0.197 | |
| ↑0.169 | ↑0.154 | ↑0.161 | ↑0.195 | ↑0.219 | ↑0.215 | |
| - | - | - | ↑0.113 | ↑0.114 | ↑0.114 | |
| Type | Models | PCHD | CCKS2017 | ||||
|---|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | ||
| Baseline | BC [24] | 0.836 ± 0.001 | 0.849 ± 0.001 | 0.842 ± 0.001 | 0.836 ± 0.001 | 0.849 ± 0.001 | 0.842 ± 0.001 |
| BERT [49] | 0.879 ± 0.003 | 0.858 ± 0.007 | 0.868 ± 0.004 | 0.879 ± 0.003 | 0.858 ± 0.007 | 0.868 ± 0.004 | |
| BBC [50] | 0.877 ± 0.009 | 0.873 ± 0.010 | 0.875 ± 0.007 | 0.877 ± 0.009 | 0.873 ± 0.010 | 0.875 ± 0.007 | |
| RSBGC [51] | 0.891 ± 0.004 | 0.884 ± 0.003 | 0.887 ± 0.004 | 0.891 ± 0.004 | 0.884 ± 0.003 | 0.887 ± 0.004 | |
| FBBCE [52] | 0.920 ± 0.003 | 0.913 ± 0.002 | 0.916 ± 0.003 | 0.920 ± 0.003 | 0.913 ± 0.002 | 0.916 ± 0.003 | |
| DABLC [27] | 0.925 ± 0.002 | 0.919 ± 0.001 | 0.922 ± 0.001 | 0.925 ± 0.002 | 0.919 ± 0.001 | 0.922 ± 0.001 | |
| DGAN [28] | 0.950 ± 0.001 | 0.954 ± 0.001 | 0.952 ± 0.001 | 0.950 ± 0.001 | 0.954 ± 0.001 | 0.952 ± 0.001 | |
| Fine-tuned LLM | 0.694 ± 0.032 | 0.730 ± 0.023 | 0.711 ± 0.025 | 0.759 ± 0.023 | 0.702 ± 0.018 | 0.729 ± 0.021 | |
| 0.576 ± 0.021 | 0.590 ± 0.018 | 0.583 ± 0.018 | 0.830 ± 0.006 | 0.861 ± 0.004 | 0.845 ± 0.005 | ||
| 0.720 ± 0.010 | 0.721 ± 0.017 | 0.720 ± 0.012 | 0.867 ± 0.012 | 0.860 ± 0.009 | 0.863 ± 0.010 | ||
| - | - | - | 0.897 ± 0.009 | 0.915 ± 0.003 | 0.906 ± 0.005 | ||
| Models | CCKS2017 | |
|---|---|---|
| Computational Costs ($) | Power Consumption (kWh) | |
| ChatGLM2-6B | 3.93 | 52.10 |
| GLM-130B | 12.45 | 1092.45 |
| GPT-3.5 | 14.32 | 1470.60 |
| GPT-4 | 58.79 | 15,125.79 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Z.; Zhao, Q.; Li, J.; Ge, Y.; Ding, X.; Gu, T.; Zou, J.; Lv, S.; Wang, S.; Yang, J.-J. Comparative Analysis of Large Language Models in Chinese Medical Named Entity Recognition. Bioengineering 2024, 11, 982. https://doi.org/10.3390/bioengineering11100982
Zhu Z, Zhao Q, Li J, Ge Y, Ding X, Gu T, Zou J, Lv S, Wang S, Yang J-J. Comparative Analysis of Large Language Models in Chinese Medical Named Entity Recognition. Bioengineering. 2024; 11(10):982. https://doi.org/10.3390/bioengineering11100982
Chicago/Turabian StyleZhu, Zhichao, Qing Zhao, Jianjiang Li, Yanhu Ge, Xingjian Ding, Tao Gu, Jingchen Zou, Sirui Lv, Sheng Wang, and Ji-Jiang Yang. 2024. "Comparative Analysis of Large Language Models in Chinese Medical Named Entity Recognition" Bioengineering 11, no. 10: 982. https://doi.org/10.3390/bioengineering11100982
APA StyleZhu, Z., Zhao, Q., Li, J., Ge, Y., Ding, X., Gu, T., Zou, J., Lv, S., Wang, S., & Yang, J.-J. (2024). Comparative Analysis of Large Language Models in Chinese Medical Named Entity Recognition. Bioengineering, 11(10), 982. https://doi.org/10.3390/bioengineering11100982








