Application of Machine Learning in the Diagnosis of Early Gastric Cancer Using the Kyoto Classification Score and Clinical Features Collected from Medical Consultations
Abstract
1. Introduction
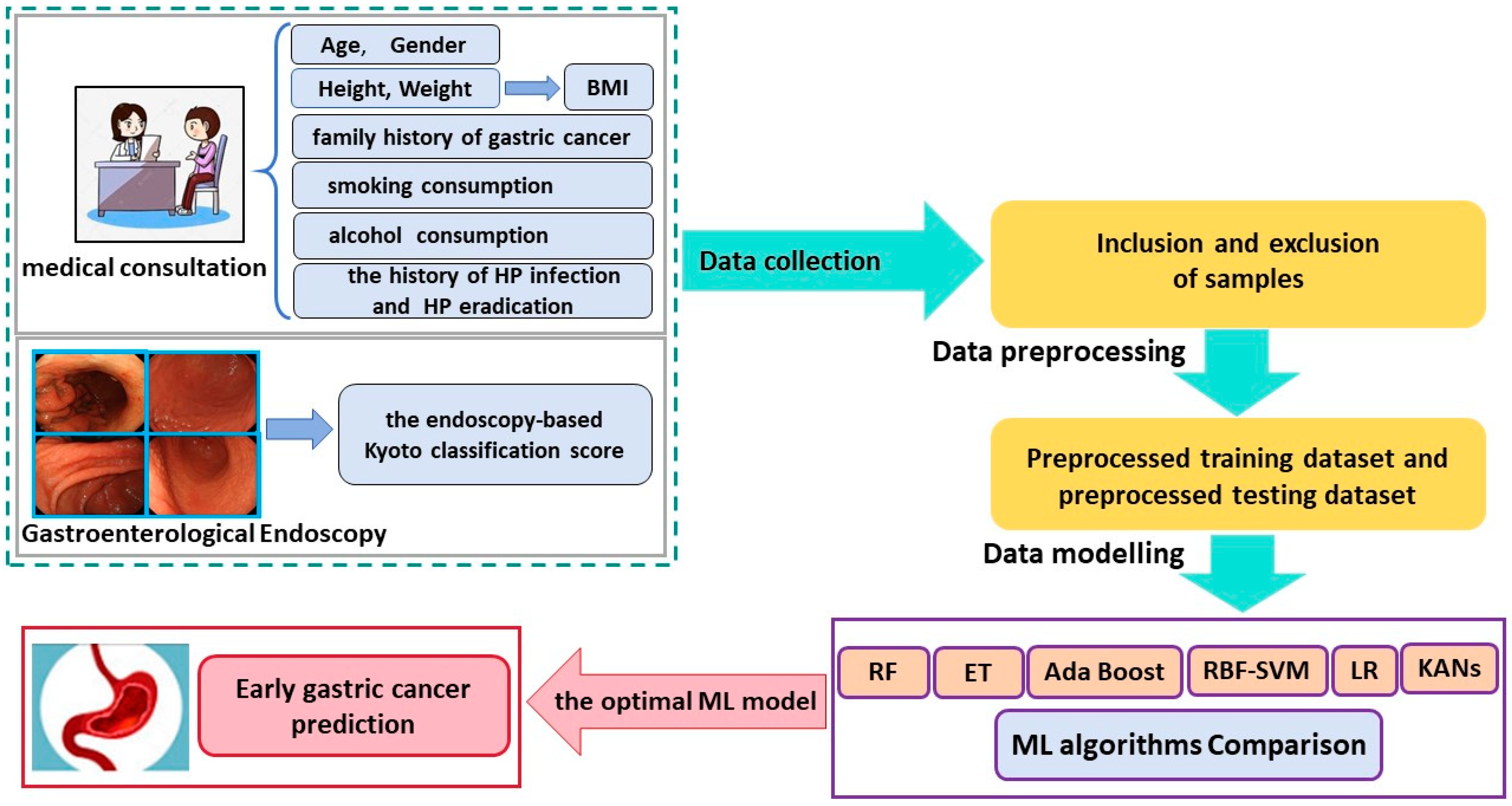
2. Materials and Methods
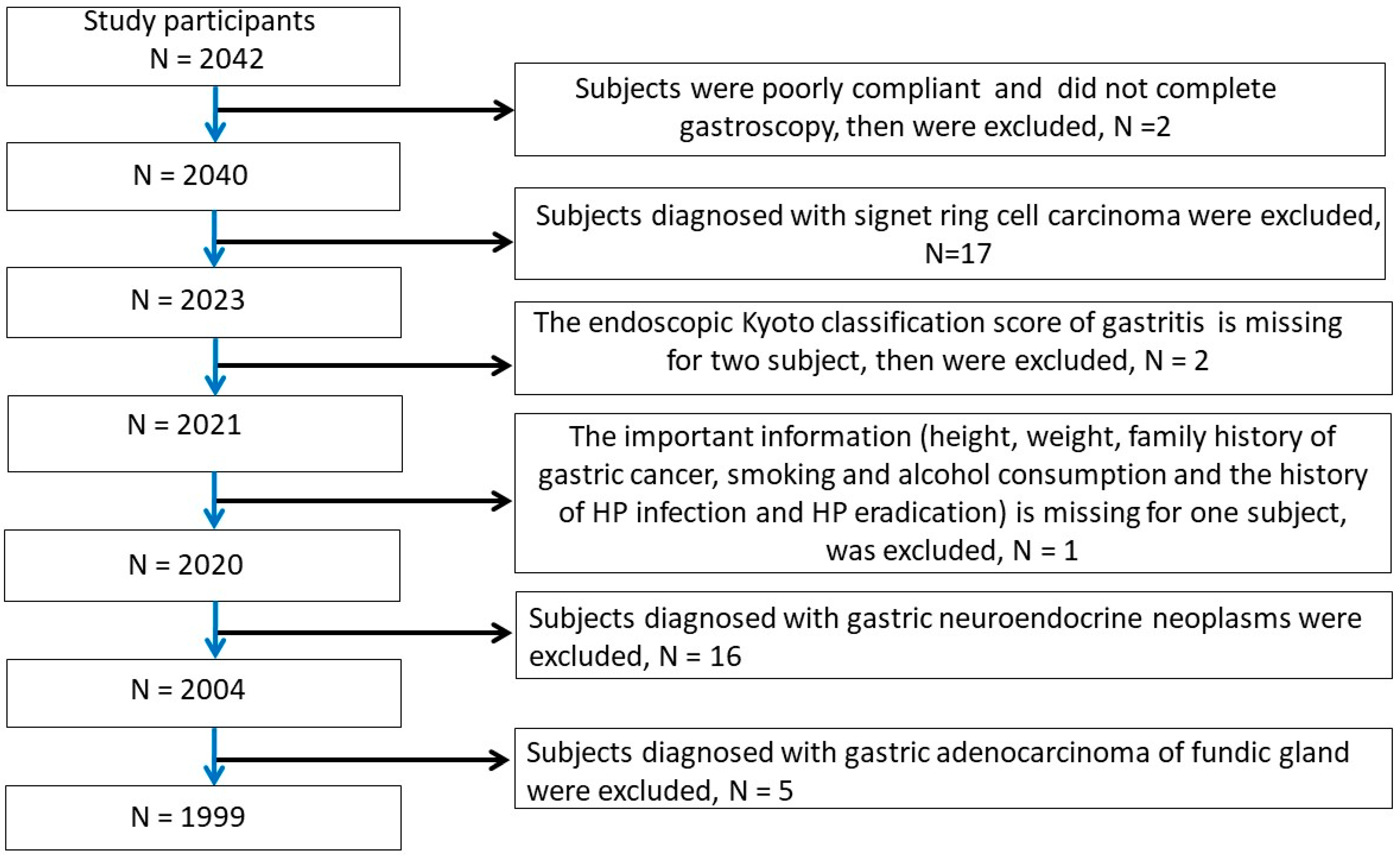
2.1. Patients and Inclusion and Exclusion Criteria
2.2. Raw Data Preprocessing
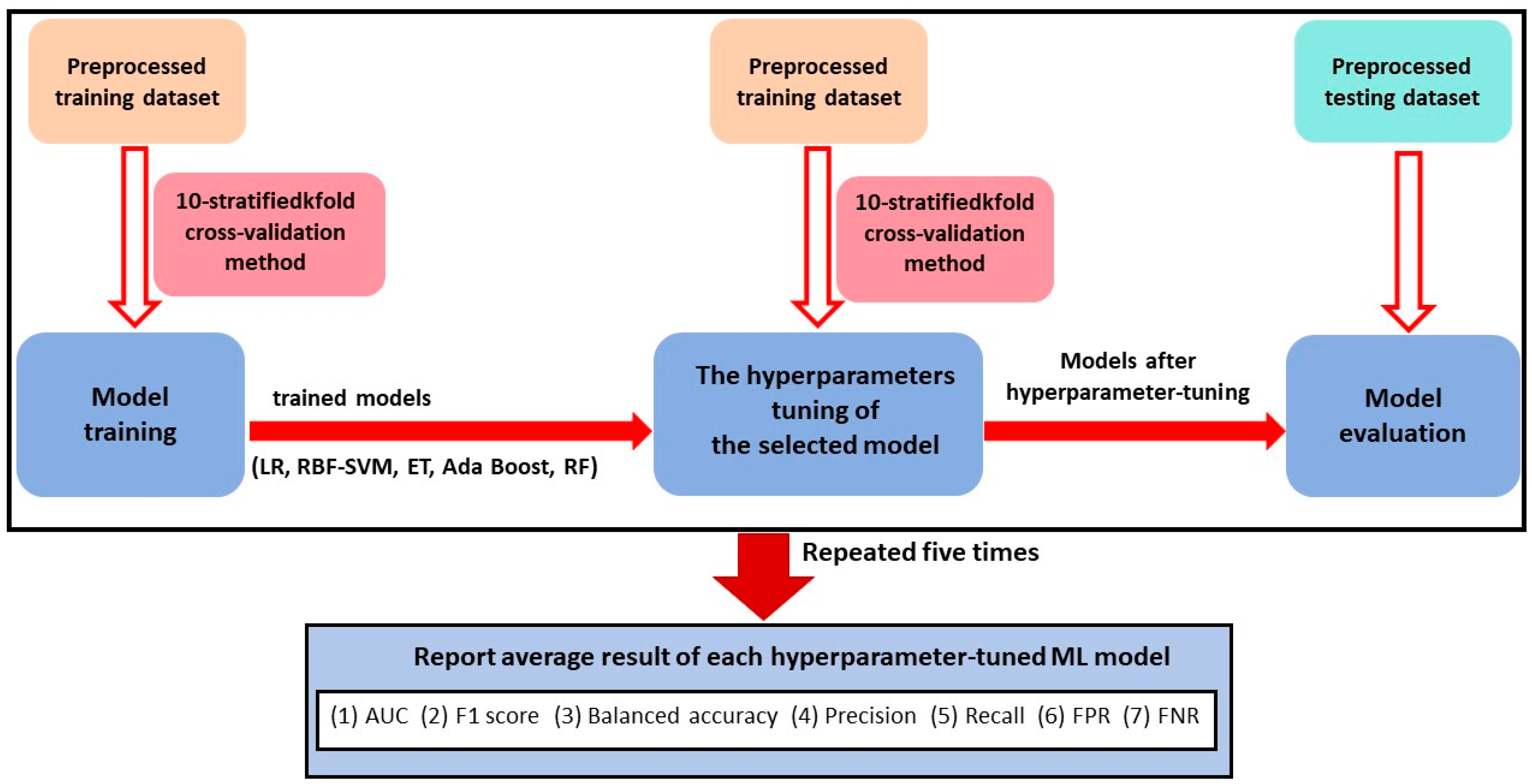
2.3. Machine Learning Methods and Evaluation Metrics for Classification
2.4. The Procedure of Data Modelling of Traditional ML Methods
2.5. The Procedure of Data Modelling of the KAN Method
2.6. The Method for Calculating Feature Importance Score of Variables
3. Results
3.1. Patient Characteristics
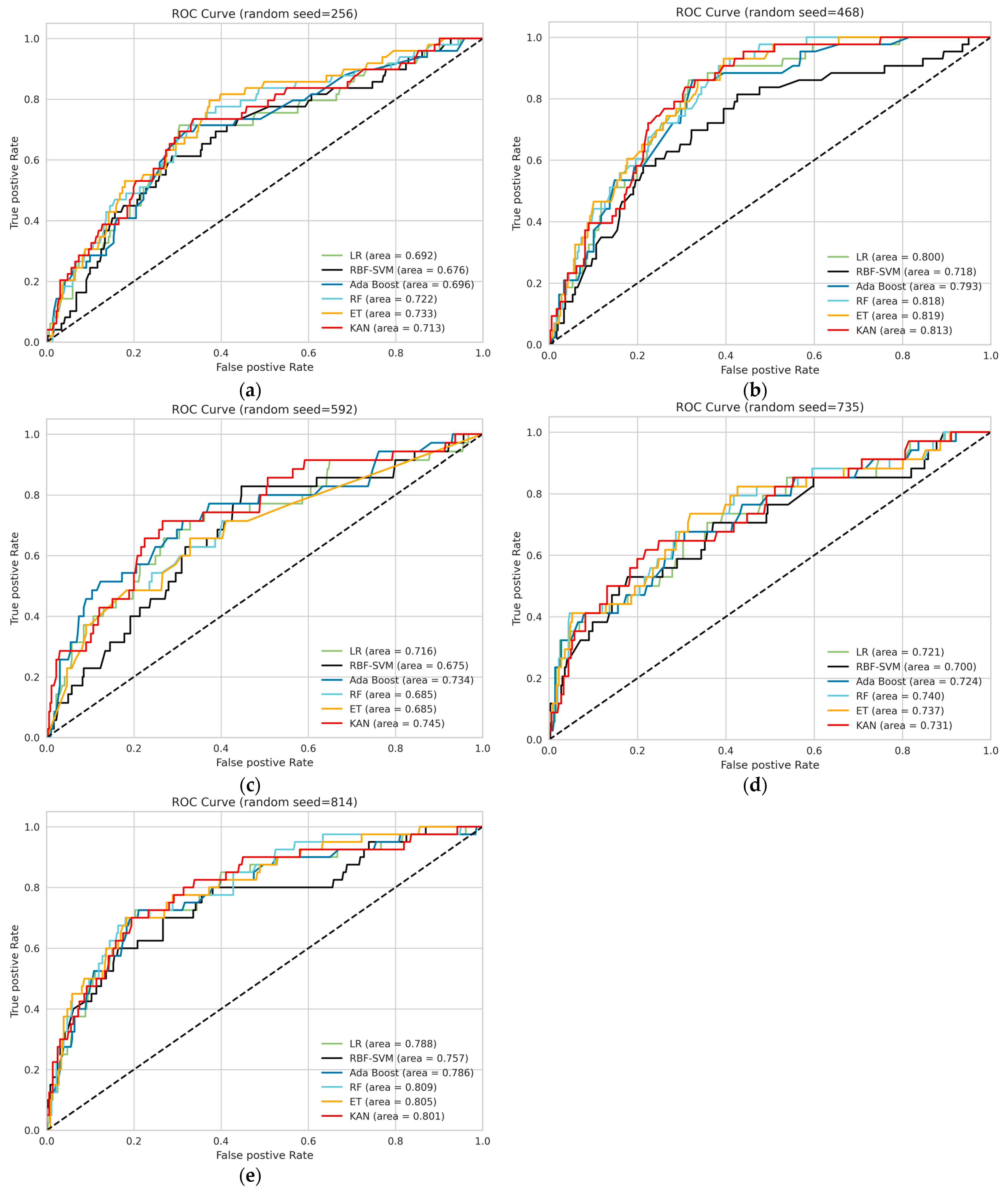
3.2. Model Performance
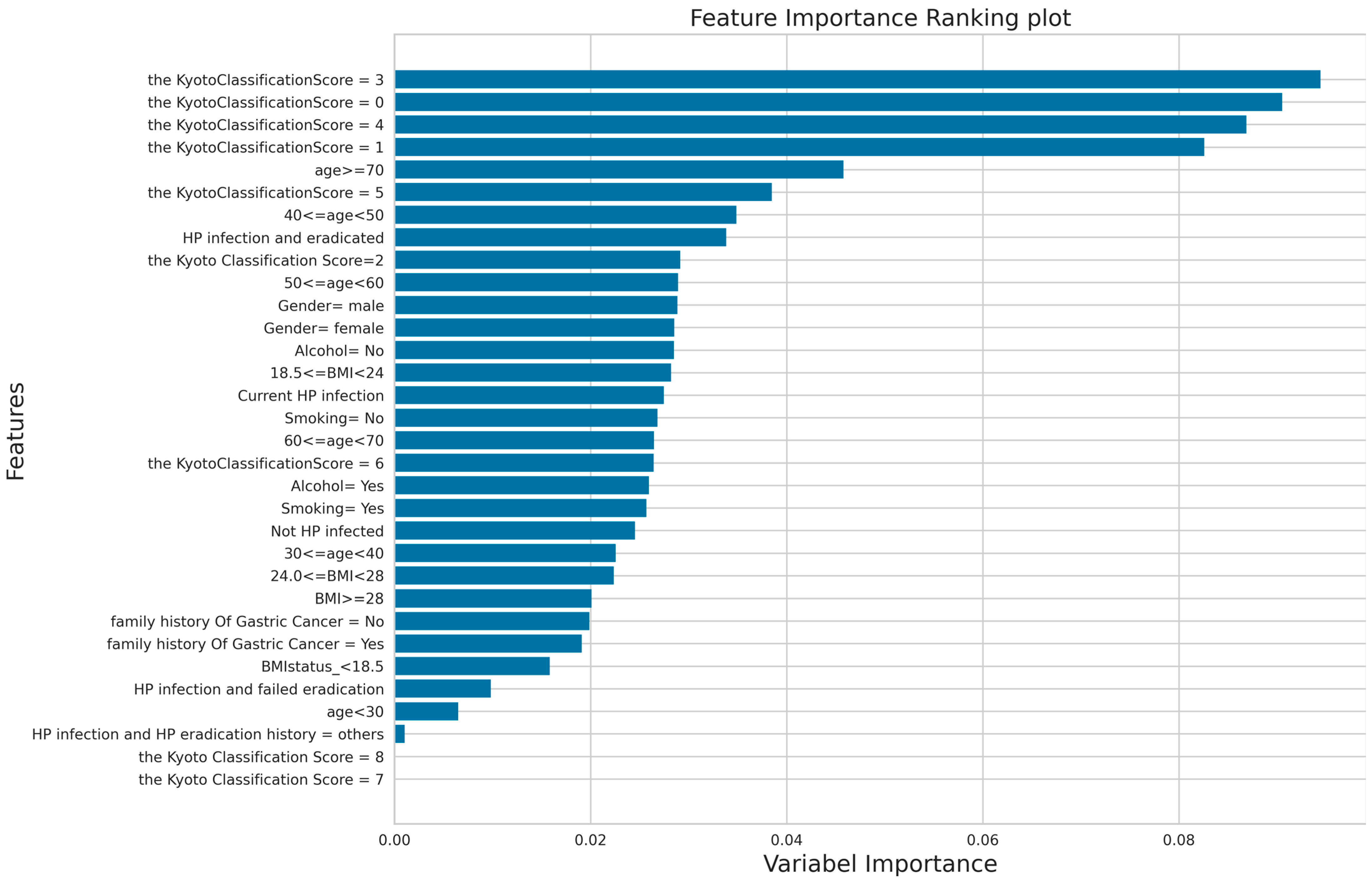
3.3. The Result of Feature Importance Score
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
Appendix A
| The Binary Classification | The Serial Number of 5 Repetitions When Using 5 Different Random Seed Values for Data Splitting | ||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| Random Seed = 256 | Random Seed = 468 | Random Seed = 592 | Random Seed = 735 | Random Seed = 814 | |
| True negative (TN) | 257 | 270 | 264 | 267 | 271 |
| False positive (FP) | 94 | 87 | 101 | 99 | 89 |
| False negative (FN) | 21 | 11 | 10 | 12 | 11 |
| True positive (TP) | 28 | 32 | 25 | 22 | 29 |
| Number of misclassified EGC samples with the Kyoto classification score < 4 | 18 | 9 | 10 | 11 | 9 |
| Number of misclassified EGC samples with the Kyoto classification score ≥ 4 | 3 | 2 | 0 | 1 | 2 |
| The ratio of misclassified EGC samples with the Kyoto classification score < 4 | 85.7% | 81.8% | 100% | 91.7% | 81.8% |
| The ratio of misclassified EGC samples with the Kyoto classification score ≥ 4 | 14.3% | 18.2% | 0% | 8.3% | 18.2% |
| False positive rate (FPR) | 26.78% | 24.36% | 27.67% | 27.04% | 24.72% |
| False negative rate (FNR) | 42.85% | 25.58% | 28.57% | 35.29% | 27.50% |
| Positive predictive value (PPV) | 22.95% | 26.89% | 19.84% | 18.18% | 24.57% |
| Negative predictive value (NPV) | 92.44% | 96.08% | 96.35% | 95.69% | 96.09% |
Appendix B
References
- He, F.; Wang, S.; Zheng, R.; Gu, J.; Zeng, H.; Sun, K.; Chen, R.; Li, L.; Han, B.; Li, X.; et al. Trends of gastric cancer burdens attributable to risk factors in China from 2000 to 2050. Lancet Reg. Health West. Pac. 2024, 44, 101003. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Zheng, R.; Zeng, H.; Wang, S.; Sun, K.; Chen, R.; Li, L.; Wei, W.; He, J. Cancer incidence and mortality in China, 2022. J. Natl. Cancer Cent. 2024, 4, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Thrift, A.P.; El-Serag, H.B. Burden of gastric cancer. Clin. Gastroenterol. Hepatol. 2020, 18, 534–542. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Su, H.; Zhou, Q.; Pan, J.; Zhou, L. Predictive value of nomogram based on kyoto classification of gastritis to diagnosis of gastric cancer. Scand. J. Gastroenterol. 2022, 57, 574–580. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Ying, X.; Liu, S.; Lyu, G.; Xu, Z.; Zhang, X.; Li, H.; Li, Q.; Wang, N.; Ji, J. Gastric cancer: Epidemiology, risk factors and prevention strategies. Chin. J. Cancer Res. 2020, 32, 695–704. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, X.; Zhang, X.; Huang, X. The significance of endoscopic kyoto classification of gastritis in the gastric cancer risk assessment: A systematic review and meta-analysis. Medicine 2023, 102, e33942. [Google Scholar] [CrossRef]
- Sun, X.; Zhao, L. RGB pixel brightness characteristics of linked color imaging in early gastric cancer: A pilot study. Gastroenterol. Res. Pract. 2020, 22, 2105874. [Google Scholar] [CrossRef]
- Dong, D.; Tang, L.; Li, Z.Y.; Fang, M.J.; Gao, J.B.; Shan, X.H.; Ying, X.J.; Sun, Y.S.; Fu, J.; Wang, X.X.; et al. Development and validation of an individualized nomogram to identify occult peritoneal metastasis in patients with advanced gastric cancer. Ann. Oncol. 2019, 30, 431–438. [Google Scholar] [CrossRef]
- Hamashima, C.; Okamoto, M.; Shabana, M.; Osaki, Y.; Kishimoto, T. Sensitivity of endoscopic screening for gastric cancer by the incidence method. Int. J. Cancer 2013, 133, 653–659. [Google Scholar] [CrossRef]
- Telford, J.J.; Enns, R.A. Endoscopic missed rates of upper gastrointestinal cancers: Parallels with colonoscopy. Am. J. Gastroenterol. 2010, 105, 1298–1300. [Google Scholar] [CrossRef]
- Fan, X.; Qin, X.; Zhang, Y.; Li, Z.; Zhou, T.; Zhang, J.; You, W.; Li, W.; Pan, K. Screening for gastric cancer in China: Advances, challenges and visions. Chin. J. Cancer Res. 2021, 33, 168–180. [Google Scholar] [CrossRef] [PubMed]
- Eom, B.W.; Joo, J.; Kim, S.; Shin, A.; Yang, H.R.; Park, J.; Choi, I.J.; Kim, Y.W.; Kim, J.; Nam, B.H. Prediction model for gastric cancer incidence in korean population. PLoS ONE 2015, 10, e0132613. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Zhu, C.; Yuan, Y.; Feng, Q.; Feng, Y.; Hao, Y.; Li, J.; Zhang, K.; Ye, G.; Ye, L.; et al. Development and validation of a prediction rule for estimating gastric cancer risk in the chinese high-risk population: A nationwide multicentre study. Gut 2019, 68, 1576–1587. [Google Scholar] [CrossRef]
- Leung, W.K.; Cheung, K.S.; Li, B.; Law, S.Y.K.; Lui, T.K.L. Applications of machine learning models in the prediction of gastric cancer risk in patients after helicobacter pylori eradication. Aliment. Pharmacol. Ther. 2021, 53, 864–872. [Google Scholar] [CrossRef]
- Mei, X.; Lee, H.C.; Diao, K.Y.; Huang, M.; Lin, B.; Liu, C.; Xie, Z.; Ma, Y.; Robson, P.M.; Chung, M.; et al. Artificial intelligence-enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020, 26, 1224–1228. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Gao, H.; He, J.; Shi, J.; Tong, Y.; Wu, J. Machine learning: A non-invasive prediction method for gastric cancer based on a survey of lifestyle behaviors. Front. Artif. Intell. 2022, 5, 956385. [Google Scholar] [CrossRef] [PubMed]
- Afrash, M.R.; Shafiee, M.; Kazemi-Arpanahi, H. Establishing machine learning models to predict the early risk of gastric cancer based on lifestyle factors. BMC Gastroenterol. 2023, 23, 6. [Google Scholar] [CrossRef]
- Brindha, S.K.; Kumar, N.S.; Chenkual, S.; Lalruatfela, S.; Zomuana, T.; Ralte, Z.; Maitra, A.; Basu, A.; Nath, P. Data mining for early gastric cancer etiological factors from diet-lifestyle characteristics. In Proceedings of the 2020 4th international conference on intelligent computing and control systems (ICICCS), Madurai, India, 13–15 May 2020; IEEE: Madurai, India, 2020; pp. 65–73. [Google Scholar]
- Jiang, F.; Lui, T.K.L.; Ju, C.; Guo, C.-G.; Cheung, K.S.; Lau, W.C.Y.; Leung, W.K. Machine learning models in predicting failure of helicobacter pylori treatment: A two country validation study. Helicobacter 2024, 29, e13051. [Google Scholar] [CrossRef]
- Arai, J.; Aoki, T.; Sato, M.; Niikura, R.; Suzuki, N.; Ishibashi, R.; Tsuji, Y.; Yamada, A.; Hirata, Y.; Ushiku, T.; et al. Machine learning-based personalized prediction of gastric cancer incidence using the endoscopic and histologic findings at the initial endoscopy. Gastrointest. Endosc. 2022, 95, 864–872. [Google Scholar] [CrossRef]
- Kolmogorov, A.N. On the Representation of Continuous Functions of Several Variables by Superpositions of Continuous Functions of a Smaller Number of Variables; American Mathematical Society: Providence, RI, USA, 1961. [Google Scholar]
- Arnold, V.I. On Functions of Three Variables. Collected Works: Representations of Functions, Celestial Mechanics and KAM Theory, 1957–1965; Springer: Berlin/Heidelberg, Germany, 2009; pp. 5–8. [Google Scholar]
- Poeta, E.; Giobergia, F.; Pastor, E.; Cerquitelli, T.; Baralis, E. A benchmarking study of kolmogorov-arnold networks on tabular data. arXiv 2024, arXiv:2406.14529. [Google Scholar]
- Lan, L.; Hu, G.; Li, R.; Wang, T.; Jiang, L.; Luo, J.; Ji, Z.; Wang, Y. Machine learning for selecting important clinical markers of imaging subgroups of cerebral small vessel disease based on a common data model. Tsinghua Sci. Technol. 2024, 29, 1495–1508. [Google Scholar] [CrossRef]
- Lin, X.; Lei, Y.; Chen, J.; Xing, Z.; Yang, T.; Wang, Q.; Wang, C. A case-finding clinical decision support system to identify subjects with chronic obstructive pulmonary disease based on public health data. Tsinghua Sci. Technol. 2023, 28, 525–540. [Google Scholar] [CrossRef]
- Hu, J.; Xu, J.; Li, M.; Jiang, Z.; Mao, J.; Feng, L.; Miao, K.; Li, H.; Chen, J.; Bai, Z.; et al. Identification and validation of an explainable prediction model of acute kidney injury with prognostic implications in critically ill children: A prospective multicenter cohort study. EClinicalMedicine 2024, 68, 102409. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Kamada, T. Endoscopic findings for risk stratification of gastric cancer. In Kyoto Classification of Gastritis, 2nd ed.; Nihon Medical Center: Tokyo, Japan, 2017; pp. 97–110. [Google Scholar]
- Shichijo, S.; Hirata, Y.; Niikura, R.; Hayakawa, Y.; Yamada, A.; Koike, K. Association between gastric cancer and the kyoto classification of gastritis. J. Gastroenterol. Hepatol. 2017, 32, 1581–1586. [Google Scholar] [CrossRef] [PubMed]
- Toyoshima, O.; Nishizawa, T.; Koike, K. Endoscopic kyoto classification of helicobacter pylori infection and gastric cancer risk diagnosis. World J. Gastroenterol. 2020, 26, 466–477. [Google Scholar] [CrossRef]
- Agawa, S.; Futagami, S.; Yamawaki, H.; Ikeda, G.; Noda, H.; Kirita, K.; Higuchi, K.; Murakami, M.; Kodaka, Y.; Ueki, N.; et al. Acylated ghrelin levels were associated with depressive status, physical quality of life, endoscopic findings based on kyoto classification in japan. J. Clin. Biochem. Nutr. 2019, 65, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Kohavi, R. A study of cross-validation and bootstrap for accuracy estimation and model selection. In Proceedings of the 14th International Joint Conference on Artificial Intelligence—Volume 2, Montreal, QC, Canada, 20–25 August 1995; Morgan Kaufmann Publishers Inc.: Montreal, QC, Canada, 1995; pp. 1137–1143. [Google Scholar]
- Li, J.; Zhu, Y.; Dong, Z.; He, X.; Xu, M.; Liu, J.; Zhang, M.; Tao, X.; Du, H.; Chen, D.; et al. Development and validation of a feature extraction-based logical anthropomorphic diagnostic system for early gastric cancer: A case-control study. EClinicalMedicine 2022, 46, 101366. [Google Scholar] [CrossRef] [PubMed]
- Ostir, G.V.; Uchida, T. Logistic regression: A nontechnical review. Am. J. Phys. Med. Rehabil. 2000, 79, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Govindarajan, M. A hybrid rbf-svm ensemble approach for data mining applications. I.J. Intell. Syst. Appl. 2014, 6, 84–95. [Google Scholar] [CrossRef]
- Boser, B.E.; Guyon, I.M.; Vapnik, V.N. A training algorithm for optimal margin classifiers. In Proceedings of the fifth Annual Workshop on Computational Learning Theory, Pittsburgh, PA, USA, 27–29 July 1992; Association for Computing Machinery: Pittsburgh, PA, USA, 1992; pp. 144–152. [Google Scholar]
- Geurts, P.; Ernst, D.; Wehenkel, L. Extremely randomized trees. Mach. Learn. 2006, 63, 3–42. [Google Scholar] [CrossRef]
- Schapire, R.E.; Freund, Y. Boosting: Foundations and Algorithms; The MIT Press: Cambridge, MA, USA, 2012; Volume 42. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, Y.; Vaidya, S.; Ruehle, F.; Halverson, J.; Soljačić, M.; Hou, T.Y.; Tegmark, M. Kan: Kolmogorov-arnold networks. arXiv 2024, arXiv:2404.19756. [Google Scholar]
- Hosmer, D.W., Jr.; Lemeshow, S.; Sturdivant, R.X. Applied Logistic Regression; John Wiley & Sons: Hoboken, NJ, USA, 2013. [Google Scholar]
- Murdoch, W.J.; Singh, C.; Kumbier, K.; Abbasi-Asl, R.; Yu, B. Definitions, methods, and applications in interpretable machine learning. Proc. Natl. Acad. Sci. USA 2019, 116, 22071–22080. [Google Scholar] [CrossRef] [PubMed]
- Hearst, M.A.; Dumais, S.T.; Osuna, E.; Platt, J.; Scholkopf, B. Support vector machines. IEEE Intell. Syst. Their Appl. 1998, 13, 18–28. [Google Scholar] [CrossRef]
- Hastie, T.; Rosset, S.; Zhu, J.; Zou, H. Multi-class adaboost. Stat Interface 2009, 2, 349–360. [Google Scholar] [CrossRef]
- Fawcett, T. An introduction to roc analysis. J. Pattern. Recogn. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Kuhn, M.; Johnson, K. Factors that can affect model performance. In Applied Predictive Modeling; Springer: New York, NY, USA, 2013; pp. 521–546. [Google Scholar]
- Blealtan, A.D. An Efficient Implementation of Kolmogorov-Arnold Network. Available online: https://github.com/Blealtan/efficient-kan (accessed on 19 July 2024).
- Ramachandran, P.; Zoph, B.; Le, Q. Searching for activation functions. arXiv 2017, arXiv:1710.05941. [Google Scholar]
- Loshchilov, I.; Hutter, F. Decoupled weight decay regularization. arXiv 2017, arXiv:1711.05101. [Google Scholar]
- Strobl, C.; Boulesteix, A.L.; Kneib, T.; Augustin, T.; Zeileis, A. Conditional variable importance for random forests. BMC Bioinform. 2008, 9, 307. [Google Scholar] [CrossRef]
- Liu, X.-M.; Ma, X.-Y.; Liu, F.; Liu, Z.-L.; Tang, X.-Y.; Ji, M.-Z.; Zheng, J.-X. Gastric cancer screening methods: A comparative study of the chinese new gastric cancer screening score and kyoto classification of gastritis. Gastroenterol. Res. Pract. 2022, 2022, 7639968. [Google Scholar] [CrossRef]
- Toyoshima, O.; Nishizawa, T.; Yoshida, S.; Aoki, T.; Nagura, F.; Sakitani, K.; Tsuji, Y.; Nakagawa, H.; Suzuki, H.; Koike, K. Comparison of endoscopic gastritis based on kyoto classification between diffuse and intestinal gastric cancer. World J. Gastrointest. Endosc. 2021, 13, 125. [Google Scholar] [CrossRef] [PubMed]
- Sakitani, K.; Nishizawa, T.; Toyoshima, A.; Yoshida, S.; Matsuno, T.; Yamada, T.; Irokawa, M.; Takahashi, Y.; Nakai, Y.; Toyoshima, O.; et al. Kyoto classification in patients who developed multiple gastric carcinomas after helicobacter pylori eradication. World J. Gastrointest. Endosc. 2020, 12, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Pinsky, P.F. Principles of cancer screening. J. Surg. Clin. 2015, 95, 953–966. [Google Scholar] [CrossRef] [PubMed]
- Ashayeri, H.; Sobhi, N.; Pławiak, P.; Pedrammehr, S.; Alizadehsani, R.; Jafarizadeh, A. Transfer learning in cancer genetics, mutation detection, gene expression analysis, and syndrome recognition. Cancers 2024, 16, 2138. [Google Scholar] [CrossRef]
| The Binary Classification | The Serial Number of 5 Repetitions When Using 5 Different Random Seed Values for Data Splitting | ||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| Random Seed = 256 | Random Seed = 468 | Random Seed = 592 | Random Seed = 735 | Random Seed = 814 | |
| Number of samples with the positive class label in a training set after data preprocessing | 841 | 775 | 790 | 809 | 793 |
| Number of samples with the negative class label in a training set after data preprocessing | 758 | 824 | 809 | 790 | 806 |
| Number of samples with the positive class label in a testing set after data preprocessing | 49 | 43 | 35 | 34 | 40 |
| Number of samples with the negative class label in a testing set after data preprocessing | 351 | 357 | 365 | 366 | 360 |
| Variable Name | Attribute and Type | Values | With Early Gastric Cancer (203) | No Early Gastric Cancer (1796) | Total |
|---|---|---|---|---|---|
| Age (years) | Categorical variable | <30 | 1 | 14 | 15 |
| 30–39 | 0 | 64 | 64 | ||
| 40–49 | 8 | 219 | 227 | ||
| 50–59 | 35 | 510 | 545 | ||
| 60–69 | 82 | 626 | 708 | ||
| ≥70 | 77 | 363 | 440 | ||
| BMI | Categorical variable | <18.5 | 15 | 106 | 121 |
| 18.5–23.9 | 88 | 796 | 884 | ||
| 24–27.9 | 78 | 729 | 807 | ||
| ≥28 | 22 | 165 | 187 | ||
| Gender | Categorical variable | Male | 132 | 980 | 1112 |
| Female | 71 | 816 | 887 | ||
| HP infection and HP eradication history | Categorical variable | Previous infection and eradicated | 90 | 668 | 758 |
| Previous infection and failed eradication | 5 | 53 | 58 | ||
| Current infection | 27 | 200 | 227 | ||
| Not HP infected | 79 | 857 | 936 | ||
| Others | 2 | 18 | 20 | ||
| Smoking | Categorical variable | No | 132 | 1324 | 1456 |
| Yes | 71 | 472 | 543 | ||
| Alcohol | Categorical variable | No | 138 | 1289 | 1427 |
| Yes | 65 | 507 | 572 | ||
| Family history of gastric cancer | Categorical variable | No | 176 | 1612 | 1788 |
| Yes | 27 | 184 | 211 | ||
| The endoscopy-based Kyoto classification score | Categorical variable | 0 | 23 | 593 | 616 |
| 1 | 25 | 567 | 592 | ||
| 2 | 39 | 329 | 368 | ||
| 3 | 49 | 177 | 226 | ||
| 4 | 39 | 68 | 107 | ||
| 5 | 18 | 48 | 66 | ||
| 6 | 9 | 13 | 22 | ||
| 7 | 1 | 0 | 1 | ||
| 8 | 0 | 1 | 1 |
| Metrics | ET | Ada Boost | LR | RF | RBF-SVM | KAN |
|---|---|---|---|---|---|---|
| AUC | 0.758 ± 0.05 | 0.744 ± 0.05 | 0.742 ± 0.05 | 0.760 ± 0.05 | 0.691 ± 0.05 | 0.760 ± 0.04 |
| F1 score | 57.87 ± 3.34 | 57.23 ± 3.95 | 58.07 ± 2.99 | 57.68 ± 3.82 | 55.05 ± 4.89 | 58.46 ± 2.73 |
| Precision | 58.29 ± 2.15 | 58.03 ± 2.13 | 58.45 ± 1.85 | 58.11 ± 2.59 | 56.33 ± 3.47 | 58.91 ± 1.91 |
| Recall | 68.84 ± 3.64 | 68.49 ± 3.53 | 69.35 ± 3.25 | 68.44 ± 5.02 | 64.52 ± 7.66 | 70.96 ± 3.99 |
| FPR | 25.32 ± 2.56 | 26.56 ± 4.83 | 25.38 ± 2.57 | 25.27 ± 2.75 | 27.28 ± 2.16 | 26.11 ± 1.47 |
| FNR | 37.01 ± 6.45 | 36.46 ± 3.59 | 35.90 ± 5.16 | 37.86 ± 8.85 | 43.69 ± 13.45 | 31.96 ± 7.10 |
| BA | 68.83 ± 3.64 | 68.48 ± 3.53 | 69.35 ± 3.25 | 68.43 ± 5.02 | 64.51 ± 7.66 | 70.96 ± 3.99 |
| Studies | No. of Patients Enrolled | Characteristics Used for Prediction | No. of Features Collected | Methodology | Result |
|---|---|---|---|---|---|
| Cai Q et al. [13] (2019) | 14929 | Age, sex, BMI, H. pylori infection, PG I, PG II, PG I/II ratio, G-17, anti-H. pylori IgG antibody, pickled food, fried food, high-salt diet, alcohol consumption and smoking consumption, etc. | 21 | Logistic regression | The prediction rule owns a good discrimination, with an AUC of 0.76. |
| Lin J et al. [4] (2022) | 2639 | Age, sex, PG I/II ratio, HP antibody, atrophy, intestinal metaplasia, enlarged fold, diffuse redness and the Kyoto classification score | 9 | Nomogram | The AUC of the nomogram to predict GC was 0.79 |
| Our study (2024) | 1999 | The Kyoto classification score, age, gender, BMI, family history of gastric cancer, the history of H. Pylori infection and H. Pylori eradication, smoking consumption and alcohol consumption | 8 | Logistic regression, extra trees, radial basis function kernel support vector machine, Ada Boost, random forest, Kolmogorov–Arnold networks | The KAN model outperformed other ML models with the highest average AUC value of 0.76, the highest average balanced accuracy of 70.96%. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, X.; Zhang, L.; Luo, Q.; Zhou, Y.; Du, J.; Fu, D.; Wang, Z.; Lei, Y.; Wang, Q.; Zhao, L. Application of Machine Learning in the Diagnosis of Early Gastric Cancer Using the Kyoto Classification Score and Clinical Features Collected from Medical Consultations. Bioengineering 2024, 11, 973. https://doi.org/10.3390/bioengineering11100973
Sun X, Zhang L, Luo Q, Zhou Y, Du J, Fu D, Wang Z, Lei Y, Wang Q, Zhao L. Application of Machine Learning in the Diagnosis of Early Gastric Cancer Using the Kyoto Classification Score and Clinical Features Collected from Medical Consultations. Bioengineering. 2024; 11(10):973. https://doi.org/10.3390/bioengineering11100973
Chicago/Turabian StyleSun, Xue, Liping Zhang, Qingfeng Luo, Yan Zhou, Jun Du, Dongmei Fu, Ziyu Wang, Yi Lei, Qing Wang, and Li Zhao. 2024. "Application of Machine Learning in the Diagnosis of Early Gastric Cancer Using the Kyoto Classification Score and Clinical Features Collected from Medical Consultations" Bioengineering 11, no. 10: 973. https://doi.org/10.3390/bioengineering11100973
APA StyleSun, X., Zhang, L., Luo, Q., Zhou, Y., Du, J., Fu, D., Wang, Z., Lei, Y., Wang, Q., & Zhao, L. (2024). Application of Machine Learning in the Diagnosis of Early Gastric Cancer Using the Kyoto Classification Score and Clinical Features Collected from Medical Consultations. Bioengineering, 11(10), 973. https://doi.org/10.3390/bioengineering11100973






