Microalgae-Based Remediation of Real Textile Wastewater: Assessing Pollutant Removal and Biomass Valorisation
Abstract
1. Introduction
2. Materials and Methods
2.1. Textile Effluents
2.2. Microalgal Cultures
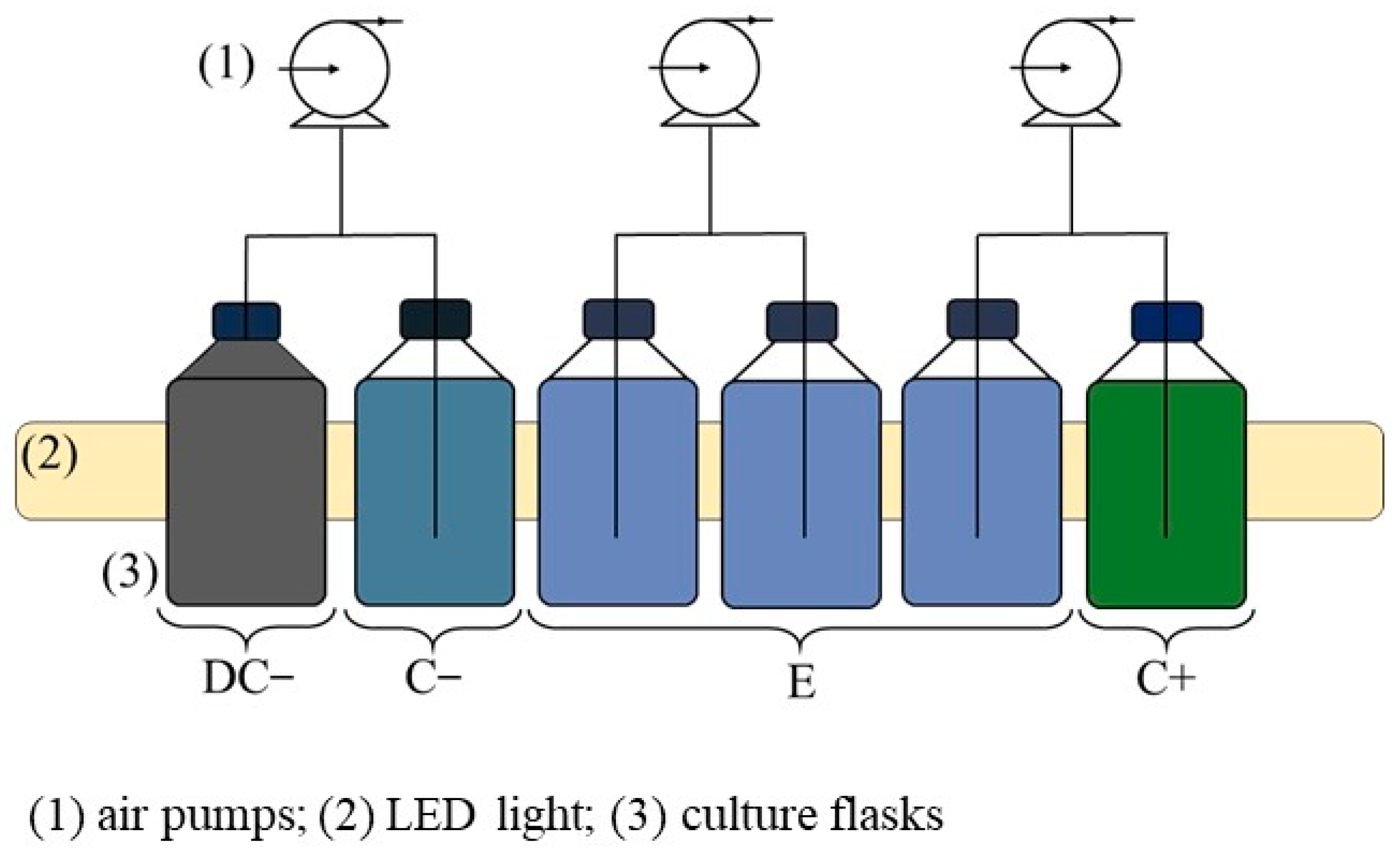
2.3. Experimental Setup
2.4. Analytical Methods
2.5. Initial Effluent Composition
2.6. Growth Kinetic Parameters and Nutrient Removal
3. Results and Discussion
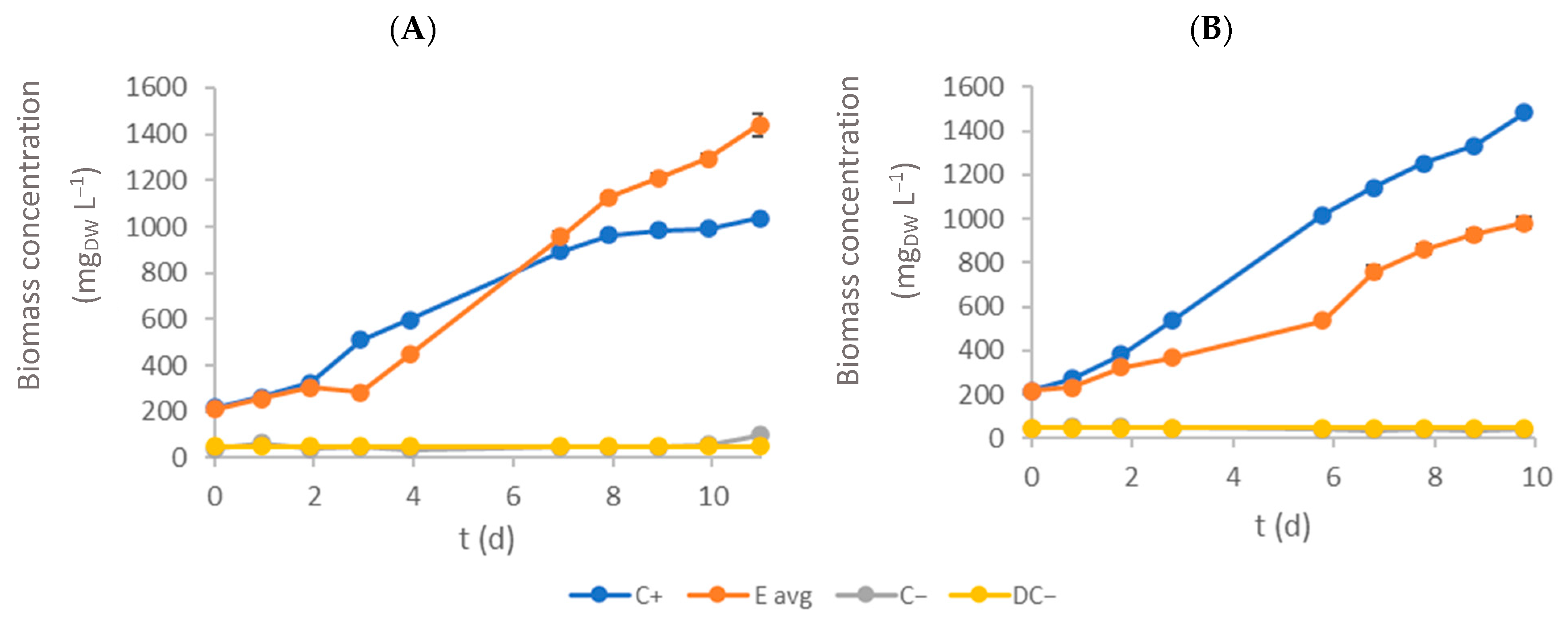
3.1. Biomass Growth Rate and Productivity
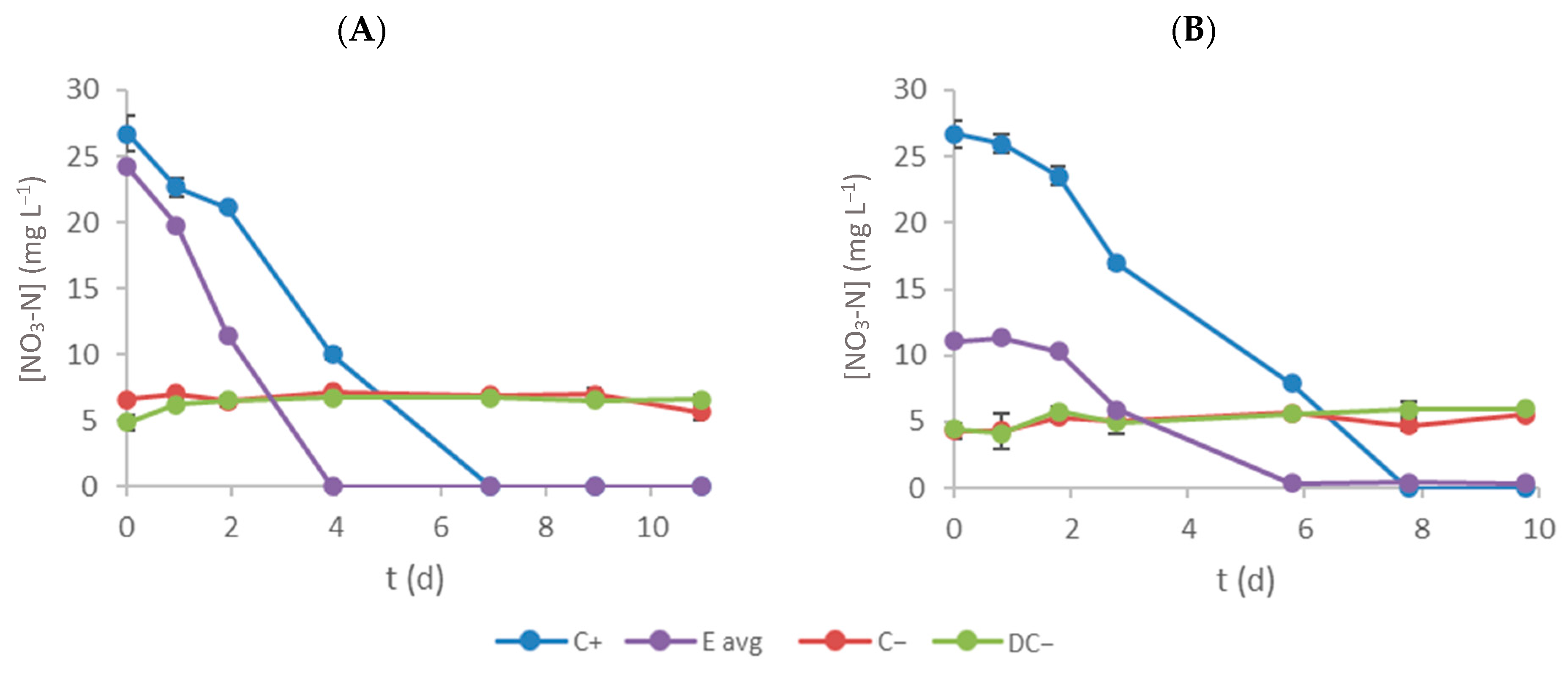
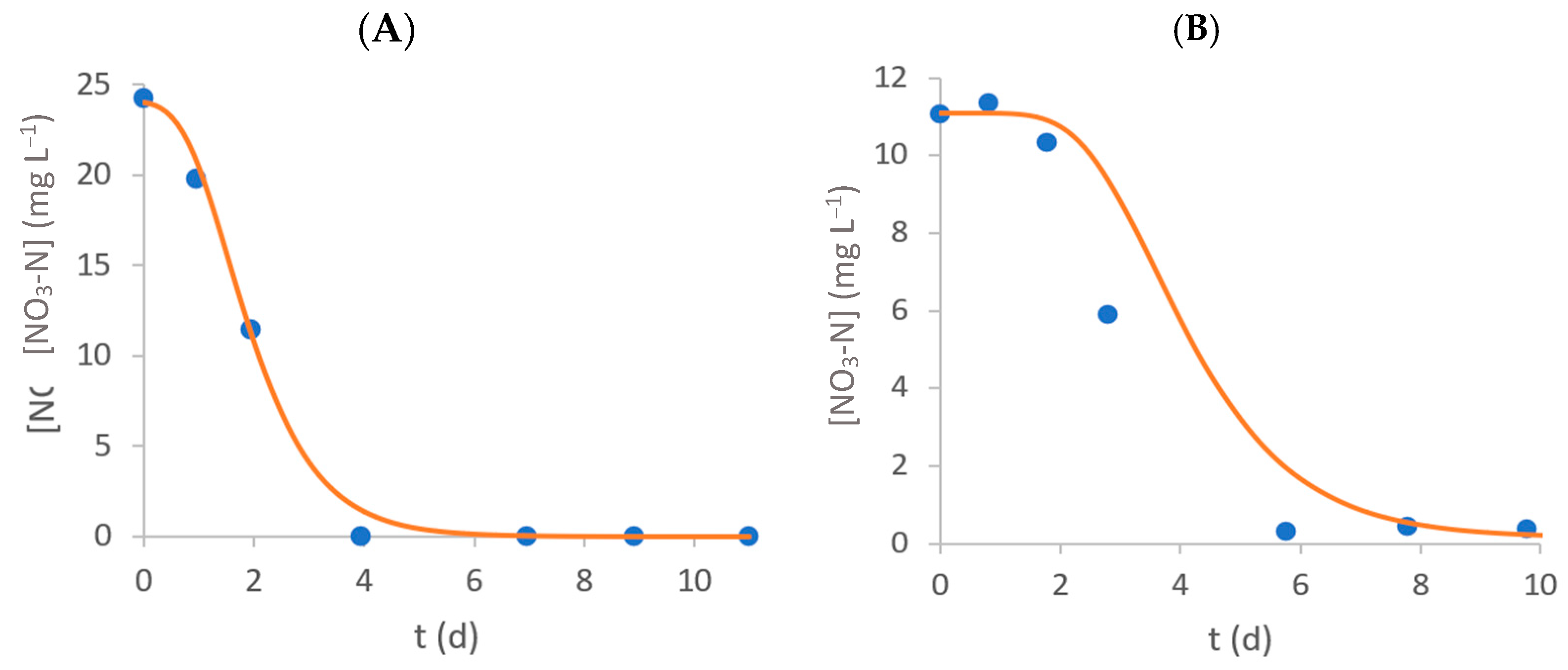
3.2. Nitrate–Nitrogen Removal
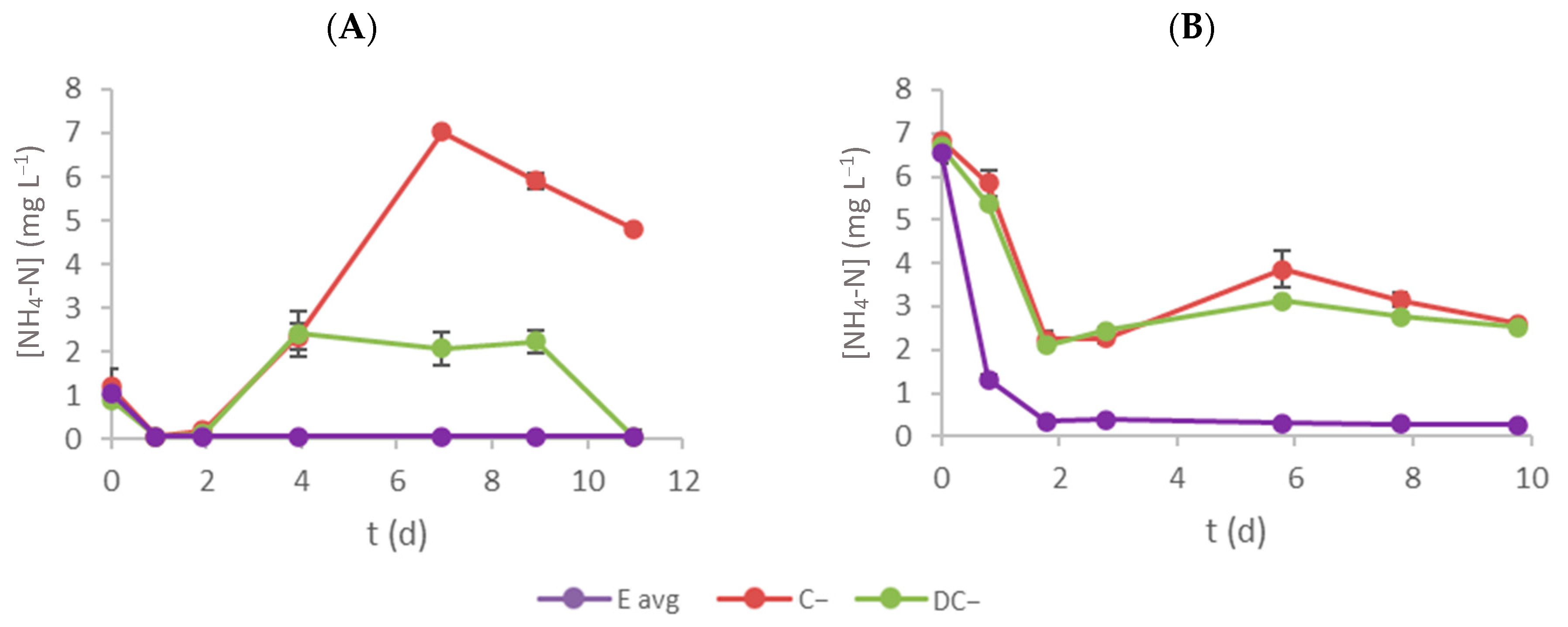
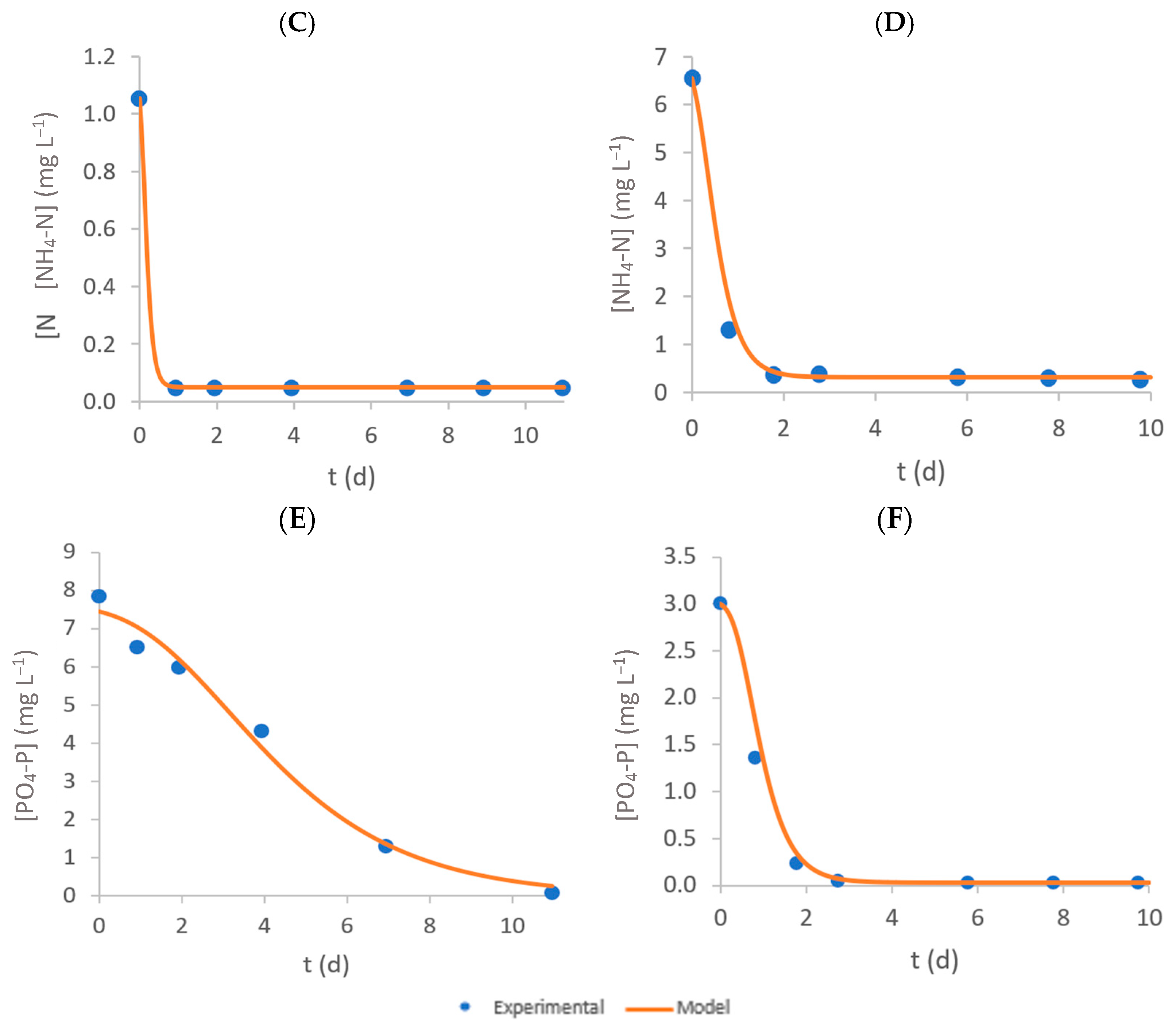
3.3. Ammonium–Nitrogen Removal
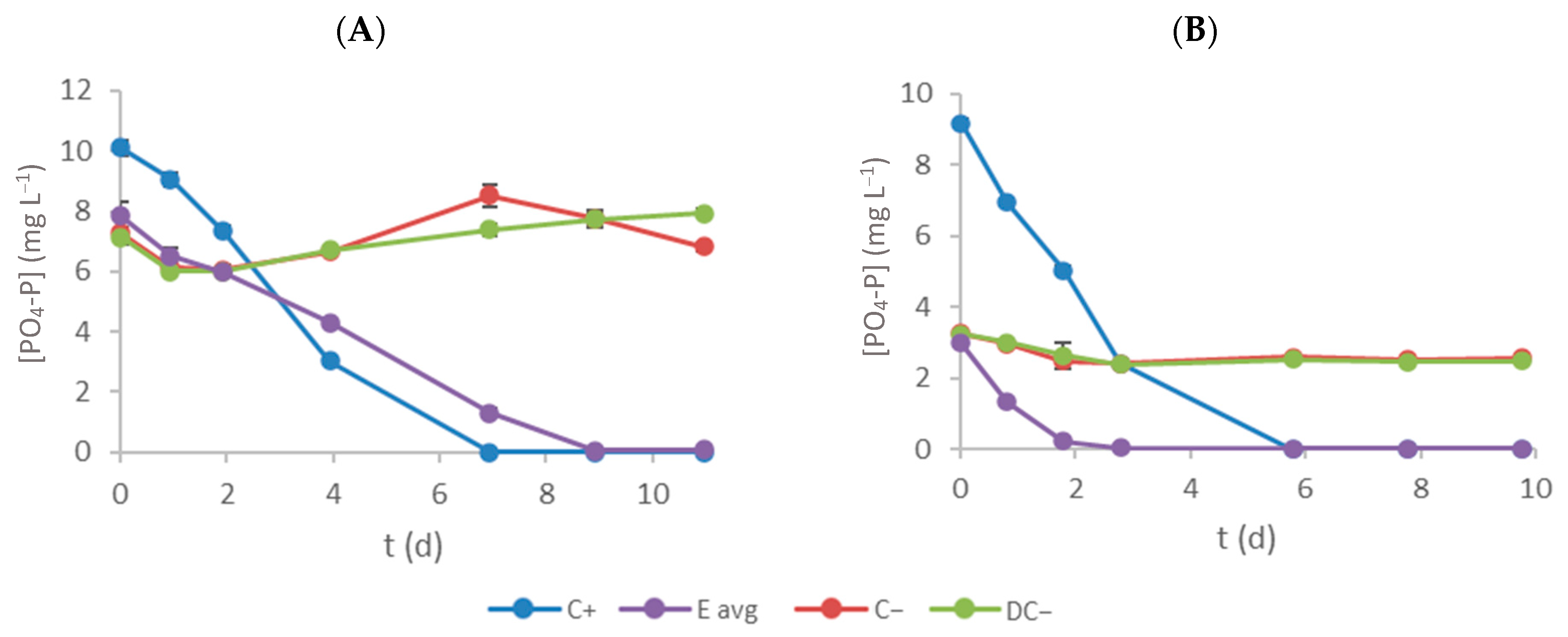
3.4. Phosphate–Phosphorus Removal
3.5. Modified Gompertz Model
3.6. Biomass Specific Yield
3.7. Chemical Oxygen Demand
3.8. Turbidity and Colour Removal
3.9. Microalgal Pigment Content
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Thakker, A.M.; Sun, D.M. Sustainable Development Goals for Textiles and Fashion. Environ. Sci. Pollut. Res. 2023, 30, 101989–102009. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, J.; Sonia. 13—Dyes and dyeing processes for natural textiles and their key sustainability issues. In Fundamentals of Natural Fibres and Textiles; Mondal, M.I.H., Ed.; Woodhead Publishing: Cambridge, UK, 2021; pp. 439–472. [Google Scholar]
- Al-Tohamy, R.; Ali, S.S.; Li, F.H.; Okasha, K.M.; Mahmoud, Y.A.G.; Elsamahy, T.; Jiao, H.X.; Fu, Y.Y.; Sun, J.Z. A critical review on the treatment of dye-containing wastewater: Ecotoxicological and health concerns of textile dyes and possible remediation approaches for environmental safety. Ecotoxicol. Environ. Saf. 2022, 231, 113160. [Google Scholar] [CrossRef] [PubMed]
- Holkar, C.R.; Jadhav, A.J.; Pinjari, D.V.; Mahamuni, N.M.; Pandit, A.B. A critical review on textile wastewater treatments: Possible approaches. J. Environ. Manag. 2016, 182, 351–366. [Google Scholar] [CrossRef] [PubMed]
- Marazzi, F.; Fornaroli, R.; Clagnan, E.; Brusetti, L.; Ficara, E.; Bellucci, M.; Mezzanotte, V. Wastewater from textile digital printing as a substrate for microalgal growth and valorization. Bioresour. Technol. 2023, 375, 128828. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.-Q.; Kabra, A.N.; Salama, E.-S. 15—Microalgae-based biotechnologies for treatment of textile wastewater. In Current Developments in Bioengineering and Biotechnology; Govindwar, S.P., Kurade, M.B., Jeon, B.-H., Pandey, A., Eds.; Elsevier: Amsterdam, The Netherlands, 2023; pp. 457–471. [Google Scholar]
- Khan, A.A.; Gul, J.; Naqvi, S.R.; Ali, I.; Farooq, W.; Liaqat, R.; AlMohamadi, H.; Stepanec, L.; Juchelkova, D. Recent progress in microalgae-derived biochar for the treatment of textile industry wastewater. Chemosphere 2022, 306, 135565. [Google Scholar] [CrossRef] [PubMed]
- Zohoorian, H.; Ahmadzadeh, H.; Molazadeh, M.; Shourian, M.; Lyon, S. Chapter 41—Microalgal bioremediation of heavy metals and dyes. In Handbook of Algal Science, Technology and Medicine; Konur, O., Ed.; Academic Press: Cambridge, MA, USA, 2020; pp. 659–674. [Google Scholar]
- Singh, G.; Singh, A.; Singh, P.; Gupta, A.; Shukla, R.; Mishra, V.K. Chapter 29—Sources, fate, and impact of pharmaceutical and personal care products in the environment and their different treatment technologies. In Microbe Mediated Remediation of Environmental Contaminants; Kumar, A., Singh, V.K., Singh, P., Mishra, V.K., Eds.; Woodhead Publishing: Cambridge, UK, 2021; pp. 391–407. [Google Scholar]
- Mustafa, S.; Bhatti, H.N.; Maqbool, M.; Iqbal, M. Microalgae biosorption, bioaccumulation and biodegradation efficiency for the remediation of wastewater and carbon dioxide mitigation: Prospects, challenges and opportunities. J. Water Process Eng. 2021, 41, 102009. [Google Scholar] [CrossRef]
- Fazal, T.; Mushtaq, A.; Rehman, F.; Khan, A.U.; Rashid, N.; Farooq, W.; Rehman, M.S.U.; Xu, J. Bioremediation of textile wastewater and successive biodiesel production using microalgae. Renew. Sustain. Energy Rev. 2018, 82, 3107–3126. [Google Scholar] [CrossRef]
- Abdelfattah, A.; Ali, S.S.; Ramadan, H.; El-Aswar, E.I.; Eltawab, R.; Ho, S.H.; Elsamahy, T.; Li, S.N.; El-Sheekh, M.M.; Schagerl, M.; et al. Microalgae-based wastewater treatment: Mechanisms, challenges, recent advances, and future prospects. Environ. Sci. Ecotechnol. 2023, 13, 100205. [Google Scholar] [CrossRef]
- Mishra, V.; Mudgal, N.; Rawat, D.; Poria, P.; Mukherjee, P.; Sharma, U.; Kumria, P.; Pani, B.; Singh, M.; Yadav, A.; et al. Integrating microalgae into textile wastewater treatment processes: Advancements and opportunities. J. Water Process Eng. 2023, 55, 104128. [Google Scholar] [CrossRef]
- Premaratne, M.; Nishshanka, G.K.S.H.; Liyanaarachchi, V.C.; Nimarshana, P.H.V.; Ariyadasa, T.U. Bioremediation of textile dye wastewater using microalgae: Current trends and future perspectives. J. Chem. Technol. Biotechnol. 2021, 96, 3249–3258. [Google Scholar] [CrossRef]
- Lim, S.L.; Chu, W.L.; Phang, S.M. Use of Chlorella vulgaris for bioremediation of textile wastewater. Bioresour. Technol. 2010, 101, 7314–7322. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Zamora, M.; Cristiani-Urbina, E.; Martínez-Jerónimo, F.; Perales-Vela, H.; Ponce-Noyola, T.; Montes-Horcasitas, M.D.; Cañizares-Villanueva, R.O. Bioremoval of the azo dye Congo Red by the microalga Chlorella vulgaris. Environ. Sci. Pollut. Res. 2015, 22, 10811–10823. [Google Scholar] [CrossRef] [PubMed]
- Oyebamiji, O.O.; Boeing, W.J.; Holguin, F.O.; Ilori, O.; Amund, O. Green microalgae cultured in textile wastewater for biomass generation and biodetoxification of heavy metals and chromogenic substances. Bioresour. Technol. Rep. 2019, 7, 100247. [Google Scholar] [CrossRef]
- Ge, S.J.; Qiu, S.; Tremblay, D.; Viner, K.; Champagne, P.; Jessop, P.G. Centrate wastewater treatment with Chlorella vulgaris: Simultaneous enhancement of nutrient removal, biomass and lipid production. Chem. Eng. J. 2018, 342, 310–320. [Google Scholar] [CrossRef]
- Esteves, A.F.; Soares, S.M.; Salgado, E.M.; Boaventura, R.A.R.; Pires, J.C.M. Microalgal Growth in Aquaculture Effluent: Coupling Biomass Valorisation with Nutrients Removal. Appl. Sci. 2022, 12, 12608. [Google Scholar] [CrossRef]
- EPA. Method 352.1: Nitrogen, Nitrate (Colorimetric, Brucine) by Spectrophotometer. In Methods for Chemical Analysis of Water and Wastes; EPA: Washington, DC, USA, 1983; pp. 365–367. [Google Scholar]
- Lee, B.; Park, S.Y.; Hae, Y.S.; Yea, S.S.; Kim, D.E. Efficient Colorimetric Assay of RNA Polymerase Activity Using Inorganic Pyrophosphatase and Ammonium Molybdate. Bull. Korean Chem. Soc. 2009, 30, 2485–2488. [Google Scholar]
- Clément-Larosière, B.; Lopes, F.; Gonçalves, A.; Taidi, B.; Benedetti, M.; Minier, M.; Pareau, D. Carbon dioxide biofixation by Chlorella vulgaris at different CO2 concentrations and light intensities. Eng. Life Sci. 2014, 14, 509–519. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1987; Volume 148, pp. 350–382. [Google Scholar]
- APHA. Standard Methods for the Examination of Water and Wastewater; American Public Health Association, American Water Works Association; Water Environment Federation: Washington, DC, USA, 2012. [Google Scholar]
- Salgado, E.M.; Esteves, A.F.; Gonçalves, A.L.; Pires, J.C.M. Microalgal cultures for the remediation of wastewaters with different nitrogen to phosphorus ratios: Process modelling using artificial neural networks. Environ. Res. 2023, 231, 116076. [Google Scholar] [CrossRef]
- Zwietering, M.H.; Jongenburger, I.; Rombouts, F.M.; van’t Riet, K. Modeling of the Bacterial Growth Curve. Appl. Environ. Microbiol. 1990, 56, 1875–1881. [Google Scholar] [CrossRef]
- Javed, F.; Rehman, F.; Khan, A.U.; Fazal, T.; Hafeez, A.; Rashid, N. Real textile industrial wastewater treatment and biodiesel production using microalgae. Biomass Bioenergy 2022, 165, 106559. [Google Scholar] [CrossRef]
- El-Kassas, H.Y.; Mohamed, L.A. Bioremediation of the textile waste effluent by Chlorella vulgaris. Egypt. J. Aquat. Res. 2014, 40, 301–308. [Google Scholar] [CrossRef]
- Pathak, V.V.; Kothari, R.; Chopra, A.K.; Singh, D.P. Experimental and kinetic studies for phycoremediation and dye removal by Chlorella pyrenoidosa from textile wastewater. J. Environ. Manag. 2015, 163, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.Y.; Lay, C.H.; Chen, C.C.; Wu, S.Y. Lipid accumulating microalgae cultivation in textile wastewater: Environmental parameters optimization. J. Taiwan Inst. Chem. Eng. 2017, 79, 1–6. [Google Scholar] [CrossRef]
- European Union. Commission Directive 98/15/EC of 27 February 1998 amending Council Directive 91/271/EEC with respect to certain requirements established in Annex I thereof. Off. J. Eur. Communities 1998, 67, 29–30. [Google Scholar]
- Ayatollahi, S.Z.; Esmaeilzadeh, F.; Mowla, D. Integrated CO2 capture, nutrients removal and biodiesel production using Chlorella vulgaris. J. Environ. Chem. Eng. 2021, 9, 104763. [Google Scholar] [CrossRef]
- Silva, N.F.P.; Gonçalves, A.L.; Moreira, F.C.; Silva, T.F.C.V.; Martins, F.G.; Alvim-Ferraz, M.C.M.; Boaventura, R.A.R.; Vilar, V.J.P.; Pires, J.C.M. Towards sustainable microalgal biomass production by phycoremediation of a synthetic wastewater: A kinetic study. Algal Res. 2015, 11, 350–358. [Google Scholar] [CrossRef]
- Maroneze, M.M.; Zepka, L.Q.; Lopes, E.J.; Pérez-Gálvez, A.; Roca, M. Chlorophyll Oxidative Metabolism during the Phototrophic and Heterotrophic Growth of. Antioxidants 2019, 8, 600. [Google Scholar] [CrossRef]
| µ (d−1) | PX,avg (mgDW L−1 d−1) | Pmax (mgDW L−1 d−1) | Xmax (mgDW L−1) | ||
|---|---|---|---|---|---|
| Effluent 1 | C+ | 0.331 ± 0.002 a | 74.6 ± 0.3 a | 184 ± 7 a | 1036 ± 3 a |
| E | 0.290 ± 0.003 a | 112.39 ± 0.07 a | 176 ± 2 b | 1440 ± 47 a | |
| Effluent 2 | C+ | 0.341 ± 0.004 a | 129 ± 1 a | 160 ± 4 a | 1483 ± 13 a |
| E | 0.234 ± 0.005 a | 78 ± 3 a | 221 ± 2 a | 1006 ± 25 a |
| Effluent 1 | Effluent 2 | |||||
|---|---|---|---|---|---|---|
| S0 (mg L−1) | RE (%) | RR (mg L−1 d−1) | S0 (mg L−1) | RE (%) | RR (mg L−1 d−1) | |
| C+ | 27 ± 1 a | 100 ± 0 a | 2.4 ± 0.2 a | 27 ± 1 a | 100 ± 0 a | 2.7 ± 0.1 a |
| E | 27 ± 5 b | 100 ± 0 b | 1.9 ± 0.4 a | 11.1 ± 0.7 a | 96.46 ± 0.01 a | 1.1 ± 0.1 a |
| S0 (mg L−1) | RE (%) | RR (mg L−1 d−1) | |
|---|---|---|---|
| Effluent 1 | 1.1 ± 0.1 | 95.1 ± 0.4 | 0.09 ± 0.02 |
| Effluent 2 | 6.6 ± 0.2 | 95.951 ± 0.003 | 0.64 ± 0.03 |
| Effluent 1 | Effluent 2 | |||||
|---|---|---|---|---|---|---|
| S0 (mg L−1) | RE (%) | RR (mg L−1 d−1) | S0 (mg L−1) | RE (%) | RR (mg L−1 d−1) | |
| C+ | 10.1 ± 0.2 a | 100 ± 0 a | 0.92 ± 0.03 a | 9.2 ± 0.1 a | 100 ± 0 a | 0.94 ± 0.02 a |
| E | 7.8 ± 0.5 a | 99.062 ± 0.005 a | 0.71 ± 0.05 a | 3.02 ± 0.04 a | 98.80 ± 0.02 b | 0.30 ± 0.01 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martins, R.A.; Salgado, E.M.; Gonçalves, A.L.; Esteves, A.F.; Pires, J.C.M. Microalgae-Based Remediation of Real Textile Wastewater: Assessing Pollutant Removal and Biomass Valorisation. Bioengineering 2024, 11, 44. https://doi.org/10.3390/bioengineering11010044
Martins RA, Salgado EM, Gonçalves AL, Esteves AF, Pires JCM. Microalgae-Based Remediation of Real Textile Wastewater: Assessing Pollutant Removal and Biomass Valorisation. Bioengineering. 2024; 11(1):44. https://doi.org/10.3390/bioengineering11010044
Chicago/Turabian StyleMartins, Rúben A., Eva M. Salgado, Ana L. Gonçalves, Ana F. Esteves, and José C. M. Pires. 2024. "Microalgae-Based Remediation of Real Textile Wastewater: Assessing Pollutant Removal and Biomass Valorisation" Bioengineering 11, no. 1: 44. https://doi.org/10.3390/bioengineering11010044
APA StyleMartins, R. A., Salgado, E. M., Gonçalves, A. L., Esteves, A. F., & Pires, J. C. M. (2024). Microalgae-Based Remediation of Real Textile Wastewater: Assessing Pollutant Removal and Biomass Valorisation. Bioengineering, 11(1), 44. https://doi.org/10.3390/bioengineering11010044










