Nuclear Factor-κB Decoy Oligodeoxynucleotide Attenuates Cartilage Resorption In Vitro
Abstract
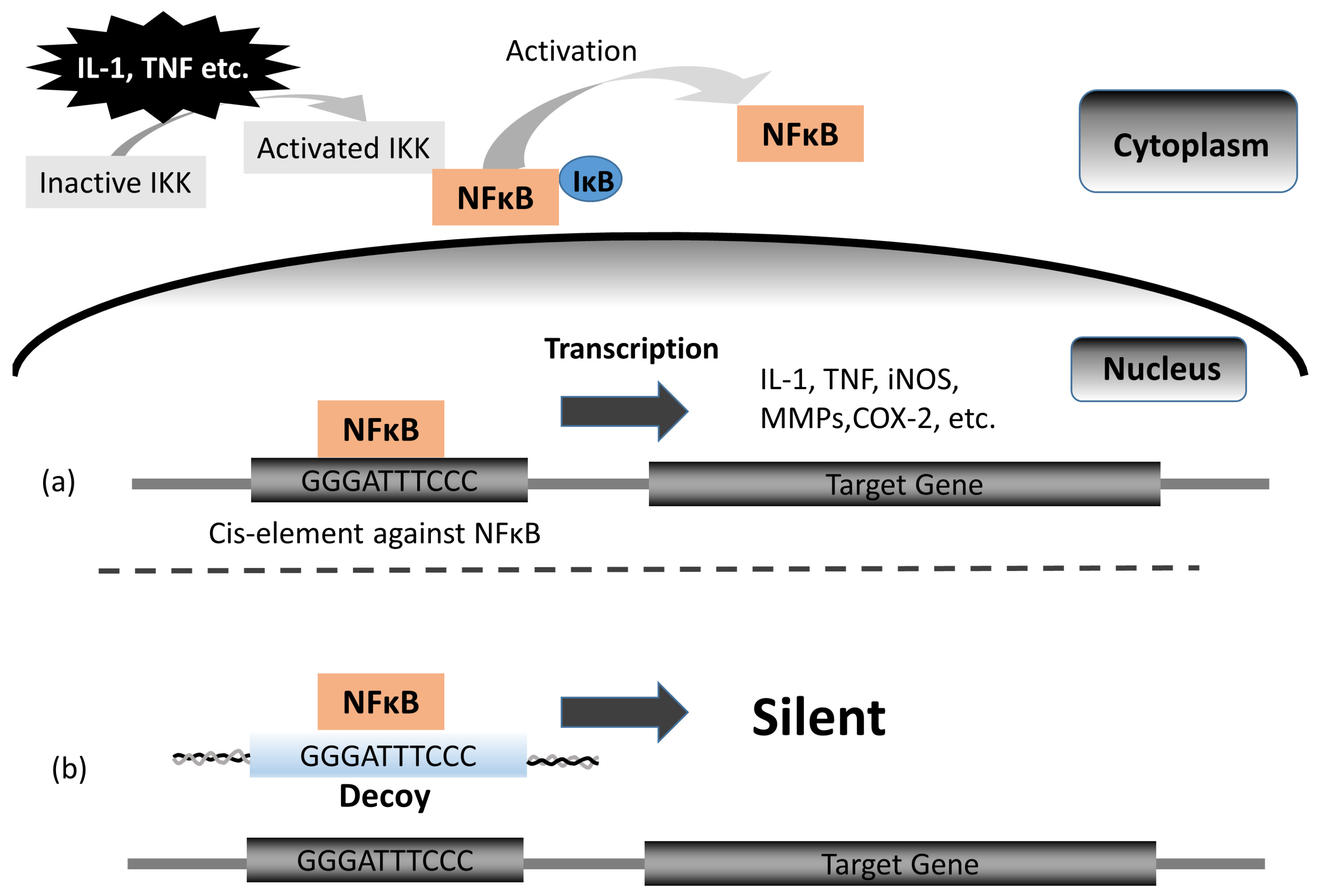
1. Background
2. Materials and Methods
2.1. rNSCh Cell Isolation and Culture in Alginate Beads
2.2. Cytotoxicity Analysis of Decoy
2.3. rNSCh PG Synthesis Assay
2.4. rNSCh PG Turnover Assay
2.5. hNSC Harvest and Tissue Culture
2.6. Efficiency of Transfection of Decoy
2.7. hNSC PG Turnover Assay
2.8. Enzyme-Linked Immunosorbent Assay (ELISA) Matrix Metalloprotease 3 (MMP-3), TNF, and IL-6
2.9. Measurement of Nitric Oxide (NO)
2.10. Statistical Analysis
3. Results
3.1. Decoy Inhibiting the Release of LDH
3.2. Decoy Not Affecting PG Synthesis
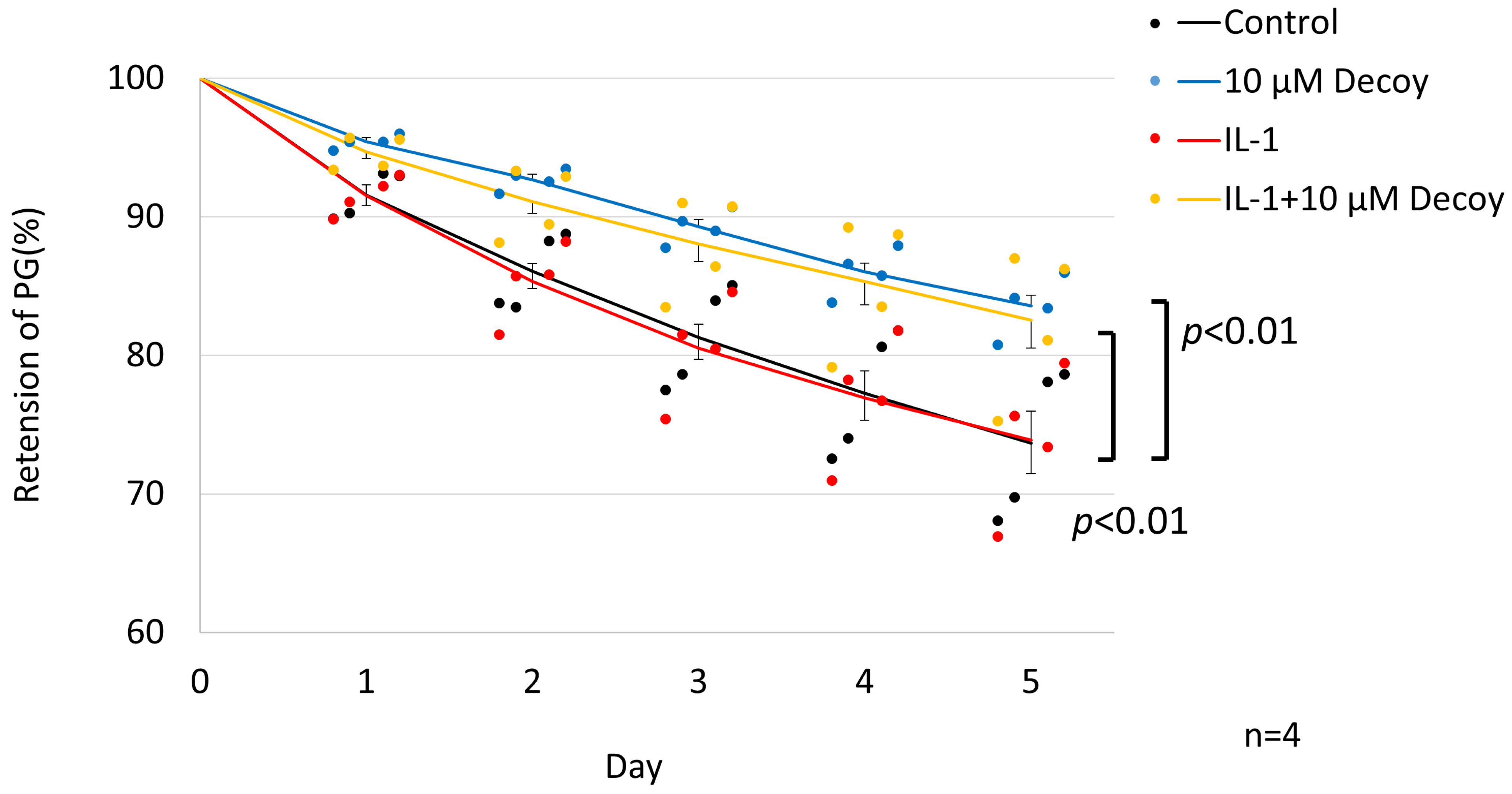
3.3. Inhibiting of PG Degradation in rNSCh by 10 μM Decoy
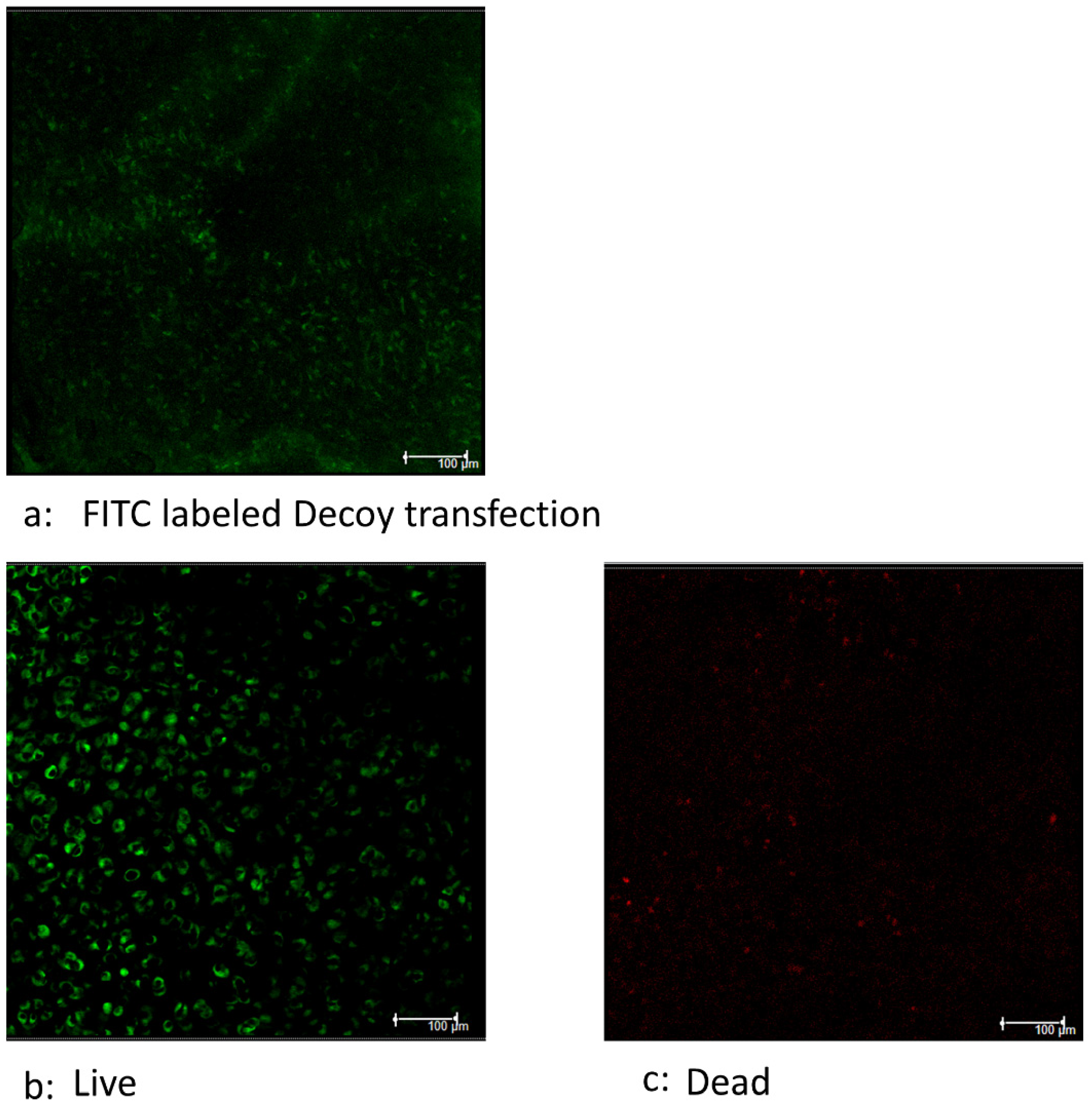
3.4. Decoy Transfected into Viable Cells
3.5. Inhibiting of PG Degradation in hNSC Tissue Culture by 10 μM Decoy
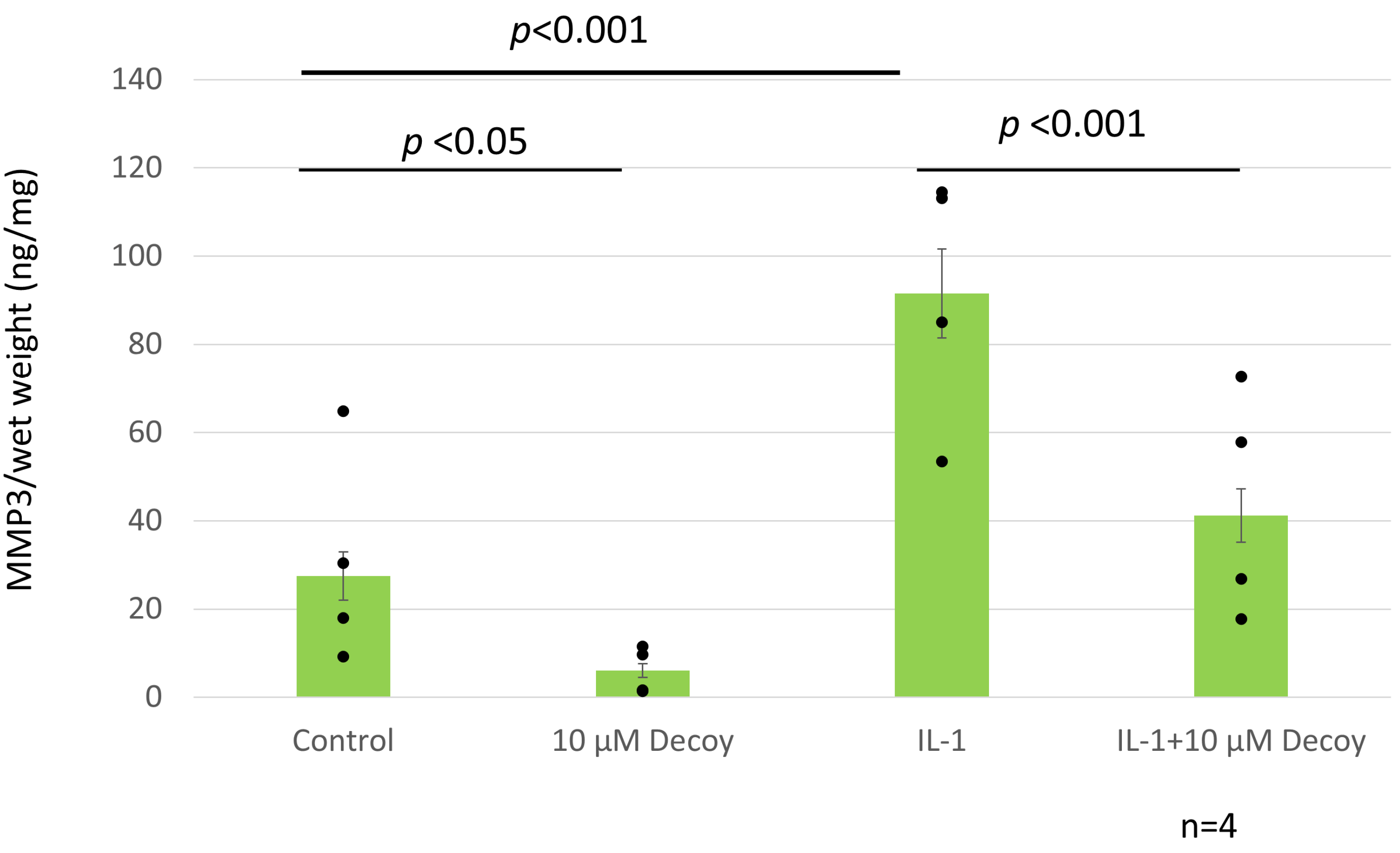
3.6. Decoy Inhibiting MMP-3 Production (ELISA and Nitrate Assay)
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fu, Y.; He, A.; Xie, Y.; Zhu, Y.; Li, C.; Zhang, T. The Effect of Fixation Materials on the Long-Term Stability of Cartilage Framework for Microtia Reconstruction. Facial Plast. Surg. Aesthetic Med. 2022, 25, 126–131. [Google Scholar] [CrossRef]
- Chen, H.; Wang, X.; Deng, Y. Complications Associated with Autologous Costal Cartilage Used in Rhinoplasty: An Updated Meta-Analysis. Aesthetic Plast. Surg. 2023, 47, 304–312. [Google Scholar] [CrossRef]
- Gentile, P.; Cervelli, V. Nasal Tip Remodeling Using Autologous Cartilage Grafts: Systematic Review. J. Craniofacial Surg. 2022, 33, 2035–2040. [Google Scholar] [CrossRef]
- Brent, B. The versatile cartilage autograft: Current trends in clinical transplantation. Clin. Plast. Surg. 1979, 6, 163–180. [Google Scholar] [CrossRef]
- De Gabory, L.; Fricain, J.C.; Stoll, D. Resorption of cartilage grafts in rhinoplasty: Fundamental basis. Rev. Laryngol.-Otol.-Rhinol. 2010, 131, 83–88. [Google Scholar]
- Watson, D.; Reuther, M.S. Tissue-engineered cartilage for facial plastic surgery. Curr. Opin. Otolaryngol. Head Neck Surg. 2014, 22, 300–306. [Google Scholar] [CrossRef]
- Bujía, J. Determination of the viability of crushed cartilage grafts: Clinical implications for wound healing in nasal surgery. Ann. Plast. Surg. 1994, 32, 261–265. [Google Scholar] [CrossRef]
- Hom, D. The wound healing response to grafted tissues. Otolaryngol. Clin. N. Am. 1994, 27, 13–24. [Google Scholar]
- Janis, J.E.; Harrison, B. Wound Healing: Part I. Basic Science. Plast. Reconstr. Surg. 2014, 133, 199e–207e. [Google Scholar] [CrossRef]
- Wu, T.H.; Hsu, S.H.; Chang, M.H.; Huang, Y.Y. Reducing scar formation by regulation of IL-1 and MMP-9 expression by using sustained release of prednisolone-loaded PDLL microspheres in a murine wound model. J. Biomed. Mater. Res. Part A 2013, 101, 1165–1172. [Google Scholar] [CrossRef]
- Haisch, A.; Gröger, A.; Radke, C.; Ebmeyer, J.; Sudhoff, H.; Grasnick, G.; Jahnke, V.; Burmester, G.; Sittinger, M. Macroencapsulation of human cartilage implants: Pilot study with polyelectrolyte complex membrane encapsulation. Biomaterials 2000, 21, 1561–1566. [Google Scholar] [CrossRef]
- Yu, H.; Lin, L.; Zhang, Z.; Zhang, H.; Hu, H. Targeting NF-κB pathway for the therapy of diseases: Mechanism and clinical study. Signal Transduct. Target. Ther. 2020, 5, 209. [Google Scholar] [CrossRef]
- Sen, R.; Baltimore, D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell 1986, 46, 705–716. [Google Scholar] [CrossRef]
- Gilmore, T.D. The Rel/NF-kappaB signal transduction pathway: Introduction. Oncogene 1999, 18, 6842. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef]
- Ahmed, A.S.; Gedin, P.; Hugo, A.; Bakalkin, G.; Kanar, A.; Hart, D.A.; Druid, H.; Svensson, C.; Kosek, E. Activation of NF-κB in Synovium versus Cartilage from Patients with Advanced Knee Osteoarthritis: A Potential Contributor to Inflammatory Aspects of Disease Progression. J. Immunol. 2018, 201, 1918–1927. [Google Scholar] [CrossRef]
- Uemura, T.; Tsujii, M.; Akeda, K.; Iino, T.; Satonaka, H.; Hasegawa, M.; Sudo, A. Transfection of nuclear factor-kappaB decoy oligodeoxynucleotide protects against ischemia/reperfusion injury in a rat epigastric flap model. J. Gene Med. 2012, 14, 623–631. [Google Scholar] [CrossRef]
- Sullenger, B.A.; Gallardo, H.F.; Ungers, G.E.; Gilboa, E. Overexpression of TAR sequences renders cells resistant to human immunodeficiency virus replication. Cell 1990, 63, 601–608. [Google Scholar] [CrossRef]
- Bielinska, A.; Shivdasani, R.A.; Zhang, L.; Nabel, G.J. Regulation of gene expression with double-stranded phosphorothioate oligonucleotides. Science 1990, 250, 997–1000. [Google Scholar] [CrossRef]
- Morishita, R.; Tomita, N.; Kaneda, Y.; Ogihara, T. Molecular therapy to inhibit NFκB activation by transcription factor decoy oligonucleotides. Curr. Opin. Pharmacol. 2004, 4, 139–146. [Google Scholar] [CrossRef]
- Kato, K.; Akeda, K.; Miyazaki, S.; Yamada, J.; Muehleman, C.; Miyamoto, K.; Asanuma, Y.A.; Asanuma, K.; Fujiwara, T.; Lenz, M.E.; et al. NF-kB decoy oligodeoxynucleotide preserves disc height in a rabbit anular-puncture model and reduces pain induction in a rat xenograft-radiculopathy model. Eur. Cells Mater. 2021, 42, 90–109. [Google Scholar] [CrossRef]
- Morishita, R.; Sugimoto, T.; Aoki, M.; Kida, I.; Tomita, N.; Moriguchi, A.; Ogihara, T. In vivo transfection of cis element “decoy” against nuclear factor-κB binding site prevents myocardial infarction. Nat. Med. 1997, 3, 894–899. [Google Scholar] [CrossRef]
- Nishimura, A.; Akeda, K.; Matsubara, T.; Kusuzaki, K.; Matsumine, A.; Masuda, K.; Gemba, T.; Uchida, A.; Sudo, A. Transfection of NF-κB decoy oligodeoxynucleotide suppresses pulmonary metastasis by murine osteosarcoma. Cancer Gene Ther. 2011, 18, 250–259. [Google Scholar] [CrossRef]
- Cao, C.C.; Ding, X.Q.; Ou, Z.L.; Liu, C.F.; Li, P.; Wang, L.; Zhu, C.F. In vivo transfection of NF-κB decoy oligodeoxynucleotides attenuate renal ischemia/reperfusion injury in rats. Kidney Int. 2004, 65, 834–845. [Google Scholar] [CrossRef]
- Xu, M.-Q.; Shuai, X.-R.; Yan, M.-L.; Zhang, M.-M.; Yan, L.-N. Nuclear factor-kappaB decoy oligodeoxynucleotides attenuates ischemia/reperfusion injury in rat liver graft. World J. Gastroenterol. 2005, 11, 6960. [Google Scholar] [CrossRef]
- Tomita, T.; Takano, H.; Tomita, N.; Morishita, R.; Kaneko, M.; Shi, K.; Takahi, K.; Nakase, T.; Kaneda, Y.; Yoshikawa, H. Transcription factor decoy for NFκB inhibits cytokine and adhesion molecule expressions in synovial cells derived from rheumatoid arthritis. Rheumatology 2000, 39, 749–757. [Google Scholar] [CrossRef]
- Abeyama, K.; Eng, W.; Jester, J.V.; Vink, A.A.; Edelbaum, D.; Cockerell, C.J.; Bergstresser, P.R.; Takashima, A. A role for NF-κB–dependent gene transactivation in sunburn. J. Clin. Investig. 2000, 105, 1751–1759. [Google Scholar] [CrossRef]
- Tahara, K.; Samura, S.; Tsuji, K.; Yamamoto, H.; Tsukada, Y.; Bando, Y.; Tsujimoto, H.; Morishita, R.; Kawashima, Y. Oral nuclear factor-κB decoy oligonucleotides delivery system with chitosan modified poly (D, L-lactide-co-glycolide) nanospheres for inflammatory bowel disease. Biomaterials 2011, 32, 870–878. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Ishida, Y.; Hosomichi, J.; Suzuki, J.-I.; Hatano, K.; Usumi-Fujita, R.; Shimizu, Y.; Kaneko, S.; Ono, T. Ultrasound microbubble-mediated transfection of NF-κB decoy oligodeoxynucleotide into gingival tissues inhibits periodontitis in rats in vivo. PLoS ONE 2017, 12, e0186264. [Google Scholar] [CrossRef]
- NIH. AMG0103 in Subjects with Chronic Discogenic Lumbar Back Pain—Full Text View—ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT03263611 (accessed on 28 January 2023).
- Anges. Notice on the Progress of Domestic Phase II Clinical Trials of NF-κB Decoy Oligo DNA for the Treatment of Chronic Intervertebral Disc Low Back Pain. Available online: https://www.anges.co.jp/pdf_news/public/GfGrjjpRguyfyO9W4mBxvpydIyJHs3fG.pdf (accessed on 30 November 2023).
- Miyake, T.; Miyake, T.; Sakaguchi, M.; Nankai, H.; Nakazawa, T.; Morishita, R. Prevention of Asthma Exacerbation in a Mouse Model by Simultaneous Inhibition of NF-κB and STAT6 Activation Using a Chimeric Decoy Strategy. Mol. Ther. Nucleic Acids 2018, 10, 159–169. [Google Scholar] [CrossRef]
- Morales, T.I.; Hascall, V.C. Factors involved in the regulation of proteoglycan metabolism in articular cartilage. Arthritis Rheum. 1989, 32, 1197–1201. [Google Scholar] [CrossRef]
- Hascall, V.C.; Handley, C.J.; McQuillan, D.J.; Hascall, G.K.; Robinson, H.C.; Lowther, D.A. The effect of serum on biosynthesis of proteoglycans by bovine articular cartilage in culture. Arch. Biochem. Biophys. 1983, 224, 206–223. [Google Scholar] [CrossRef]
- Campbell, M.A.; Handley, C.J.; Hascall, V.C.; Campbell, R.A.; Lowther, D.A. Turnover of proteoglycans in cultures of bovine articular cartilage. Arch. Biochem. Biophys. 1984, 234, 275–289. [Google Scholar] [CrossRef]
- Luyten, F.P.; Hascall, V.C.; Nissley, S.P.; Morales, T.I.; Reddi, A.H. Insulin-like growth factors maintain steady-state metabolism of proteoglycans in bovine articular cartilage explants. Arch. Biochem. Biophys. 1988, 267, 416–425. [Google Scholar] [CrossRef]
- Fell, H.B.; Jubb, R.W. The effect of synovial tissue on the breakdown of articular cartilage in organ culture. Arthritis Rheum. 1977, 20, 1359–1371. [Google Scholar] [CrossRef]
- Saklatvala, J.; Pilsworth, L.M.; Sarsfield, S.J.; Gavrilovic, J.; Heath, J.K. Pig catabolin is a form of interleukin 1. Cartilage and bone resorb, fibroblasts make prostaglandin and collagenase, and thymocyte proliferation is augmented in response to one protein. Biochem. J. 1984, 224, 461–466. [Google Scholar] [CrossRef]
- Morales, T.I.; Hascall, V.C. Effects of interleukin-1 and lipopolysaccharides on protein and carbohydrate metabolism in bovine articular cartilage organ cultures. Connect. Tissue Res. 1989, 19, 255–275. [Google Scholar] [CrossRef]
- Jenei-Lanzl, Z.; Meurer, A.; Zaucke, F. Interleukin-1β signaling in osteoarthritis—Chondrocytes in focus. Cell. Signal. 2019, 53, 212–223. [Google Scholar] [CrossRef]
- Bonassar, L.J.; Frank, E.H.; Murray, J.C.; Paguio, C.G.; Moore, V.L.; Lark, M.W.; Sandy, J.D.; Wu, J.J.; Eyre, D.R.; Grodzinsky, A.J. Changes in cartilage composition and physical properties due to stromelysin degradation. Arthritis Rheum. 1995, 38, 173–183. [Google Scholar] [CrossRef]
- Temple, M.M.; Xue, Y.; Chen, M.Q.; Sah, R.L. Interleukin-1alpha induction of tensile weakening associated with collagen degradation in bovine articular cartilage. Arthritis Rheum. 2006, 54, 3267–3276. [Google Scholar] [CrossRef] [PubMed]
- Furman, B.D.; Mangiapani, D.S.; Zeitler, E.; Bailey, K.N.; Horne, P.H.; Huebner, J.L.; Kraus, V.B.; Guilak, F.; Olson, S.A. Targeting pro-inflammatory cytokines following joint injury: Acute intra-articular inhibition of interleukin-1 following knee injury prevents post-traumatic arthritis. Arthritis Res. Ther. 2014, 16, R134. [Google Scholar] [CrossRef] [PubMed]
- Sawa, Y.; Morishita, R.; Suzuki, K.; Kagisaki, K.; Kaneda, Y.; Maeda, K.; Kadoba, K.; Matsuda, H. A novel strategy for myocardial protection using in vivo transfection of cis element ‘decoy’ against NFkappaB binding site: Evidence for a role of NFkappaB in ischemia-reperfusion injury. Circulation 1997, 96, II-280–II-284; discussion II-285. [Google Scholar]
- Yoshimura, S.; Morishita, R.; Hayashi, K.; Yamamoto, K.; Nakagami, H.; Kaneda, Y.; Sakai, N.; Ogihara, T. Inhibition of intimal hyperplasia after balloon injury in rat carotid artery model using cis-element ‘decoy’of nuclear factor-kB binding site as a novel molecular strategy. Gene Ther. 2001, 8, 1635–1642. [Google Scholar] [CrossRef] [PubMed]
- Chiba, K.; Andersson, G.B.; Masuda, K.; Thonar, E.J.A. Metabolism of the extracellular matrix formed by intervertebral disc cells cultured in alginate. Spine 1997, 22, 2885–2893. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, D.; Yin, L.; Zhang, C.; Qu, H.; Xu, J. Neuroglobin protects against cerebral ischemia/reperfusion injury in rats by suppressing mitochondrial dysfunction and endoplasmic reticulum stress-mediated neuronal apoptosis through synaptotagmin-1. Environ. Toxicol. 2023, 38, 1891–1904. [Google Scholar] [CrossRef]
- Masuda, K.; Shirota, H.; Thonar, E.J.A. Quantification of 35 S-Labeled Proteoglycans Complexed to Alcian Blue by Rapid Filtration in Multiwell Plates. Anal. Biochem. 1994, 217, 167–175. [Google Scholar] [CrossRef]
- Mok, S.S.; Masuda, K.; Häuselmann, H.; Aydelotte, M.B.; Thonar, E. Aggrecan synthesized by mature bovine chondrocytes suspended in alginate. Identification of two distinct metabolic matrix pools. J. Biol. Chem. 1994, 269, 33021–33027. [Google Scholar] [CrossRef]
- Levin, A.A. A review of issues in the pharmacokinetics and toxicology of phosphorothioate antisense oligonucleotides. Biochim. Biophys. Acta (BBA)-Gene Struct. Expr. 1999, 1489, 69–84. [Google Scholar] [CrossRef]
- Xue, L.; Wang, D.; Zhang, X.; Xu, S.; Zhang, N. Targeted and triple therapy-based liposomes for enhanced treatment of rheumatoid arthritis. Int. J. Pharm. 2020, 586, 119642. [Google Scholar] [CrossRef]
- Tomita, T.; Takeuchi, E.; Tomita, N.; Morishita, R.; Kaneko, M.; Yamamoto, K.; Nakase, T.; Seki, H.; Kato, K.; Kaneda, Y. Suppressed severity of collagen-induced arthritis by in vivo transfection of nuclear factor κB decoy oligodeoxynucleotides as a gene therapy. Arthritis Rheum. Off. J. Am. Coll. Rheumatol. 1999, 42, 2532–2542. [Google Scholar] [CrossRef]
- Tan, Y.; Zhang, J.-S.; Huang, L. Codelivery of NF-κB Decoy-Related Oligodeoxynucleotide Improves LPD-Mediated Systemic Gene Transfer. Mol. Ther. 2002, 6, 804–812. [Google Scholar] [CrossRef] [PubMed]
- Neuman, M.K.; Briggs, K.K.; Masuda, K.; Sah, R.L.; Watson, D. A compositional analysis of cadaveric human nasal septal cartilage. Laryngoscope 2013, 123, 2120–2124. [Google Scholar] [CrossRef] [PubMed]
- Allaith, S.; Tew, S.R.; Hughes, C.E.; Clegg, P.D.; Canty-Laird, E.G.; Comerford, E.J. Characterisation of key proteoglycans in the cranial cruciate ligaments (CCLs) from two dog breeds with different predispositions to CCL disease and rupture. Vet. J. 2021, 272, 105657. [Google Scholar] [CrossRef] [PubMed]
- Hascall, V.; Luyten, F.; Plaas, A.; Sandy, J. Steady-state metabo-lism of proteoglycans in bovine articular cartilage explants. In Methods in Cartilage Research; Maroudas, A., Kuettner, K.E., Eds.; Academic Press: London, UK, 1990; pp. 108–112. [Google Scholar]
- Peng, Z.; Sun, H.; Bunpetch, V.; Koh, Y.; Wen, Y.; Wu, D.; Ouyang, H. The regulation of cartilage extracellular matrix homeostasis in joint cartilage degeneration and regeneration. Biomaterials 2021, 268, 120555. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Sun, C.; He, B.; Zhang, X.; Hao, H.; Hou, Y.; Li, A.; Wang, Y.; Wang, Y. Macrophage Migration Inhibitory Factor Promotes Expression of Matrix Metalloproteinases 1 and 3 in Spinal Cord Astrocytes following Gecko Tail Amputation. J. Integr. Neurosci. 2023, 22, 29. [Google Scholar] [CrossRef] [PubMed]
- Tyler, J.; Bolis, S.; Dingle, J.; Middleson, J.F.S. Mediators of matrix metabolism. In Articular Cartilage and Osteoarthritis; Kuettner, K., Schleyerbach, R., Peyron, J., Eds.; Raven Press: New York, NY, USA, 1992; pp. 251–264. [Google Scholar]
- Cawston, T.; Curry, V.; Summers, C.; Clark, I.; Riley, G.; Life, P.; Spaull, J.; Goldring, M.; Koshy, P.; Rowan, A. The role of oncostatin M in animal and human connective tissue collagen turnover and its localization within the rheumatoid joint. Arthritis Rheum. 1998, 41, 1760–1771. [Google Scholar] [CrossRef] [PubMed]
- Okada, Y. Regulation of Matrix Metalloprotease. Jikken Igaku 1992, 10, 256–262. (In Japanese) [Google Scholar]
- Pichika, R.; Akeda, K.; Gemba, T.; Miyamoto, K.; An, H.S.; Masuda, K. Transcription Factor Decoy for Nfkb Inhibits the Appearance of Active Mmps and Adamts4 in the Medium of Human Intervertebral Disc Cells Cultured in Alginate. In Proceedings of the 2005 Annual Meeting of the Orthopaedic Research Society, Chicago, IL, USA, 20–23 May 2005. [Google Scholar]
- Perkins, N.D. Achieving transcriptional specificity with NF-κB. Int. J. Biochem. Cell Biol. 1997, 29, 1433–1448. [Google Scholar] [CrossRef]
- Somma, D.; Kok, F.O.; Kerrigan, D.; Wells, C.A.; Carmody, R.J. Defining the Role of Nuclear Factor (NF)-κB p105 Subunit in Human Macrophage by Transcriptomic Analysis of NFKB1 Knockout THP1 Cells. Front. Immunol. 2021, 12, 669906. [Google Scholar] [CrossRef]
- Guan, H.; Hou, S.; Ricciardi, R.P. DNA binding of repressor nuclear factor-kappaB p50/p50 depends on phosphorylation of Ser337 by the protein kinase A catalytic subunit. J. Biol. Chem. 2005, 280, 9957–9962. [Google Scholar] [CrossRef]
- Nelson, H.S.; Leung, D.Y.; Bloom, J.W. Update on glucocorticoid action and resistance. J. Allergy Clin. Immunol. 2003, 111, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.W.; Zhang, T.; Wang, W.B. Gluococorticoid could influence extracellular matrix synthesis through Sox9 via p38 MAPK pathway. Rheumatol. Int. 2012, 32, 3669–3673. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.A.; Reuther, M.S.; Briggs, K.K.; Schumacher, B.L.; Williams, G.M.; Corr, M.; Sah, R.L.; Watson, D. In vivo implantation of tissue-engineered human nasal septal neocartilage constructs: A pilot study. Otolaryngol.—Head Neck Surg. 2012, 146, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Asnaghi, M.A.; Power, L.; Barbero, A.; Haug, M.; Köppl, R.; Wendt, D.; Martin, I. Biomarker Signatures of Quality for Engineering Nasal Chondrocyte-Derived Cartilage. Front. Bioeng. Biotechnol. 2020, 8, 283. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nemoto, H.; Sakai, D.; Watson, D.; Masuda, K. Nuclear Factor-κB Decoy Oligodeoxynucleotide Attenuates Cartilage Resorption In Vitro. Bioengineering 2024, 11, 46. https://doi.org/10.3390/bioengineering11010046
Nemoto H, Sakai D, Watson D, Masuda K. Nuclear Factor-κB Decoy Oligodeoxynucleotide Attenuates Cartilage Resorption In Vitro. Bioengineering. 2024; 11(1):46. https://doi.org/10.3390/bioengineering11010046
Chicago/Turabian StyleNemoto, Hitoshi, Daisuke Sakai, Deborah Watson, and Koichi Masuda. 2024. "Nuclear Factor-κB Decoy Oligodeoxynucleotide Attenuates Cartilage Resorption In Vitro" Bioengineering 11, no. 1: 46. https://doi.org/10.3390/bioengineering11010046
APA StyleNemoto, H., Sakai, D., Watson, D., & Masuda, K. (2024). Nuclear Factor-κB Decoy Oligodeoxynucleotide Attenuates Cartilage Resorption In Vitro. Bioengineering, 11(1), 46. https://doi.org/10.3390/bioengineering11010046







