Cells-in-Touch: 3D Printing in Reconstruction and Modelling of Microscopic Biological Geometries for Education and Future Research Applications
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Cell Culture and Staining Procedures
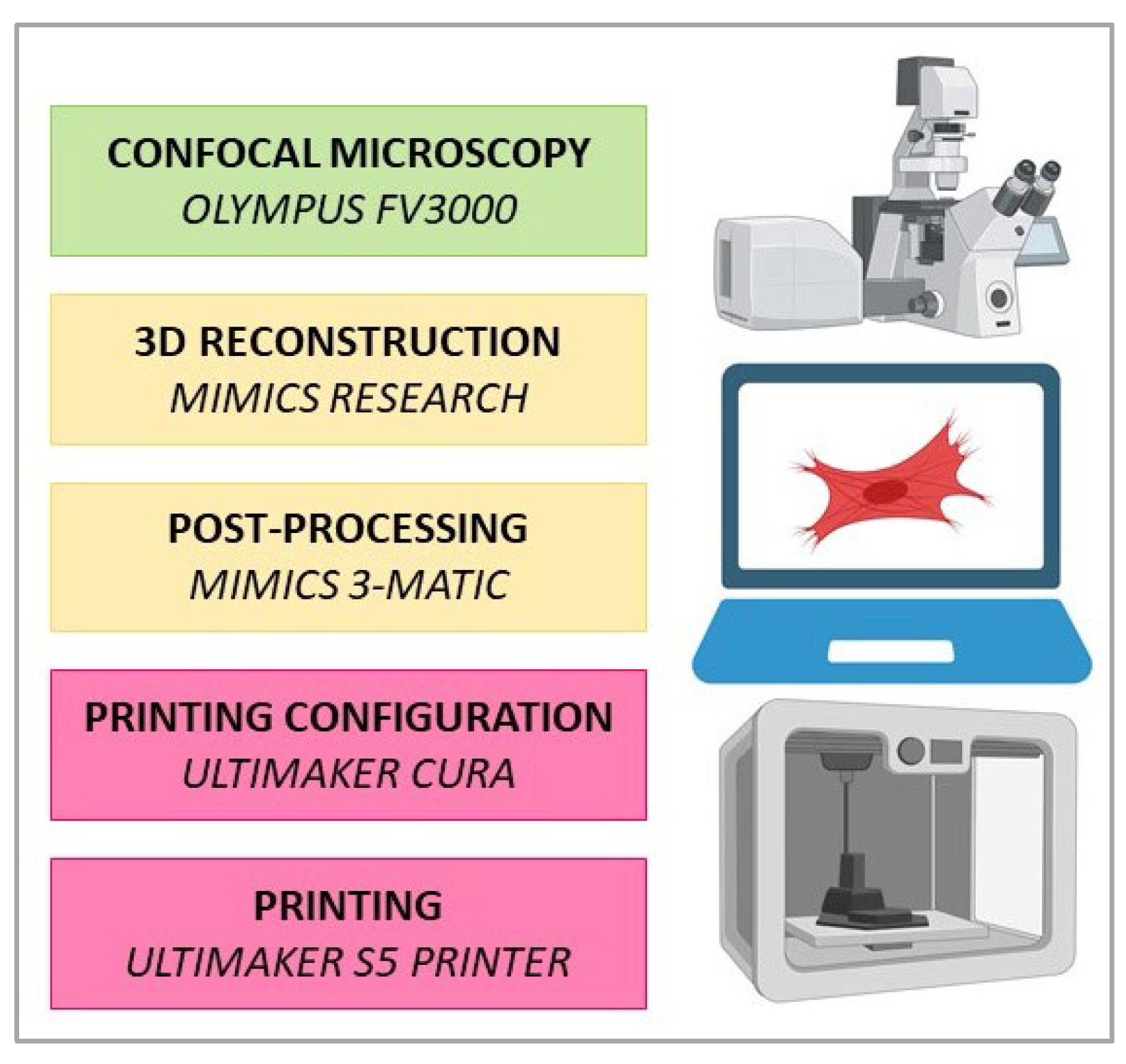
2.3. Image Acquisition and 3D Reconstruction
2.4. Post-Processing
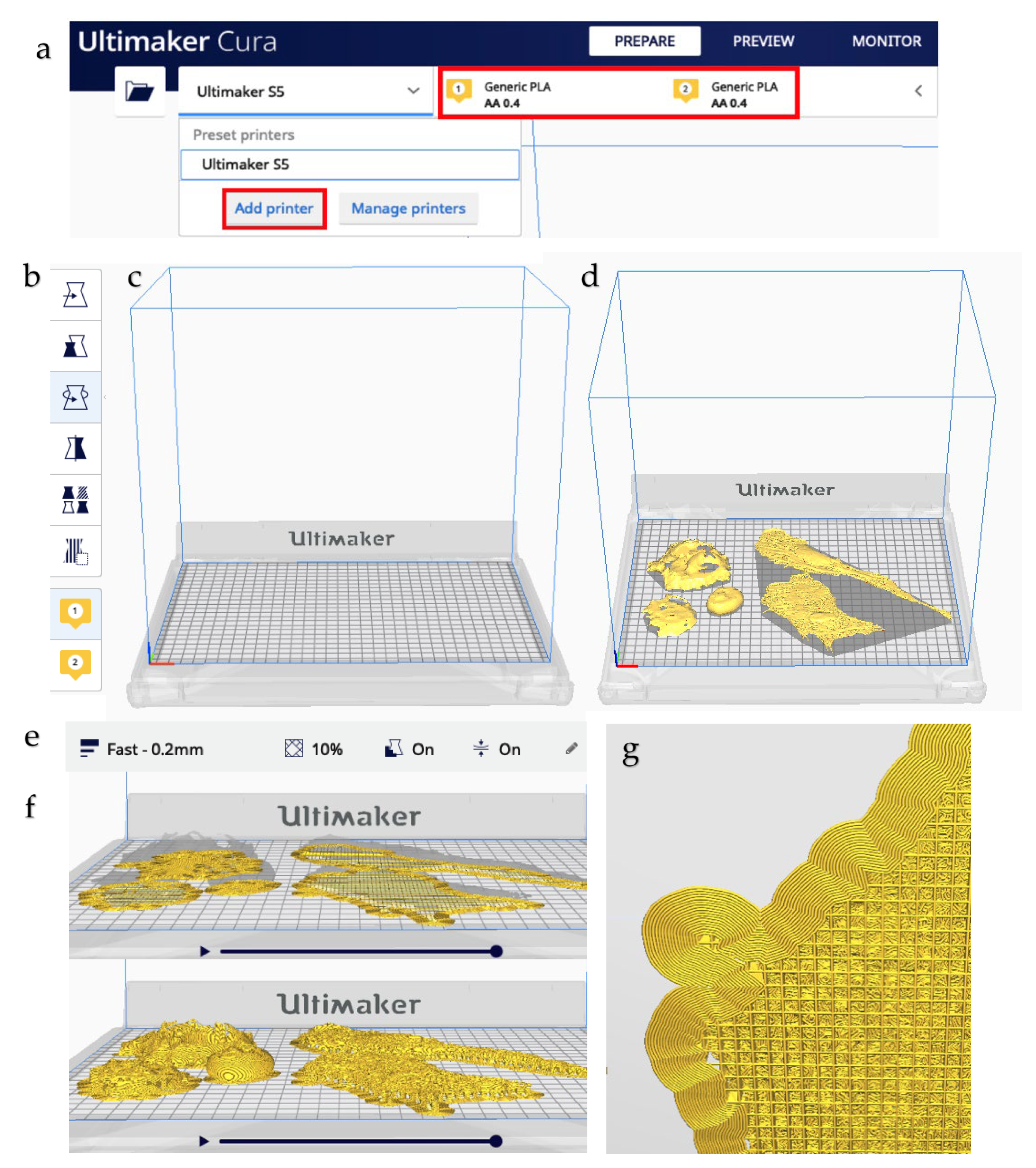
2.5. Printing
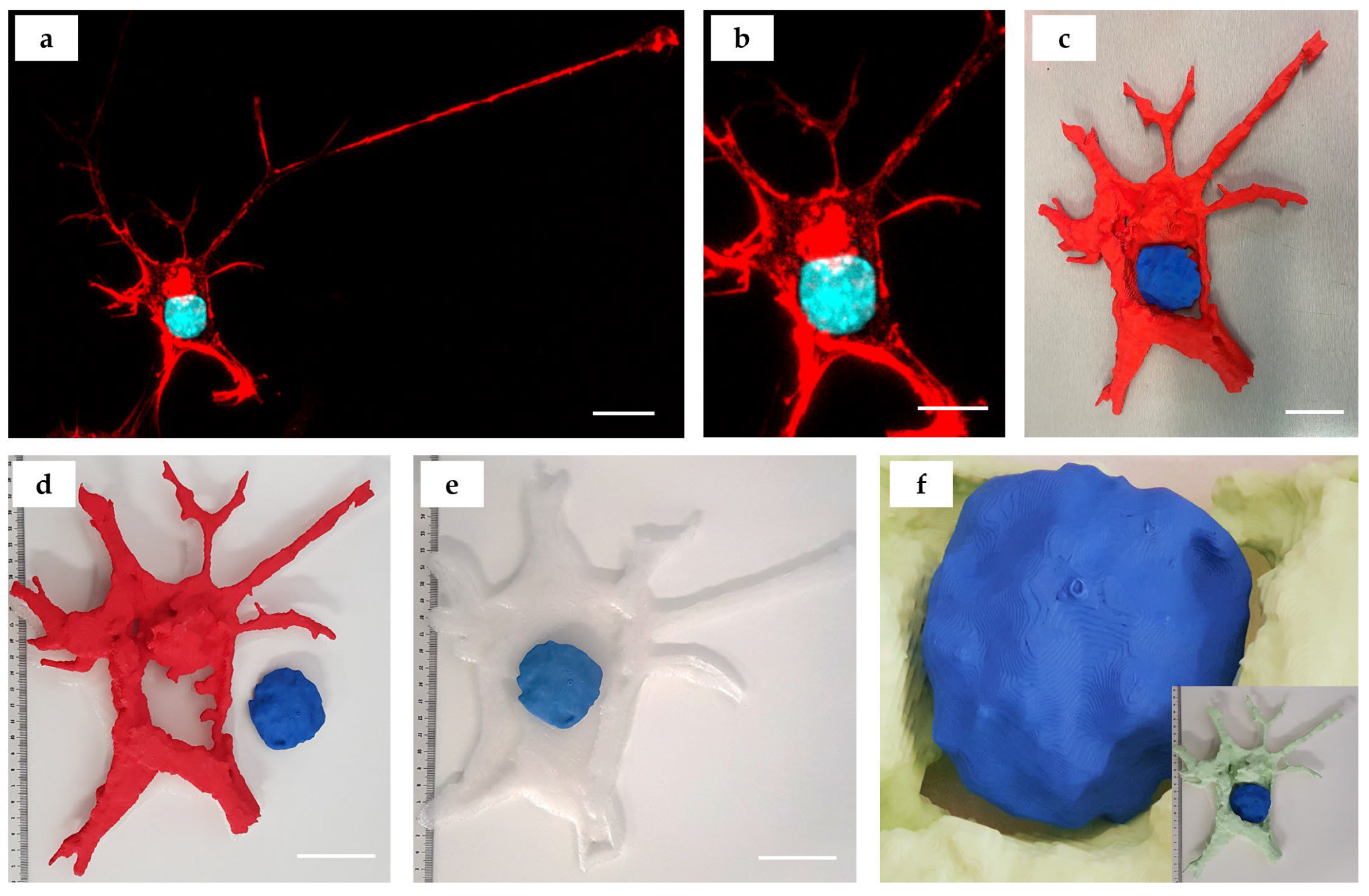
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| AM Parameter | Biological Object | |||||
|---|---|---|---|---|---|---|
| Natural Killer Cell | Blood Cells | C. elegans Embryo and Distal Tip Cell, a Part of a Plant Cell | Pollen, Blood Cells, Plant Root, Insect Eye, Zebrafish Larva | Nucleus of a Hela Cell, Pollen Shells, Fruit Fly | Plant Golgi Stacks | |
| References | [4] | [21] | [20] | [5] | [22] | [23] |
| Imaging modality 1 | STED | TEM | MPM, SCF, and TEM | SCF (all samples, except zebrafish larva) and LSHM (zebrafish larva) | SCF | TEM |
| Detected signals 2 | Phalloidin-Alexa Fluor 488 (f-actin) | SET | AF, gGFP | AF (all samples, except zebrafish larva), Phalloidin-FITC (f-actin), DAPI (omitted) * (zebrafish larva only) | DAPI (cell nucleus), AF (other objects) | SET |
| 3D reconstruction | Huygens, Imaris, Blender | AMIRA, IMOD, Rhinoceros 3D | Mimics, Geomagics | Bitplane Imaris, MeshLab | ImageJ 3D Viewer Plugin | IMOD, 3dmod |
| 3D printer | MakerBot Replicator 2X | FELIX 3.0 Dual Extruder | Spectrum Z510 | Zortrax M200 | UP Plus 2 | Lulzbot Mini and Form1+ |
| Printing software 3 | Makerware | Repetier-Host for FELIX Printers | Zprint Software | Zortrax Z-Suite | N.D. | N.D. |
| 3D printing technology 4 | FDM | FDM | IPT | FDM | FDM | FDM and SLA |
| Printing material 5 | ABS filament | PLA, PVA (support) | Plaster Powder | ABS filament | TP | TP and UR |
Appendix A.1. The Biomedical Educational and Research Context
- Cell culture terminology and methodology: linear (immortalized) cells vs. primary cells [33]. The technical article by Merck explains cell culture protocols applicable both to linear cells (presented by cancer PANC-1 cells) and primary cells (human dermal fibroblasts, HDF).
- Healthy cells (HDF) vs. cancer cells (PANC-1). Fibroblasts are the main cell type in connective tissues, responsible for the production and degradation of collagen and other components of the extracellular matrix. A specialized form of fibroblasts, myofibroblasts, can exert strong contraction of tissue (particularly important for wound healing and regeneration). The source of PANC-1 cells in pancreatic ductal adenocarcinoma is deadly cancer with limited treatment options [34,35]. This malignant tumor commonly contains large amounts of collagen and fibroblasts, which together contribute to its treatment resistance [36].
- Embryonic origin of cells and tissues. Pancreatic adenocarcinoma originates from pancreatic glandular epithelium which has an ectodermal embryonic origin. Fibroblasts are cells of mesodermal origin. Neuron-like cells SH-SY5Y are derived from human neuroblastoma, a malignant tumor that originates from the neural crest cells.
- Cell shape and phenotype. Untreated PANC-1 cells are characterized by epithelioid phenotype; the HDFs have a mesenchymal-like phenotype, while SH-SY5Y cells may have varying phenotypes, depending on the cell culture conditions. In the current study, neuronal-like differentiation was maintained in these cells. Among several classifying features, morphology is one of the most prominent and obvious signatures of cellular phenotype. Epithelioid cells have a rounded shape, and their nucleus is usually centrally located. The signature of mesenchymal cells is a more elongated shape, quite often spindle-like, and the nucleus of the cell is usually more eccentric [33]. Neurons feature a clearly discernible cell body with centrally located round nuclei and various types of cytoplasmatic processes (the branching ones are termed dendrites, and the long, non-branching processes are named axons). Cell shape is a recognized feature associated with adhesion and motility potential, as well as their differentiation commitment [37,38,39].
- EMT and MET. One of the critical hallmarks of cancer progression is the so-called epithelial-to-mesenchymal transition (EMT) and the reverse (MET) process, reflecting the adaptation of cancer cells to new environments, for example, during the metastatic colonization of distant organs. The signature for EMT is a loss of epithelioid phenotype in epithelial (healthy or malignant) cells and the acquisition of a mesenchymal phenotype. MET presents the opposite transition. EMT/MET phenotype changes are reflected, in particular, in cell shape [40,41].
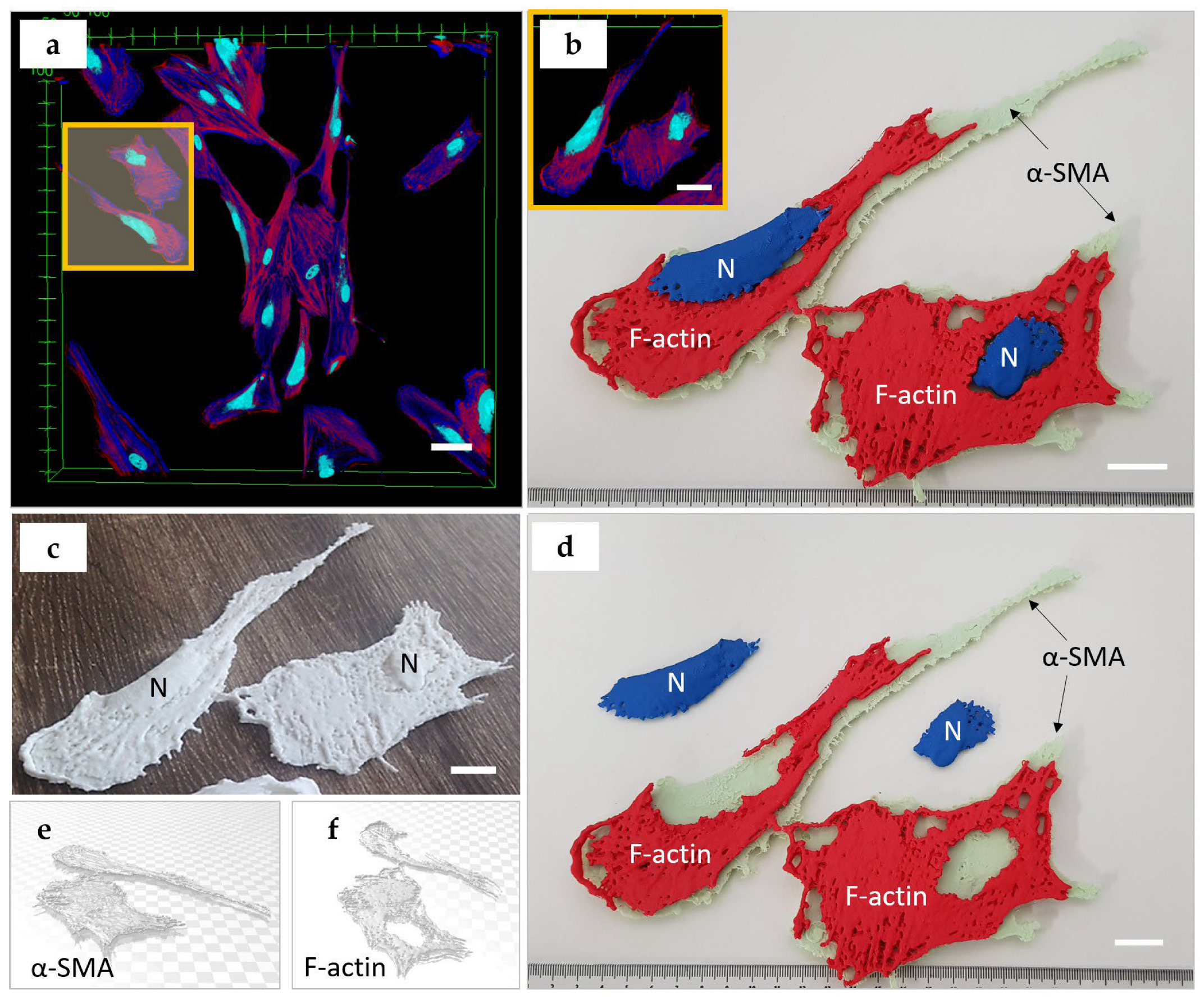
- Fibrosis. Fibrosis is the scarring of tissues and organs, characterized by excessive accumulation of extracellular matrix. At certain stages, it is also associated with the rapid proliferation of fibroblasts and their transformation into myofibroblasts. The signature of myofibroblasts is an expression of α-smooth muscle actin (α-SMA). The fibroblast-to-myofibroblast transdifferentiation, as well as the transformation of other cells into myofibroblasts, is a typical sign of fibrosis [42,43,44,45,46].
- Cytoskeleton. Three types of subcellular structures were imaged in the current study: cell nuclei, polymerized f-actin filaments representing the cytoskeleton component defining the shape of cells [37,38,39], and the specialized form of actin, known as α-smooth muscle actin (α-SMA), which is recognized as a phenotypical marker of cells bearing mechanical stress, such as smooth muscle cells or myofibroblasts [47]. Both the shape of the cytoskeleton and the level of α-SMA expression are key indicators of the cell’s functional state. Cell cytoskeleton and nucleus shape are dynamic characteristics that can reflect the phase of the mitotic cycle and the migration pattern [48]. In standard two-dimensional cell culture models, larger mean surface area and proportion of contractile α-SMA fibers indicate myofibroblast transdifferentiation of fibroblasts followed by excess synthesis of collagen [49]. This phenotypical transition reflects cellular fibrotic response on the tissue level, e.g., in skin scar or peri-implant connective tissue capsule formation [50]. These parameters are important to monitor in in vitro studies of cancer treatment, drug testing, and all the areas where cells are responding to external factors.
- The research and bioengineering applications of the CiTo-3DP methodology are dependent on the choice of the cells and subcellular structures. For the printed cellular models presented in this study, we envisage a scope of analytical tasks related to the relationship between the nucleus and cytoskeleton. For example, the data on the mass vs. the volume of the organelles, the surface texture of the organelles, the architecture of the intracellular space, and their reorganization in response to the experimental stimuli could serve for more biologically accurate bioengineering simulations such as, for instance, computational fluid dynamics research and analysis of the intracellular mechanical microenvironment. Further development of the proposed approach with the development of multi-material models or layered multi-material coatings, may be useful in cognitive and rehabilitation sciences.
| Settings | Subcellular Structures | ||
|---|---|---|---|
| Nuclei (DNA) | F-Actin | α-SMA | |
| Staining | DAPI | TRITC | Alexa Fluor 647 |
| Excitation laser, nm | 405 | 561 | 640 |
| Emission Filter (range), nm | 430–470 (DAPI) | 572–612 | 670–770 |
| Excitation maximum | 357 | 552 | 658 |
| Emission maximum, nm | 463 | 576 | 675 |
| Settings | Cell Type | ||
|---|---|---|---|
| PANC-1 | HDF | SH-SY5Y | |
| Image Size (pixels) | 1024 × 1024 | 1024 × 1024 | 703 × 880 |
| Objective | UPLSAPO 60× | UPLSAPO 20× | UAPON 100 × OTIRF 100× |
| Numerical Aperture | 1.35 | 0.75 | 1.49 |
| Immersion media | PBS | PBS | PBS |
| Image Size (XY) (µm) | 212.132 × 212.132 | 636.396 × 636.396 | 87.380 × 109.381 |
| Voxel resolution (XYZ) (µm/pixel3) | 0.207 × 0.207 × 1.000 | 0.621 × 0.621 × 0.500 | 0.240 × 0.240 × 0.500 |
| Number of z-slices | 16 | 33 | 36 |

| Parameters | Cell Type | |
|---|---|---|
| PANC-1, SH-SY5Y | HDF | |
| Printing scale, % | 100 (colored) and 200 (white) | 100 |
| Print scale (µm: mm) | 1 (colored) and 0.5 (white) | 1 |
| Layer height (mm) | 0.2 | 0.2 |
| Resolution (µm) | 0.1 | 0.2 |
Appendix B
Appendix B.1. Detailed Protocol Notes
- In the proposed workflow, 2D confocal microscopy image z-stacks were spliced together using GV-intensity thresholding in Mimics Research 21.0 software. PANC-1, HDF, and SH-SY5Y cells were separately cultured and stained for immunofluorescent visualization of nuclei and cytoskeletal structures under a confocal laser scanning microscope. Respective .tiff z-stack files and corresponding image .txt metadata files were created as follows: 16 PANC-1 culture XY-planar images were sequentially captured and stacked under 60× oil-submerged magnification, generating an XYZ voxel resolution of 0.207 × 0.207 × 1.000 (µm/pixel3). Similarly, 33 HDF culture XY-planar images were sequentially captured and stacked under 20× magnification, generating an XYZ voxel resolution of with XYZ voxel resolution of 0.621 × 0.621 × 0.500 (µm/pixel3). Similarly, the imaging parameters for the SH-SY5Y cells are shown in Table A3 (Appendix B). All sets of images were optimized by adjusting laser intensity and voltage gain and offset of the PMT-amplified signal.
- Note that voxel resolution was identified as the principal determinant of image quality and hence 3D reconstruction accuracy.
- The open-source Ultimaker software CURA v4.7.0 was used to prepare the .stl files for 3D printing and configure the print settings of the selected printer. All cell types were printed simultaneously on a dual-extrusion Ultimaker S5 FFF-technology printer with a 330 × 240 × 300 mm build volume.
- White PLA material was extruded at 205 °C through a 0.4 mm extruder head onto a build plate surface at 65 °C. Fast printing settings were chosen to minimize printing time, which came to approximately 12 h. In particular, a 10% infill and 60° support angle were chosen. The printer was allowed to cool prior to removing the printed models from the build plate. Supports were removed by hand and with the aid of plyers.
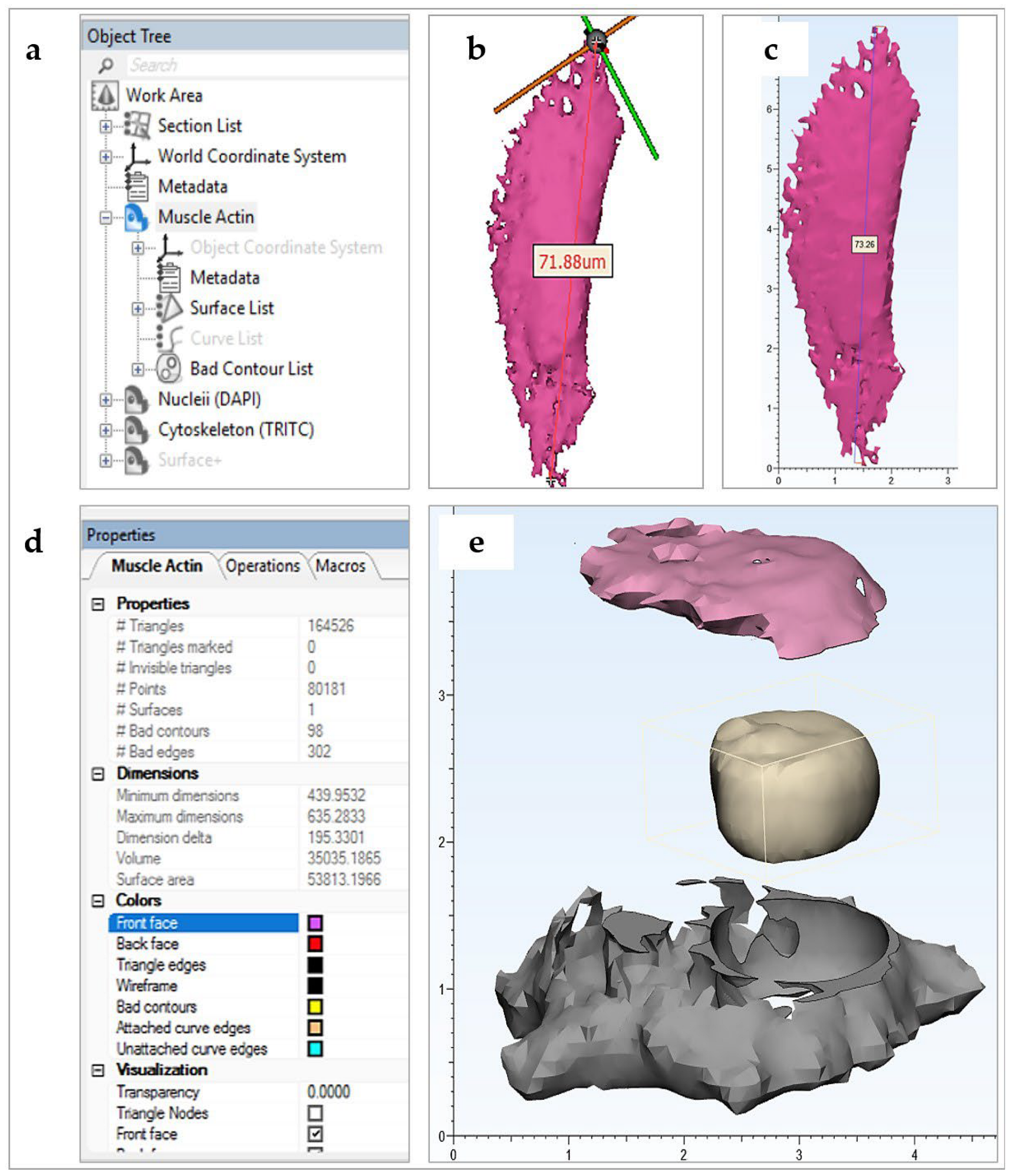
- Cropped 3D reconstructions of cellular structures were generated in Mimics Research 21.0 software from the imported z-stacks using grey-value thresholding. Single PANC-1 and SH-SY5Y cells and two connected HDF cells were, respectively, isolated. Minor edits were made to masks to better represent the cellular components imaged. Specifically, the Smart Fill tool was used to fill small holes between reconstructed voxels.
- This was particularly important in generating close-to-solid nucleus structures. The initial length measurement of a selected nucleus object was taken in µm for scale verification throughout the workflow. The resultant objects were exported directly into Mimics 3-Matic for post-processing. A second length measurement of the previously selected nucleus object was taken in mm, which verified the import rescaling from µm to mm automatically performed by the Mimics software.
- The 3D objects were optimized for 3D printing using various editing tools. A 1 mm external uniform offset was applied to the meshed surface geometries, followed by iterations of the smoothing, wrapping and remeshing tools. The models were designed such that the nucleus could be extracted from the rest of the cell body model. To achieve this, an XY-plane trim was performed to slice the cytoskeleton geometry in half. The aligned nucleus geometry was then Boolean-subtracted with a 1 mm clearance factor from the trimmed cytoskeleton geometries.
- Finally, the quality of the resultant surface meshes was checked using the Fix Wizard tool. The surfaces (nucleus, cytoskeleton upper, cytoskeleton lower) were exported as separate .stl files. Mimics and 3-Matic (Materialise) are already readily used in biomedical research applications as design-orientated software.
- As previously noted, Mimics have been used by Liu et al. [14] to improve surgical planning and performance, as well as by McMenamin et al. [13] as a cheaper and more ethically neutral alternative teaching aid to cadavers in medical education. In comparison to other commercial software, Martin et al. [7] showed that Mimics possessed more powerful image manipulation, visualization, and editing functions. 3-matic, also part of Materialise and often packaged with Mimics, allows for further design iterations and is well-suited to optimizing meshes for FDM 3D printing as STL files.
- Notably, neither software has seen significant uptake in areas of micro-scale biology. In comparison to open-access image-processing software such as 3D Slicer and ImageJ, commercial software provides a faster, more powerful, and more versatile user experience. In terms of CAD, commercial software, such as 3-Matic, allows for greater interactivity to be easily built into printable models. Although this design power was not fully explored in this methodology, its effect was demonstrated by the interactivity of the PANC-1 cytoskeleton-nucleus cell model. To achieve similar results using free software would require a transfer between software, which is often cumbersome in terms of file formatting and file sizes. Considering that Materialise provides both image-processing and CAD, and is already used in biology-related sciences, it was chosen for this project. It should be noted that Materialise also offers a variety of online tutorial resources, making it far easier to learn the software.
References
- Ngo, T.D.; Kashani, A.; Imbalzano, G.; Nguyen, K.T.Q.; Hui, D. Additive Manufacturing (3D Printing): A Review of Materials, Methods, Applications and Challenges. Compos. Part B Eng. 2018, 143, 172–196. [Google Scholar] [CrossRef]
- Ahangar, P.; Cooke, M.E.; Weber, M.H.; Rosenzweig, D.H. Current Biomedical Applications of 3D Printing and Additive Manufacturing. Appl. Sci. 2019, 9, 1713. [Google Scholar] [CrossRef]
- Attaran, M. The rise of 3-D printing: The advantages of additive manufacturing over traditional manufacturing. Bus. Horiz. 2017, 60, 677–688. [Google Scholar] [CrossRef]
- Mace, E.M.; Moon, J.; Orange, J.S. Three-Dimensional Printing of Super-Resolution Microscopy Images. Microsc. Today 2015, 23, 26–29. [Google Scholar] [CrossRef]
- Perry, I.; Szeto, J.-Y.; Isaacs, M.; Gealy, E.; Rose, R.; Scofield, S.; Watson, P.; Hayes, A. Production of 3D Printed Scale Models from Microscope Volume Datasets for use in STEM Education. EMS Eng. Sci. J. 2017, 1, 002. [Google Scholar]
- Del Rosario, M.; Heil, H.S.; Mendes, A.; Saggiomo, V.; Henriques, R. The Field Guide to 3D Printing in Optical Microscopy for Life Sciences. Adv Biol. 2022, 6, e2100994. [Google Scholar] [CrossRef]
- Martin, C.M.; Roach, V.; Nguyen, N.; Rice, C.L.; Wilson, T.D. Comparison of 3D reconstructive technologies used for morphometric research and the translation of knowledge using a decision matrix. Anat. Sci. Educ. 2013, 6, 393–403. [Google Scholar] [CrossRef]
- Werz, S.M.; Zeichner, S.J.; Berg, B.-I.; Zeilhofer, H.-F.; Thieringer, F. 3D Printed Surgical Simulation Models as educational tool by maxillofacial surgeons. Eur. J. Dent. Educ. 2018, 22, e500–e505. [Google Scholar] [CrossRef]
- Wojtyła, S.; Klama, P.; Baran, T. Is 3D printing safe? Analysis of the thermal treatment of thermoplastics: ABS, PLA, PET, and nylon. J. Occup. Environ. Hyg. 2017, 14, D80–D85. [Google Scholar] [CrossRef]
- Tack, P.; Victor, J.; Gemmel, P.; Annemans, L. 3D-printing techniques in a medical setting: A systematic literature review. Biomed. Eng. Online 2016, 15, 115. [Google Scholar] [CrossRef]
- Manero, A.; Smith, P.; Sparkman, J.; Dombrowski, M.; Courbin, D.; Kester, A.; Womack, I.; Chi, A. Implementation of 3D Printing Technology in the Field of Prosthetics: Past, Present, and Future. Int. J. Environ. Res. Public Health 2019, 16, 1641. [Google Scholar] [CrossRef] [PubMed]
- Zadpoor, A.A.; Malda, J. Additive Manufacturing of Biomaterials, Tissues, and Organs. Ann. Biomed. Eng. 2017, 45, 1–11. [Google Scholar] [CrossRef] [PubMed]
- McMenamin, P.G.; Quayle, M.R.; McHenry, C.R.; Adams, J.W. The Production of Anatomical Teaching Resources Using Three-Dimensional (3D) Printing Technology. Anat. Sci. Educ. 2014, 7, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Hu, Z.; Huang, S.; Wang, P.; Dong, Y.; Cheng, P.; Xu, H.; Tang, B.; Zhu, J. Application of 3D Printed Models of Complex Hypertrophic Scars for Preoperative Evaluation and Surgical Planning. Front. Bioeng. Biotechnol. 2020, 8, 115. [Google Scholar] [CrossRef]
- Bernhard, J.-C.; Isotani, S.; Matsugasumi, T.; Duddalwar, V.; Hung, A.J.; Suer, E.; Baco, E.; Satkunasivam, R.; Djaladat, H.; Metcalfe, C.; et al. Personalized 3D printed model of kidney and tumor anatomy: A useful tool for patient education. World J. Urol. 2015, 34, 337–345. [Google Scholar] [CrossRef]
- Witowski, J.S.; Pędziwiatr, M.; Major, P.; Budzyński, A. Cost-effective, personalized, 3D-printed liver model for preoperative planning before laparoscopic liver hemihepatectomy for colorectal cancer metastases. Int. J. Comput. Assist. Radiol. Surg. 2017, 12, 2047–2054. [Google Scholar] [CrossRef]
- Hachulla, A.-L.; Noble, S.; Guglielmi, G.; Agulleiro, D.; Müller, H.; Vallée, J.-P. 3D-printed heart model to guide LAA closure: Useful in clinical practice? Eur. Radiol. 2018, 29, 251–258. [Google Scholar] [CrossRef]
- Jin, Z.; Li, Y.; Yu, K.; Liu, L.; Fu, J.; Yao, X.; Zhang, A.; He, Y. 3D Printing of Physical Organ Models: Recent Developments and Challenges. Adv. Sci. 2021, 8, e2101394. [Google Scholar] [CrossRef]
- Jang, J.; Yi, H.-G.; Cho, D.-W. 3D Printed Tissue Models: Present and Future. ACS Biomater. Sci. Eng. 2016, 2, 1722–1731. [Google Scholar] [CrossRef]
- Cox, B.L.; Schumacher, N.; Konieczny, J.; Reifschneider, I.; Mackie, T.R.; Otegui, M.S.; Eliceiri, K.W. Fabrication approaches for the creation of physical models from microscopy data. 3D Print. Med. 2017, 3, 2. [Google Scholar] [CrossRef]
- Augusto, I.; Monteiro, D.; Girard-Dias, W.; dos Santos, T.O.; Belmonte, S.L.R.; de Oliveira, J.P.; Mauad, H.; Pacheco, M.D.S.; Lenz, D.; Bittencourt, A.S.; et al. Virtual Reconstruction and Three-Dimensional Printing of Blood Cells as a Tool in Cell Biology Education. PLoS ONE 2016, 11, e0161184. [Google Scholar] [CrossRef] [PubMed]
- Holt, K.; Savoian, M. Epi-fluorescence microscopy and 3D printing: An easily implemented approach for producing accurate physical models of micro-and macro-scopic biological samples. In Microscopy and Imaging Science: Practical Approaches to Applied Research and Education; Méndez-Vilas, A., Ed.; Formatex Research Centre: Badajoz, Spain, 2017; pp. 697–702. [Google Scholar]
- Mai, K.K.K.; Kang, M.J.; Kang, B.-H. 3D Printing of Plant Golgi Stacks from Their Electron Tomographic Models. In Plant Protein Secretion: Methods and Protocols; Jiang, L., Ed.; Springer New York: New York, NY, USA, 2017; pp. 105–113. [Google Scholar]
- Baghaie, A.; Tafti, A.P.; Owen, H.A.; D’souza, R.M.; Yu, Z. Three-dimensional reconstruction of highly complex microscopic samples using scanning electron microscopy and optical flow estimation. PLoS ONE 2017, 12, e0175078. [Google Scholar] [CrossRef] [PubMed]
- Merck. Human Dermal Fibroblasts (HDF) Culture Protocol. Available online: https://www.sigmaaldrich.com/AU/en/technical-documents/protocol/cell-culture-and-cell-culture-analysis/primary-cell-culture/human-dermal-fibroblasts (accessed on 15 December 2021).
- Beltrame, E.D.V.; Tyrwhitt-Drake, J.; Roy, I.; Shalaby, R.; Suckale, J.; Krummel, D.P. 3D Printing of Biomolecular Models for Research and Pedagogy. J. Vis. Exp. 2017, 121, e55427. [Google Scholar] [CrossRef]
- Rocheva, V.V.; Koroleva, A.V.; Savelyev, A.G.; Khaydukov, K.V.; Generalova, A.N.; Nechaev, A.V.; Guller, A.E.; Semchishen, V.A.; Chichkov, B.N.; Khaydukov, E.V. High-resolution 3D photopolymerization assisted by upconversion nanoparticles for rapid prototyping applications. Sci. Rep. 2018, 8, 3663. [Google Scholar] [CrossRef] [PubMed]
- Togni, P.; Cifra, M.; Drizdal, T. COMSOL Multiphysics in Undergraduate Education of Electromagnetic Field Biological Interactions. In Proceedings of the 14th Nordic-Baltic Conference on Biomedical Engineering and Medical Physics, Riga, Latvia, 16–20 June 2008; Volume 20, pp. 433–436. [Google Scholar] [CrossRef]
- Tang, G.; Galluzzi, M.; Zhang, B.; Shen, Y.-L.; Stadler, F.J. Biomechanical Heterogeneity of Living Cells: Comparison between Atomic Force Microscopy and Finite Element Simulation. Langmuir 2018, 35, 7578–7587. [Google Scholar] [CrossRef]
- Calì, C.; Kare, K.; Agus, M.; Castillo, M.F.V.; Boges, D.; Hadwiger, M.; Magistretti, P. A Method for 3D Reconstruction and Virtual Reality Analysis of Glial and Neuronal Cells. J. Vis. Exp. 2019, 151, e59444. [Google Scholar] [CrossRef]
- Alessandri, K.; Andrique, L.; Feyeux, M.; Bikfalvi, A.; Nassoy, P.; Recher, G. All-in-one 3D printed microscopy chamber for multidimensional imaging, the UniverSlide. Sci. Rep. 2017, 7, 42378. [Google Scholar] [CrossRef]
- Kaplan, H.; Pyayt, A. Tactile Visualization and 3D Printing for Education. In Encyclopedia of Computer Graphics and Games; Lee, N., Ed.; Springer: Cham, Switzerland, 2015; pp. 1–8. [Google Scholar] [CrossRef]
- Merck. Cell Types & Culture Characteristics. Available online: https://www.sigmaaldrich.com/AU/en/technical-documents/technical-article/cell-culture-and-cell-culture-analysis/mammalian-cell-culture/cell-types-culture (accessed on 21 December 2021).
- Garrido-Laguna, I.; Hidalgo, M. Pancreatic cancer: From state-of-the-art treatments to promising novel therapies. Nat. Rev. Clin. Oncol. 2015, 12, 319–334. [Google Scholar] [CrossRef]
- Raimondi, S.; Maisonneuve, P.; Lowenfels, A.B. Epidemiology of pancreatic cancer: An overview. Nat. Rev. Gastroenterol. Hepatol. 2009, 6, 699–708. [Google Scholar] [CrossRef]
- Awaji, M.; Futakuchi, M.; Heavican, T.; Iqbal, J.; Singh, R.K. Cancer-Associated Fibroblasts Enhance Survival and Progression of the Aggressive Pancreatic Tumor Via FGF-2 and CXCL8. Cancer Microenviron. Off. J. Int. Cancer Microenviron. Soc. 2019, 12, 37–46. [Google Scholar] [CrossRef]
- Mendez, M.G.; Kojima, S.; Goldman, R.D. Vimentin induces changes in cell shape, motility, and adhesion during the epithelial to mesenchymal transition. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2010, 24, 1838–1851. [Google Scholar] [CrossRef] [PubMed]
- McBeath, R.; Pirone, D.M.; Nelson, C.M.; Bhadriraju, K.; Chen, C.S. Cell Shape, Cytoskeletal Tension, and RhoA Regulate Stem Cell Lineage Commitment. Dev. Cell 2004, 6, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Lecuit, T.; Lenne, P.-F. Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat. Rev. Mol. Cell Biol. 2007, 8, 633–644. [Google Scholar] [CrossRef]
- Rozova, V.S.; Anwer, A.G.; Guller, A.E.; Es, H.A.; Khabir, Z.; Sokolova, A.I.; Gavrilov, M.U.; Goldys, E.M.; Warkiani, M.E.; Thiery, J.P.; et al. Machine learning reveals mesenchymal breast carcinoma cell adaptation in response to matrix stiffness. PLOS Comput. Biol. 2021, 17, e1009193. [Google Scholar] [CrossRef] [PubMed]
- Antony, J.; Thiery, J.P.; Huang, R.Y. Epithelial-to-mesenchymal transition: Lessons from development, insights into cancer and the potential of EMT-subtype based therapeutic intervention. Phys. Biol. 2019, 16, 041004. [Google Scholar] [CrossRef] [PubMed]
- Schulz, J.N.; Plomann, M.; Sengle, G.; Gullberg, D.; Krieg, T.; Eckes, B. New developments on skin fibrosis—Essential signals emanating from the extracellular matrix for the control of myofibroblasts. Matrix Biol. J. Int. Soc. Matrix Biol. 2018, 68–69, 522–532. [Google Scholar] [CrossRef]
- Pakshir, P.; Hinz, B. The big five in fibrosis: Macrophages, myofibroblasts, matrix, mechanics, and miscommunication. Matrix Biol. J. Int. Soc. Matrix Biol. 2018, 68–69, 81–93. [Google Scholar] [CrossRef]
- Bochaton-Piallat, M.-L.; Gabbiani, G.; Hinz, B. The myofibroblast in wound healing and fibrosis: Answered and unanswered questions. F1000Research 2016, 5, 752. [Google Scholar] [CrossRef]
- Willis, B.C.; duBois, R.M.; Borok, Z. Epithelial origin of myofibroblasts during fibrosis in the lung. Proc. Am. Thorac. Soc. 2006, 3, 377–382. [Google Scholar] [CrossRef]
- Radisky, D.C.; Kenny, P.A.; Bissell, M.J. Fibrosis and cancer: Do myofibroblasts come also from epithelial cells via EMT? J. Cell. Biochem. 2007, 101, 830–839. [Google Scholar] [CrossRef]
- Jones, C.; Ehrlich, H.P. Fibroblast expression of α-smooth muscle actin, α2β1 integrin and αvβ3 integrin: Influence of surface rigidity. Exp. Mol. Pathol. 2011, 91, 394–399. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, D.A.; Mullins, R.D. Cell mechanics and the cytoskeleton. Nature 2010, 463, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Shinde, A.V.; Humeres, C.; Frangogiannis, N.G. The role of α-smooth muscle actin in fibroblast-mediated matrix contraction and remodeling. Biochim. et Biophys. Acta (BBA) Mol. Basis Dis. 2016, 1863, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Fayzullin, A.; Churbanov, S.; Ignatieva, N.; Zakharkina, O.; Tokarev, M.; Mudryak, D.; Khristidis, Y.; Balyasin, M.; Kurkov, A.; Golubeva, E.N.; et al. Local Delivery of Pirfenidone by PLA Implants Modifies Foreign Body Reaction and Prevents Fibrosis. Biomedicines 2021, 9, 853. [Google Scholar] [CrossRef]

| Cell Type | Morphometric Characteristics, Mean ± St. Dev., µm | ||||
|---|---|---|---|---|---|
| Cell Body, D1 | Cell Body, D2 | Nuclei, D1 | Nuclei, D2 | Nuclei, Davg | |
| Epithelial (PANC-1) | 44.6 ± 6.2 | 35.9 ± 3.8 | 18.8 ± 2.3 | 13.7 ± 0.8 | 16.2 ± 3.2 |
| Mesenchymal (HDF) | 195.5 ± 70.5 | 43.4 ± 13.4 | 44.1 ± 25.3 | 17.2 ± 4.7 | N.A. |
| Neuron-like (SH-SY5Y) | 16.5 ± 0.9 | 9.7 ± 2.0 | 6.4 ± 0.8 | 5.9 ± 0.3 | 6.1 ± 0.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fitzpatrick, X.; Fayzullin, A.; Wang, G.; Parker, L.; Dokos, S.; Guller, A. Cells-in-Touch: 3D Printing in Reconstruction and Modelling of Microscopic Biological Geometries for Education and Future Research Applications. Bioengineering 2023, 10, 687. https://doi.org/10.3390/bioengineering10060687
Fitzpatrick X, Fayzullin A, Wang G, Parker L, Dokos S, Guller A. Cells-in-Touch: 3D Printing in Reconstruction and Modelling of Microscopic Biological Geometries for Education and Future Research Applications. Bioengineering. 2023; 10(6):687. https://doi.org/10.3390/bioengineering10060687
Chicago/Turabian StyleFitzpatrick, Xavier, Alexey Fayzullin, Gonglei Wang, Lindsay Parker, Socrates Dokos, and Anna Guller. 2023. "Cells-in-Touch: 3D Printing in Reconstruction and Modelling of Microscopic Biological Geometries for Education and Future Research Applications" Bioengineering 10, no. 6: 687. https://doi.org/10.3390/bioengineering10060687
APA StyleFitzpatrick, X., Fayzullin, A., Wang, G., Parker, L., Dokos, S., & Guller, A. (2023). Cells-in-Touch: 3D Printing in Reconstruction and Modelling of Microscopic Biological Geometries for Education and Future Research Applications. Bioengineering, 10(6), 687. https://doi.org/10.3390/bioengineering10060687








