A New Breakpoint to Classify 3D Voxels in MRI: A Space Transform Strategy with 3t2FTS-v2 and Its Application for ResNet50-Based Categorization of Brain Tumors
Abstract
:1. Introduction
- 3D-based HGG/LGG categorization arises as a remarkable issue to design the classification part of a fully automated CAD.
- Except for 3t2FTS, a novel feature transform strategy is not available in the literature to applicate 2D-based transfer learning models and to find the 2D-ID image of a 3D voxel.
- The design of a state-of-the-art feature transform strategy (3t2FTS-v2) to transform the 3D space information to the 2D space.
- The applicable transform strategy to be considered not only for 3D-defined tumors but also for the whole brain defined in 3D MRI (disease classification).
- A case study using FOS, GLRLM, and normalization analyses to discover 2D-ID images of 3D voxels.
- A comprehensive research obtaining promising results on 3D-based HGG/LGG categorization.
- An extensive study about ResNet50 and its hyperparameter adjustments on brain tumor classification in 3D MRI.
2. Materials and Methods
2.1. First-Order Statistics
2.2. Gray Level Run Length Matrix
2.3. Design of 3t2FTS-v2
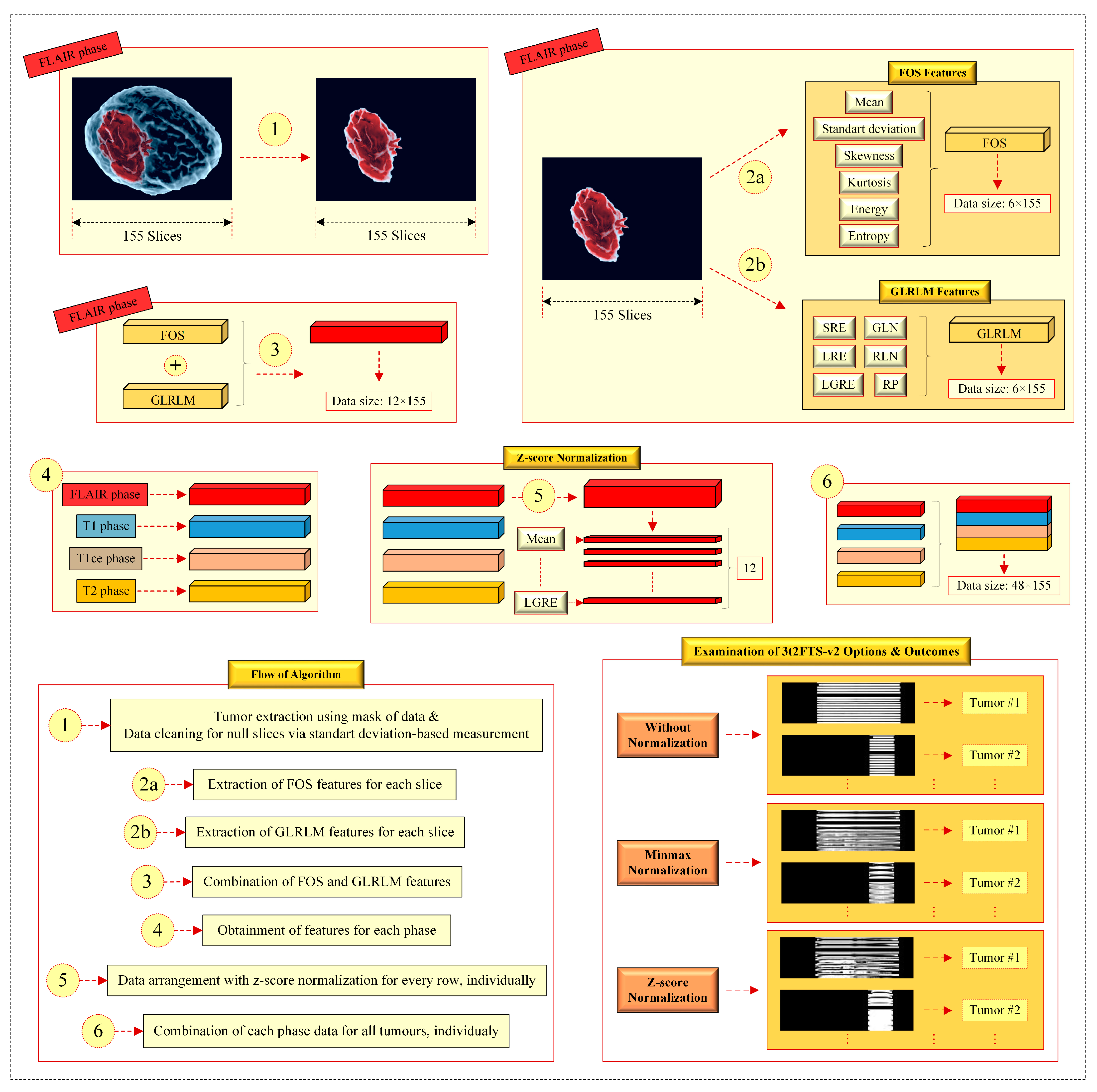
- In item (1), the tumor area is obtained by multiplying the tumor mask of BraTS 2017/2018 with the 3D MRI voxel as in 3t2FTS. However, 3t2FTS-v2 operates an additional part providing data cleaning in null slices that all pixels own some non-zero (close to zero) values. Herein, this situation can change the meaningful information in the 2D-ID image. Concerning this, the non-zero null slices are converted to the matrixes including zero values by considering the standard deviation along the image.
- In item (2a), six FOS features (mean, standard deviation, skewness, kurtosis, energy, and entropy) are generated for each slice. Regarding this, meaningful information is produced at the size of 6 × 155.
- In item (2b), six GLRLM features (SRE, LRE, GLN, RLN, RP, and LGRE) are evaluated for every slice. Concerning this, distinctive information is generated at the size of 6 × 155.
- In items (2a) and (2b), it should be reminded that location information is processed in addition to the intensity-based, size-based, and shape-based features.
- In item (3), the outcomes of items (2a) and (2b) are combined to form the information at the size of 12 × 155.
- In item (4), the previous items (1, 2a, 2b, and 3) are respectively applied for every MRI sequence. Consequently, four information matrixes belonging to all MRI phases are independently obtained at the size of 12 × 155 for one tumor, individually.
- In item (5), z-score normalization is fulfilled for every row in the data, independently. This process yields the normalization of every feature in itself and the feature transform is performed more robustly. Herein, item (5) is performed separately for all the 12 × 155 information in all phases.
- In item (6), the normalized information matrixes at the size of 12 × 155 are combined to discover the 2D-ID image of a 3D tumor.
2.4. ResNet50 Architecture
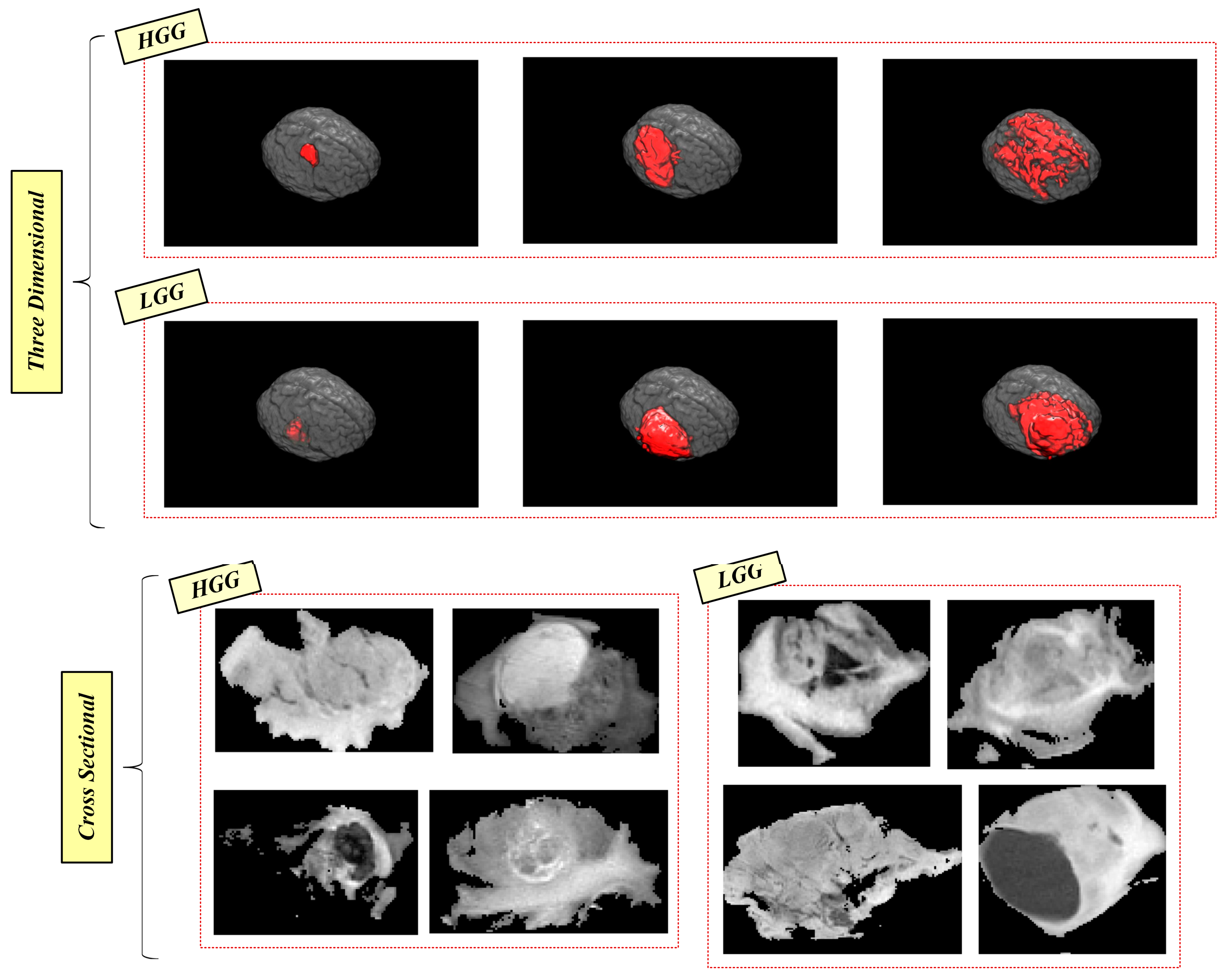
2.5. Dataset Information and Handicaps
- A tumor type (LGG or HGG) can have very different size and shape features if the examination is performed inside one type. On the contrary, if HGG and LGG-type tumors are examined together, the shape-based and size-based features can be similar.
- A tumor type can have very different intensity features inside the tumor, which can be similar to the intensity features of the opposite tumor type.
3. Experimental Analyses and Interpretations
4. Discussions
- Among average accuracy-based experiments, ResNet50 generally inclines to operate with the adam optimizer and LRDF value of ‘0.2’ (especially for normalization-available trials). Moreover, there is no discriminative adjustment for other hyperparameters in average performance-based analyses.
- Regarding the z-score normalization-based and average accuracy-based evaluations, the appropriate preferences of mini-batch size, learning rate, LRDF, and optimizer are, respectively, ‘16’, ‘0.001’, ‘0.2’, and adam.
- Concerning the highest scores observed, ResNet50 usually utilizes the mini-batch size of ‘32’, LRDF of ‘0.8’, and an adam or rmsprop optimizer.
- In relation to the z-score normalization-based and highest scores-based assessments, there is no discriminative information about the three adjustments. However, a mini-batch size of ‘32’, a learning rate of ‘0.001’, an LRDF of ‘0.8’, and adam optimizer are used twice for the obtainment of the highest scores.
- Z-score normalization reveals the most appropriate preference on the highest accuracy-based considerations by yielding 17.19% and 21.05% more accuracy than the minmax normalization and non-normalization choices, respectively.
- By means of average accuracy-based evaluations, the z-score normalization keeps its superiority by providing 22.50% and 22.82% more accuracy than the minmax normalization and non-normalization preferences, respectively.
5. Conclusions
- A comprehensive survey about 3t2FTS-v2 and its application for 3D brain-based disease categorization by using traditional machine learning algorithms or deep learning-based architectures
- An in-depth study utilizing 3t2FTS-v2 to classify various kinds of brain tumors on a large dataset by utilizing traditional machine learning algorithms or deep learning-based architectures
- The design of a novel deep learning architecture to operate with the 2D-ID identity images generated using 3t2FTS-v2
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- AboElenein, N.M.; Piao, S.; Noor, A.; Ahmed, P.N. MIRAU-Net: An improved neural network based on U-Net for gliomas segmentation. Signal Process.-Image 2022, 101, 116553. [Google Scholar] [CrossRef]
- Samee, N.A.; Ahmad, T.; Mahmoud, N.F.; Atteia, G.; Abdallah, H.A.; Rizwan, A. Clinical decision support framework for segmentation and classification of brain tumor MRIs using a U-Net and DCNN cascaded learning algorithm. Healthcare 2022, 10, 2340. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Li, L.; Sui, A.; Wu, G.; Wang, Y.; Yu, J. An efficient R-Transformer network with dual encoders for brain glioma segmentation in MR images. Biomed. Signal Proces. 2023, 79, 104034. [Google Scholar] [CrossRef]
- Gumaei, A.; Hassan, M.M.; Hassan, M.R.; Alelaiwi, A.; Fortino, G. A hybrid feature extraction method with regularized extreme learning machine for brain tumor classification. IEEE Access 2019, 7, 36266–36273. [Google Scholar] [CrossRef]
- Alshayeji, M.; Al-Buloushi, J.; Ashkanani, A.; Abed, S.E. Enhanced brain tumor classification using an optimized multi-layered convolutional neural network architecture. Multimed. Tools Appl. 2021, 80, 28897–28917. [Google Scholar] [CrossRef]
- Kumar, R.L.; Kakarla, J.; Isunuri, B.V.; Singh, M. Multi-class brain tumor classification using residual network and global average pooling. Multimed. Tools Appl. 2021, 80, 13429–13438. [Google Scholar] [CrossRef]
- Tummala, S.; Kadry, S.; Bukhari, S.A.C.; Rauf, H.T. Classification of brain tumor from magnetic resonance imaging using vision transformers ensembling. Curr. Oncol. 2022, 29, 7498–7511. [Google Scholar] [CrossRef]
- Ullah, N.; Khan, J.A.; Khan, M.S.; Khan, W.; Hassan, I.; Obayya, M.; Negm, N.; Salama, A.S. An effective approach to detect and identify brain tumors using transfer learning. Appl. Sci. 2022, 12, 5645. [Google Scholar] [CrossRef]
- Patil, D.O.; Hamde, S.T. A new enhanced local optimal-oriented pattern descriptor using steerable wavelet transform for MRI brain tumor classification. Iran. J. Sci. Technol. Trans. Electr. Eng. 2022, 47, 369–384. [Google Scholar] [CrossRef]
- Satyanarayana, G.; Naidu, P.A.; Desanamukula, V.S.; Rao, B.C. A mass correlation based deep learning approach using deep convolutional neural network to classify the brain tumor. Biomed. Signal Proces. 2023, 81, 104395. [Google Scholar] [CrossRef]
- Cristin, D.R.; Kumar, D.K.S.; Anbhazhagan, D.P. Severity level classification of brain tumor based on MRI images using fractional-chicken swarm optimization algorithm. Comput. J. 2021, 64, 1514–1530. [Google Scholar] [CrossRef]
- Mohan, P.; Veerappampalayam Easwaramoorthy, S.; Subramani, N.; Subramanian, M.; Meckanzi, S. Handcrafted deep-feature-based brain tumor detection and classification using MRI images. Electronics 2022, 11, 4178. [Google Scholar] [CrossRef]
- Mishra, L.; Verma, S. Graph attention autoencoder inspired CNN based brain tumor classification using MRI. Neurocomputing 2022, 503, 236–247. [Google Scholar] [CrossRef]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 2014, 34, 1993–2024. [Google Scholar] [CrossRef] [PubMed]
- Bakas, S.; Reyes, M.; Jakab, A.; Bauer, S.; Rempfler, M.; Crimi, A.; Shinohara, R.T.; Berger, C.; Ha, S.M.; Rozycki, M.; et al. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv 2018, arXiv:1811.02629. [Google Scholar]
- Koyuncu, H.; Barstuğan, M.; Öziç, M.Ü. A comprehensive study of brain tumour discrimination using phase combinations, feature rankings, and hybridised classifiers. Med. Biol. Eng. Comput. 2020, 58, 2971–2987. [Google Scholar] [CrossRef]
- Mzoughi, H.; Njeh, I.; Wali, A.; Slima, M.B.; BenHamida, A.; Mhiri, C.; Mahfoudhe, K.B. Deep multi-scale 3D convolutional neural network (CNN) for MRI gliomas brain tumor classification. J. Digit. Imaging 2020, 33, 903–915. [Google Scholar] [CrossRef]
- Tripathi, P.C.; Bag, S. A computer-aided grading of glioma tumor using deep residual networks fusion. Comput. Methods Programs Biomed. 2022, 215, 106597. [Google Scholar] [CrossRef]
- Montaha, S.; Azam, S.; Rafid, A.R.H.; Hasan, M.Z.; Karim, A.; Islam, A. TD-CNN-LSTM: A Hybrid approach combining CNN and LSTM to classify brain tumor on 3D MRI scans performing ablation study. IEEE Access 2022, 10, 60039–60059. [Google Scholar] [CrossRef]
- Jeong, S.W.; Cho, H.H.; Lee, S.; Park, H. Robust multimodal fusion network using adversarial learning for brain tumor grading. Comput. Methods Programs Biomed. 2022, 226, 107165. [Google Scholar] [CrossRef]
- Bhatele, K.R.; Bhadauria, S.S. Multiclass classification of central nervous system brain tumor types based on proposed hybrid texture feature extraction methods and ensemble learning. Multimed. Tools Appl. 2023, 82, 3831–3858. [Google Scholar] [CrossRef]
- Demir, F.; Akbulut, Y.; Taşcı, B.; Demir, K. Improving brain tumor classification performance with an effective approach based on new deep learning model named 3ACL from 3D MRI data. Biomed. Signal Process. 2023, 81, 104424. [Google Scholar] [CrossRef]
- Hajmohamad, A.; Koyuncu, H. 3t2FTS: A novel feature transform strategy to classify 3D MRI voxels and its application on HGG/LGG classification. Mach. Learn. Knowl. Extr. 2023, 5, 359–383. [Google Scholar] [CrossRef]
- Samee, N.A.; Mahmoud, N.F.; Atteia, G.; Abdallah, H.A.; Alabdulhafith, M.; Al-Gaashani, M.S.; Ahmad, S.; Muthanna, M.S.A. Classification framework for medical diagnosis of brain tumor with an effective hybrid transfer learning model. Diagnostics 2022, 12, 2541. [Google Scholar] [CrossRef]
- Koyuncu, H.; Barstuğan, M. COVID-19 discrimination framework for X-ray images by considering radiomics, selective information, feature ranking, and a novel hybrid classifier. Signal Process.-Image 2021, 97, 116359. [Google Scholar] [CrossRef]
- Sohail, A.S.M.; Bhattacharya, P.; Mudur, S.P.; Krishnamurthy, S. Local relative GLRLM-based texture feature extraction for classifying ultrasound medical images. In Proceedings of the 2011 24th Canadian Conference on Electrical and Computer Engineering (CCECE), Niagara Falls, ON, Canada, 8–11 May 2011; pp. 001092–001095. [Google Scholar]
- Sakallı, G.; Koyuncu, H. Identification of asynchronous motor and transformer situations in thermal images by utilizing transfer learning-based deep learning architectures. Measurement 2023, 207, 112380. [Google Scholar] [CrossRef]
- Sakallı, G.; Koyuncu, H. Categorization of asynchronous motor situations in infrared images: Analyses with ResNet50. In Proceedings of the 2022 International Conference on Data Analytics for Business and Industry (ICDABI), Sakheer, Bahrain, 25–26 October 2022; pp. 114–118. [Google Scholar]
- Militello, C.; Rundo, L.; Vitabile, S.; Russo, G.; Pisciotta, P.; Marletta, F.; Ippolito, M.; D’arrigo, C.; Midiri, M.; Gilardi, M.C. Gamma Knife treatment planning: MR brain tumor segmentation and volume measurement based on unsupervised Fuzzy C-Means clustering. Int. J. Imaging Syst. Technol. 2015, 25, 213–225. [Google Scholar] [CrossRef]
- Rundo, L.; Militello, C.; Tangherloni, A.; Russo, G.; Vitabile, S.; Gilardi, M.; Mauri, G. NeXt for neuro-radiosurgery: A fully automatic approach for necrosis extraction in brain tumor MRI using an unsupervised machine learning technique. Int. J. Imaging Syst. Technol. 2018, 28, 21–37. [Google Scholar] [CrossRef]

| Hyperparameter | Value/Preference |
|---|---|
| Epoch | 100 |
| Mini-batch size | 16, 32 |
| Learning rate | 0.001, 0.0001 |
| Learning Rate Drop Factor (LRDF) | 0.2, 0.4, 0.6, 0.8 |
| Optimizer | Adam, Rmsprop, Sgdm |
| Mini-Batch Size | Learning Rate | LRDF | Optimizer | Accuracy | Learning Rate | LRDF | Optimizer | Accuracy |
|---|---|---|---|---|---|---|---|---|
| 16 | 0.001 | 0.2 | Adam | 77.19 | 0.0001 | 0.2 | Adam | 74.73 |
| Sgdm | 72.63 | Sgdm | 74.03 | |||||
| Rmsprop | 73.70 | Rmsprop | 73.33 | |||||
| 0.4 | Adam | 75.08 | 0.4 | Adam | 70.90 | |||
| Sgdm | 77.19 | Sgdm | 71.92 | |||||
| Rmsprop | 74.73 | Rmsprop | 71.22 | |||||
| 0.6 | Adam | 73.68 | 0.6 | Adam | 76.14 | |||
| Sgdm | 75.43 | Sgdm | 72.98 | |||||
| Rmsprop | 74.03 | Rmsprop | 72.98 | |||||
| 0.8 | Adam | 77.54 | 0.8 | Adam | 76.14 | |||
| Sgdm | 71.92 | Sgdm | 75.78 | |||||
| Rmsprop | 76.14 | Rmsprop | 72.98 | |||||
| 32 | 0.001 | 0.2 | Adam | 76.49 | 0.0001 | 0.2 | Adam | 74.73 |
| Sgdm | 75.08 | Sgdm | 73.68 | |||||
| Rmsprop | 75.78 | Rmsprop | 77.19 | |||||
| 0.4 | Adam | 77.89 | 0.4 | Adam | 76.14 | |||
| Sgdm | 73.68 | Sgdm | 74.03 | |||||
| Rmsprop | 75.78 | Rmsprop | 74.73 | |||||
| 0.6 | Adam | 70.87 | 0.6 | Adam | 76.14 | |||
| Sgdm | 75.78 | Sgdm | 74.73 | |||||
| Rmsprop | 76.49 | Rmsprop | 76.84 | |||||
| 0.8 | Adam | 72.63 | 0.8 | Adam | 74.38 | |||
| Sgdm | 74.03 | Sgdm | 74.38 | |||||
| Rmsprop | 76.49 | Rmsprop | 78.59 |
| Mini-Batch Size | Learning Rate | LRDF | Optimizer | Accuracy | Learning Rate | LRDF | Optimizer | Accuracy |
|---|---|---|---|---|---|---|---|---|
| 16 | 0.001 | 0.2 | Adam | 75.78 | 0.0001 | 0.2 | Adam | 77.54 |
| Sgdm | 75.08 | Sgdm | 74.03 | |||||
| Rmsprop | 69.47 | Rmsprop | 75.43 | |||||
| 0.4 | Adam | 79.64 | 0.4 | Adam | 78.24 | |||
| Sgdm | 76.49 | Sgdm | 72.98 | |||||
| Rmsprop | 64.56 | Rmsprop | 75.78 | |||||
| 0.6 | Adam | 76.14 | 0.6 | Adam | 76.14 | |||
| Sgdm | 77.89 | Sgdm | 73.33 | |||||
| Rmsprop | 73.68 | Rmsprop | 76.49 | |||||
| 0.8 | Adam | 73.68 | 0.8 | Adam | 75.78 | |||
| Sgdm | 77.54 | Sgdm | 75.43 | |||||
| Rmsprop | 73.68 | Rmsprop | 76.14 | |||||
| 32 | 0.001 | 0.2 | Adam | 77.89 | 0.0001 | 0.2 | Adam | 79.29 |
| Sgdm | 73.33 | Sgdm | 73.33 | |||||
| Rmsprop | 74.73 | Rmsprop | 79.29 | |||||
| 0.4 | Adam | 76.14 | 0.4 | Adam | 78.59 | |||
| Sgdm | 78.59 | Sgdm | 72.98 | |||||
| Rmsprop | 71.92 | Rmsprop | 75.08 | |||||
| 0.6 | Adam | 74.57 | 0.6 | Adam | 75.08 | |||
| Sgdm | 78.24 | Sgdm | 72.63 | |||||
| Rmsprop | 62.10 | Rmsprop | 82.45 | |||||
| 0.8 | Adam | 70.52 | 0.8 | Adam | 80.35 | |||
| Sgdm | 75.43 | Sgdm | 72.63 | |||||
| Rmsprop | 66.31 | Rmsprop | 81.75 |
| Mini-Batch Size | Learning Rate | LRDF | Optimizer | Accuracy | Learning Rate | LRDF | Optimizer | Accuracy |
|---|---|---|---|---|---|---|---|---|
| 16 | 0.001 | 0.2 | Adam | 98.95 | 0.0001 | 0.2 | Adam | 98.24 |
| Sgdm | 98.95 | Sgdm | 97.19 | |||||
| Rmsprop | 94.40 | Rmsprop | 98.59 | |||||
| 0.4 | Adam | 96.80 | 0.4 | Adam | 98.94 | |||
| Sgdm | 98.59 | Sgdm | 96.14 | |||||
| Rmsprop | 95.40 | Rmsprop | 98.94 | |||||
| 0.6 | Adam | 98.59 | 0.6 | Adam | 99.29 | |||
| Sgdm | 98.59 | Sgdm | 96.49 | |||||
| Rmsprop | 91.57 | Rmsprop | 99.29 | |||||
| 0.8 | Adam | 99.29 | 0.8 | Adam | 97.54 | |||
| Sgdm | 98.95 | Sgdm | 95.08 | |||||
| Rmsprop | 99.64 | Rmsprop | 99.29 | |||||
| 32 | 0.001 | 0.2 | Adam | 98.59 | 0.0001 | 0.2 | Adam | 99.64 |
| Sgdm | 98.59 | Sgdm | 92.98 | |||||
| Rmsprop | 99.29 | Rmsprop | 98.94 | |||||
| 0.4 | Adam | 99.29 | 0.4 | Adam | 99.29 | |||
| Sgdm | 98.24 | Sgdm | 91.92 | |||||
| Rmsprop | 98.59 | Rmsprop | 98.59 | |||||
| 0.6 | Adam | 97.54 | 0.6 | Adam | 98.94 | |||
| Sgdm | 98.59 | Sgdm | 94.03 | |||||
| Rmsprop | 94.73 | Rmsprop | 98.94 | |||||
| 0.8 | Adam | 99.64 | 0.8 | Adam | 98.94 | |||
| Sgdm | 98.59 | Sgdm | 95.08 | |||||
| Rmsprop | 91.57 | Rmsprop | 98.94 |
| Study | Year | Classification System | Dataset | Task | Accuracy (%) |
|---|---|---|---|---|---|
| Koyuncu et al. [16] | 2020 | The framework including three phase information (T1, T2, FLAIR), FOS, Wilcoxon ranking, and GM-CPSO-NN | 210 HGG/75 LGG (BraTS 2017/2018) | 3D-based classification (HGG vs. LGG) | 90.18 |
| Mzoughi et al. [17] | 2020 | A model operating 3D Deep CNN, data augmentation, and T1ce phase information | 210 HGG/75 LGG (BraTS 2017/2018) | 3D-based classification (HGG vs. LGG) | 96.49 |
| Tripathi and Bag [18] | 2022 | A model utilizing ResNets fusion with a novel DST and T2 phase information | 2304 HGG/5088 LGG (TCIA) | 2D-based classification (HGG vs. LGG) | 95.87 |
| Montaha et al. [19] | 2022 | A model using TD-CNN-LSTM and all phase information | 234 HGG/74 LGG (BraTS 2020) | 3D-based classification (HGG vs. LGG) | 98.90 |
| Jeong et al. [20] | 2022 | A model determining multimodal fusion network with adversarial learning and all phase information | 210 HGG/75 LGG (BraTS 2017/2018) | 2.5D-based classification (HGG vs. LGG) | 90.91 |
| Bhatele and Bhadauria [21] | 2023 | A model comprising DWT, GGLCM, LBP, GLRLM, morphological features, PCA, ensemble classifier, and all phase information | Not clearly defined (BraTS 2013) | 2D-based classification (HGG vs. LGG) | 100 |
| 220 HGG/54 LGG (BraTS 2015) | 99.52 | ||||
| Demir et al. [22] | 2023 | A model considering 3ACL and all phase information | 220 HGG/54 LGG (BraTS 2015) | 3D-based classification (HGG vs. LGG) | 98.90 |
| 210 HGG/75 LGG (BraTS 2017/2018) | 99.29 | ||||
| Hajmohamad and Koyuncu [23] | 2023 | A model evaluating 3t2FTS and ResNet50 | 210 HGG/75 LGG (BraTS 2017/2018) | 3D-based classification (HGG vs. LGG) | 80 |
| This study | 2023 | A model examining 3t2FTS-v2 and ResNet50 | 210 HGG/75 LGG (BraTS 2017/2018) | 3D-based classification (HGG vs. LGG) | 99.64 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koyuncu, H.; Barstuğan, M. A New Breakpoint to Classify 3D Voxels in MRI: A Space Transform Strategy with 3t2FTS-v2 and Its Application for ResNet50-Based Categorization of Brain Tumors. Bioengineering 2023, 10, 629. https://doi.org/10.3390/bioengineering10060629
Koyuncu H, Barstuğan M. A New Breakpoint to Classify 3D Voxels in MRI: A Space Transform Strategy with 3t2FTS-v2 and Its Application for ResNet50-Based Categorization of Brain Tumors. Bioengineering. 2023; 10(6):629. https://doi.org/10.3390/bioengineering10060629
Chicago/Turabian StyleKoyuncu, Hasan, and Mücahid Barstuğan. 2023. "A New Breakpoint to Classify 3D Voxels in MRI: A Space Transform Strategy with 3t2FTS-v2 and Its Application for ResNet50-Based Categorization of Brain Tumors" Bioengineering 10, no. 6: 629. https://doi.org/10.3390/bioengineering10060629
APA StyleKoyuncu, H., & Barstuğan, M. (2023). A New Breakpoint to Classify 3D Voxels in MRI: A Space Transform Strategy with 3t2FTS-v2 and Its Application for ResNet50-Based Categorization of Brain Tumors. Bioengineering, 10(6), 629. https://doi.org/10.3390/bioengineering10060629