PPG2ECGps: An End-to-End Subject-Specific Deep Neural Network Model for Electrocardiogram Reconstruction from Photoplethysmography Signals without Pulse Arrival Time Adjustments
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset
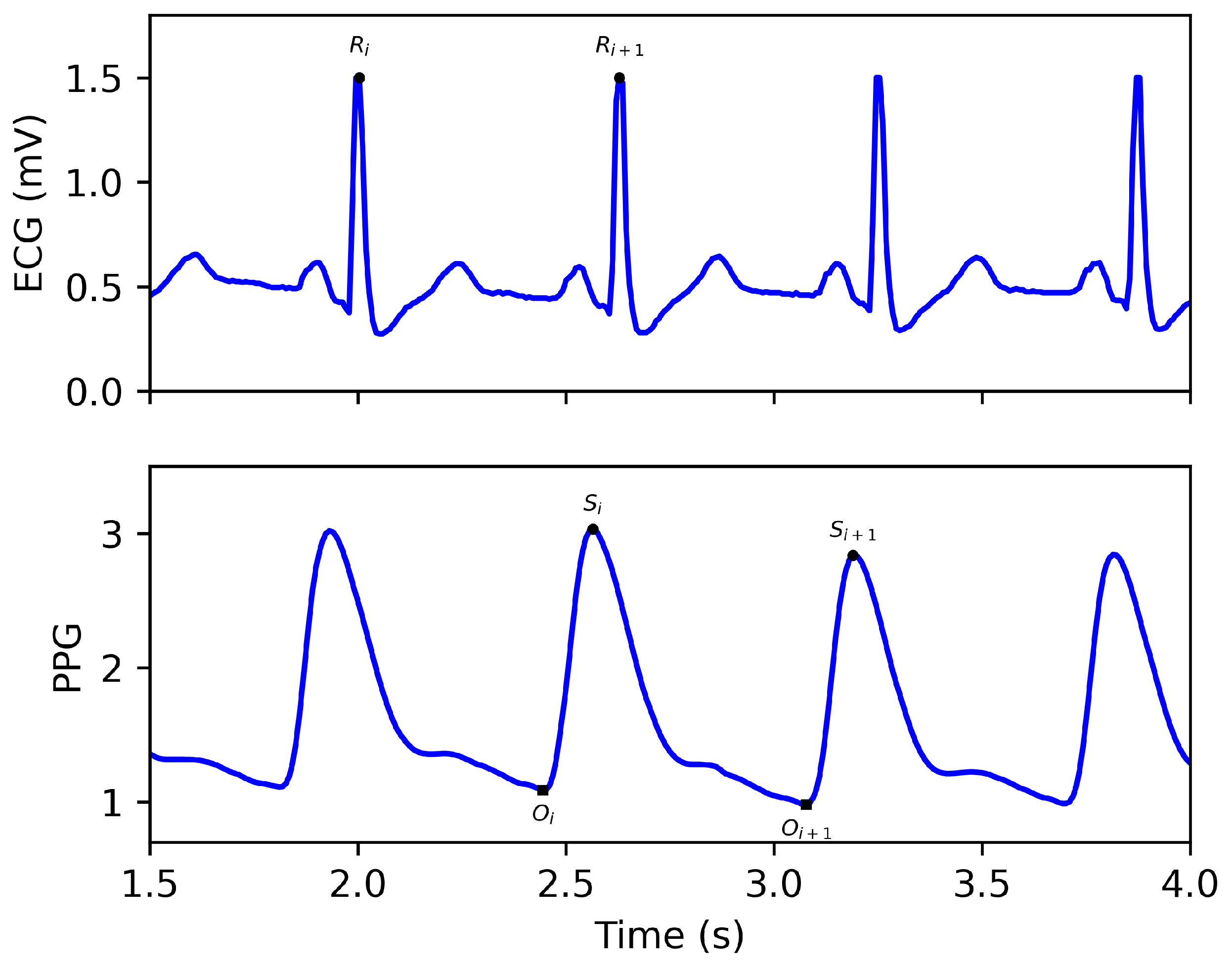
2.2. Preprocessing
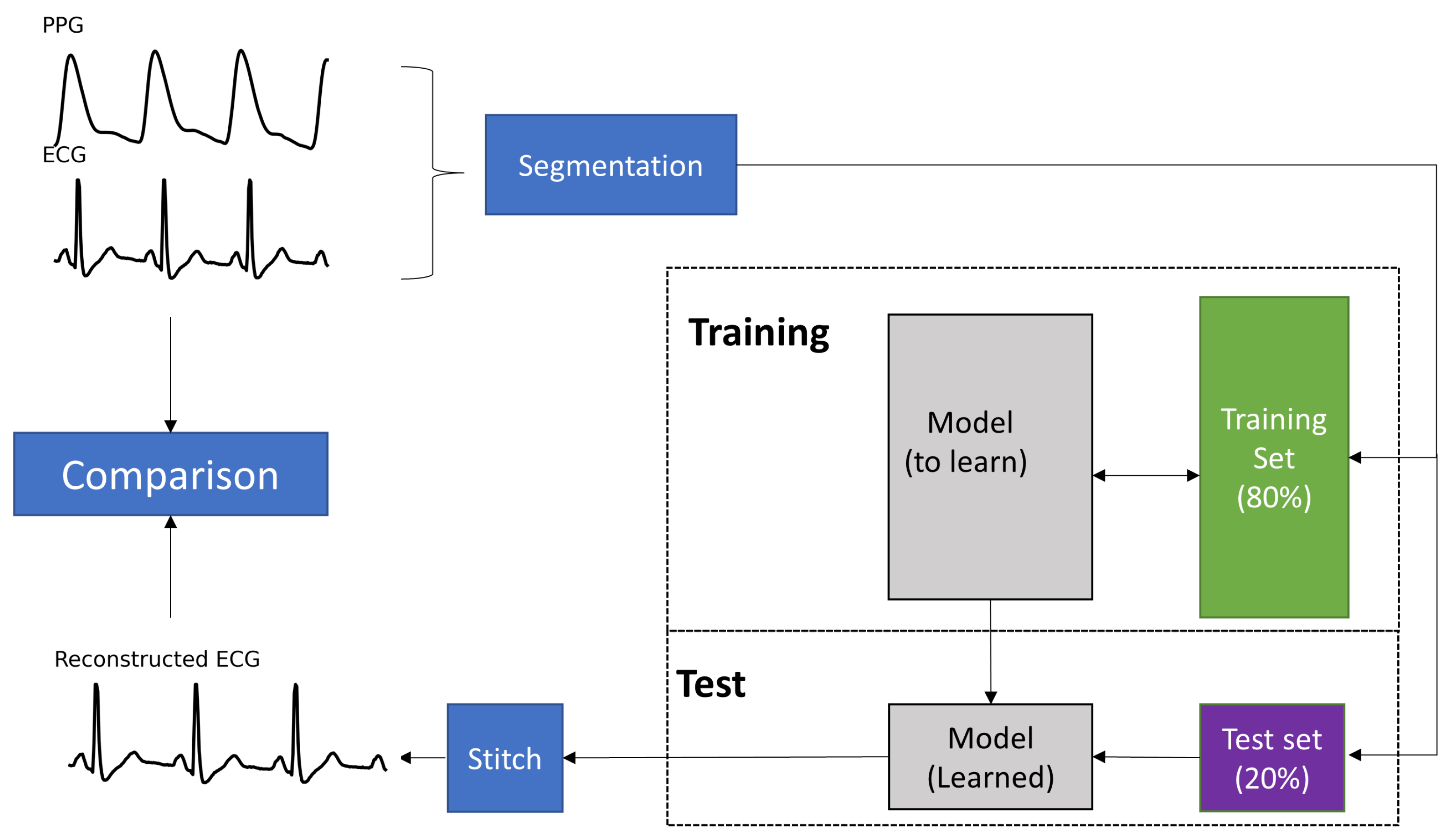
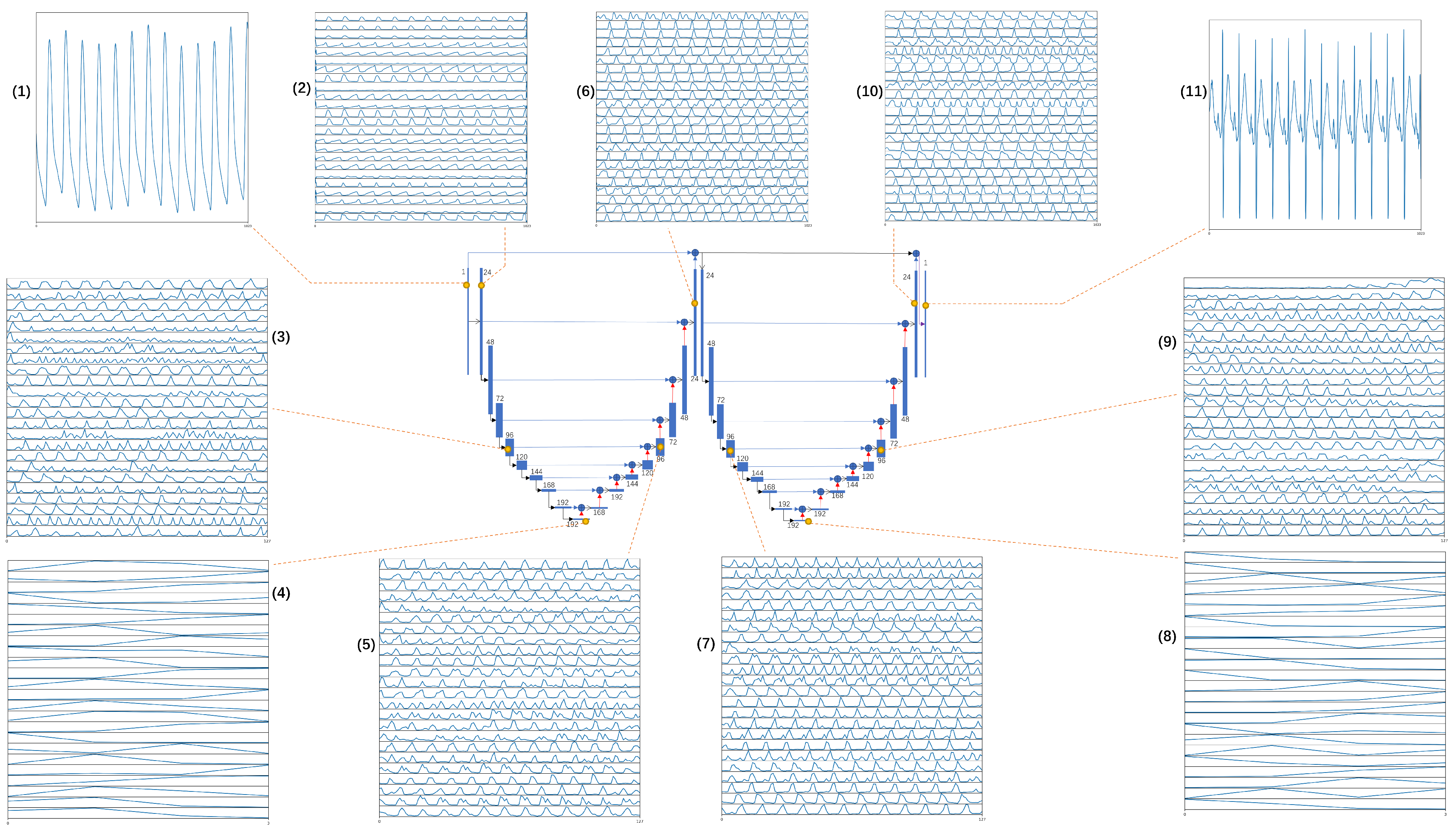
2.3. Model Architecture
2.4. Stitching the Reconstructed ECG Segments
2.5. Training Options
2.6. Performance Evaluation
- Calculate the Euclidean distance between every sample from the reconstructed ECG and every sample from the reference ECG. For the ith sample of reconstructed ECG and the jth sample of the reference ECG, the Euclidean distance is defined as follows:where i and j are the indices of the samples of the reference ECG and reconstructed ECG, respectively. Suppose that the number of samples of the reference ECG signal is N. This step will create an matrix A.
- Look for paths in matrix A that start at and end at . For any point on the path, the next point can only be one of its right, upper, or upper-right corners. Calculate the sum of the distance along the paths. The minimum sum along the paths is the DTW distance, and this path is defined as the warping path.
3. Results
4. Discussion
- 1.
- Variations in PAT signals: The model proposed in this study is subject-specific, meaning that it captures the PAT of a specific individual during the training phase. Consequently, applying the model directly to multiple subjects presents a significant challenge due to variations in PATs between individuals, making the problem different and requiring the development of an inter-subject model.
- 2.
- Variability in PPG signals: PPG signals are susceptible to variability due to factors such as skin pigmentation, motion artifacts, and changes in blood volume. This variability can affect the accuracy of the reconstructed ECG signal.
- 3.
- Limited availability of training data: The availability of subject-specific training data for PPG-based ECG reconstruction is limited. This can make it difficult to train an accurate model that can generalize well to new subjects.
- 1.
- Data augmentation: Using data augmentation techniques can help mitigate the variability in PPG signals. Techniques such as adding noise, jittering, and randomizing the signal’s amplitude and frequency can increase the model’s robustness to signal variability.
- 2.
- Transfer learning: Transfer learning can help overcome the limited availability of training data by leveraging pre-trained models on similar tasks. For example, a pre-trained model on PPG-based heart rate estimation can be fine-tuned on the ECG reconstruction task.
- 3.
- Model optimization: Optimizing the model architecture and hyperparameters can reduce the computational requirements of the end-to-end model. Techniques such as pruning, quantization, and compression can reduce the model’s size and improve its efficiency.
- 4.
- Validation on large and diverse datasets: To ensure the model can generalize well to new subjects, it is crucial to validate its performance on a wide range of diverse datasets. This validation process can help uncover any biases in the model and ultimately improve its overall performance.
- 5.
- Deployment considerations: Considerations such as hardware requirements, power consumption, and real-time performance should be taken into account when deploying the model in real-world applications. For example, deploying the model on a mobile device with limited resources may require additional optimization techniques.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PPG | photoplethysmography |
| ECG | electrocardiogram |
| PAT | pulse arrival time |
References
- Shelley, K.H. Photoplethysmography: Beyond the calculation of arterial oxygen saturation and heart rate. Anesth. Analg. 2007, 105, S31–S36. [Google Scholar] [CrossRef]
- Elgendi, M.; Fletcher, R.; Liang, Y.; Howard, N.; Lovell, N.H.; Abbott, D.; Lim, K.; Ward, R. The use of photoplethysmography for assessing hypertension. NPJ Digit. Med. 2019, 2, 1. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Pickwell-Macpherson, E.; Liang, Y.P.; Zhang, Y.T. Noninvasive cardiac output estimation using a novel photoplethysmogram index. In Proceedings of the 2009 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Minneapolis, MN, USA, 3–6 September 2009. [Google Scholar]
- Yang, C.; Veiga, C.; Rodriguez-Andina, J.J.; Farina, J.; Iniguez, A.; Yin, S. Using PPG Signals and Wearable Devices for Atrial Fibrillation Screening. IEEE Trans. Ind. Electron. 2019, 66, 8832–8842. [Google Scholar] [CrossRef]
- Neha Sardana, H.K.; Kanwade, R.; Tewary, S. Arrhythmia detection and classification using ECG and PPG techniques: A review. Phys. Eng. Sci. Med. 2021, 44, 1027–1048. [Google Scholar] [CrossRef]
- Bers, D.M. Cardiac excitation–contraction coupling. Nature 2002, 415, 198–205. [Google Scholar] [CrossRef]
- Weinschenk, S.W.; Beise, R.D.; Lorenz, J. Heart rate variability (HRV) in deep breathing tests and 5-min short-term recordings: Agreement of ear photoplethysmography with ECG measurements, in 343 subjects. Eur. J. Appl. Physiol. 2016, 116, 1527–1535. [Google Scholar] [CrossRef] [PubMed]
- Paradkar, N.; Chowdhury, S.R. Cardiac arrhythmia detection using photoplethysmography. In Proceedings of the 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Jeju, Republic of Korea, 11–15 July 2017. [Google Scholar]
- Polanía, L.F.; Mestha, L.K.; Huang, D.T.; Couderc, J. Method for classifying cardiac arrhythmias using photoplethysmography. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015. [Google Scholar]
- Banerjee, R.; Sinha, A.; Choudhury, A.D.; Visvanathan, A. PhotoECG: Photoplethysmography to estimate ECG parameter. In Proceedings of the 2014 IEEE International Conference on Acoustics, Speech and Signal Processing International Conference on Acoustics Speech and Signal Processing (ICASSP), Florence, Italy, 4–9 May 2014. [Google Scholar]
- Rai, H.M.; Trivedi, A.; Shukla, S. ECG signal processing for abnormalities detection using multi-resolution wavelet transform and Artificial Neural Network classifier. Measurement 2013, 46, 3238–3246. [Google Scholar] [CrossRef]
- Joshi, A.K.; Tomar, A.; Tomar, M. A review paper on analysis of electrocardiograph (ECG) signal for the detection of arrhythmia abnormalities. Int. J. Adv. Res. Electr. Electron. Instrum. Eng. 2014, 3, 12466–12475. [Google Scholar]
- Elgendi, M. PPG Signal Analysis: An Introduction Using MATLAB; CRC Press: Boca Raton, FL, USA, 2020. [Google Scholar]
- Elgendi, M. On the analysis of fingertip photoplethysmogram signals. Curr. Cardiol. Rev. 2012, 8, 14–25. [Google Scholar] [CrossRef]
- Elgendi, M.; Liang, Y.; Ward, R. Toward generating more diagnostic features from photoplethysmogram waveforms. Diseases 2018, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Hosanee, M.; Chan, G.; Welykholowa, K.; Cooper, R.; Kyriacou, P.A.; Zheng, D.; Allen, J.; Abbott, D.; Menon, C.; Lovell, N.H.; et al. Cuffless Single-Site Photoplethysmography for blood pressure monitoring. J. Clin. Med. 2020, 9, 723. [Google Scholar] [CrossRef]
- Liang, Y.; Abbott, D.; Howard, N.; Lim, K.; Ward, R.; Elgendi, M. How effective is pulse arrival time for evaluating blood pressure? Challenges and recommendations from a study using the MIMIC database. J. Clin. Med. 2019, 8, 337. [Google Scholar] [CrossRef]
- Zhu, Q.; Tian, X.; Wong, C.W.; Wu, M. Learning your heart actions from pulse: ECG waveform reconstruction from PPG. IEEE Internet Things J. 2021, 8, 16734–16748. [Google Scholar] [CrossRef]
- Tian, X.; Zhu, Q.; Li, Y.; Wu, M. Cross-domain joint dictionary learning for ECG reconstruction from PPG. In Proceedings of the 2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020. [Google Scholar]
- Tang, Q.; Chen, Z.; Guo, Y.; Liang, Y.; Ward, R.; Menon, C.; Elgendi, M. Robust reconstruction of electrocardiogram using photoplethysmography: A subject-based Model. Front. Physiol. 2022, 13, 859763. [Google Scholar] [CrossRef]
- Luo, X.; Miao, F.; Li, Y. PPG and ECG feature comparison for healthy people and hypertensive patients. In Proceedings of the 2012 IEEE-EMBS International Conference on Biomedical and Health Informatics, Hong Kong, China, 5–7 January 2012. [Google Scholar]
- Sarkar, P.; Etemad, A. CardioGAN: Attentive Generative Adversarial Network with Dual Discriminators for Synthesis of ECG from PPG. In Proceedings of the AAAI Conference on Artificial Intelligence, Delhi, India, 2–9 February 2021; Volume 35, pp. 488–496. [Google Scholar]
- Vo, K.; Naeini, E.K.; Naderi, A.; Jilani, D.; Rahmani, A.M.; Dutt, N.; Cao, H. P2E-WGAN: ECG waveform synthesis from PPG with conditional wasserstein generative adversarial networks. In Proceedings of the 36th Annual ACM Symposium on Applied Computing, Virtual Event, 22–26 March 2021; pp. 1030–1036. [Google Scholar]
- Chiu, H.Y.; Shuai, H.H.; Chao, P.C.P. Reconstructing QRS complex from PPG by transformed attentional neural networks. IEEE Sens. J. 2020, 20, 12374–12383. [Google Scholar] [CrossRef]
- Kachuee, M.; Kiani, M.M.; Mohammadzade, H.; Shabany, M. Cuff-less high-accuracy calibration-free blood pressure estimation using pulse transit time. In Proceedings of the 2015 IEEE International Symposium on Circuits and Systems (ISCAS), Lisbon, Portugal, 24–27 May 2015; pp. 1006–1009. [Google Scholar]
- Saeed, M.; Villarroel, M.; Reisner, A.T.; Clifford, G.; Lehman, L.; Moody, G.; Heldt, T.; Kyaw, T.H.; Moody, B.; Mark, R.G. Multiparameter Intelligent Monitoring in Intensive Care II: A public-access intensive care unit database. Crit. Care Med. 2011, 39, 952–960. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Chen, Z.; Ward, R.; Menon, C.; Elgendi, M. Subject-based model for reconstructing arterial blood pressure from Photoplethysmogram. Bioengineering 2022, 9, 402. [Google Scholar] [CrossRef]
- Stoller, D.; Ewert, S.; Dixon, S. Wave-u-net: A multi-scale neural network for end-to-end audio source separation. arXiv 2018, arXiv:1806.03185. [Google Scholar]
- Cheng, J.; Xu, Y.; Song, R.; Liu, Y.; Li, C.; Chen, X. Prediction of arterial blood pressure waveforms from photoplethysmogram signals via fully convolutional neural networks. Comput. Biol. Med. 2021, 138, 104877. [Google Scholar] [CrossRef] [PubMed]
- Gare, G.R.; Li, J.; Joshi, R.; Magar, R.; Vaze, M.P.; Yousefpour, M.; Rodriguez, R.L.; Galeotti, J.M. W-Net: Dense and diagnostic semantic segmentation of subcutaneous and breast tissue in ultrasound images by incorporating ultrasound RF waveform data. Med. Image Anal. 2022, 76, 102326. [Google Scholar] [CrossRef]
- Gargiulo, M.; Dell’Aglio, D.A.G.; Iodice, A.; Riccio, D.; Ruello, G. Integration of sentinel-1 and sentinel-2 data for land cover mapping using W-Net. Sensors 2020, 20, 2969. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Liu, J.; Tang, W.; Chen, G.; Lu, Y.; Feng, C. Correlation and agreement: Overview and clarification of competing concepts and measures. Shanghai Arch. Psychiatry 2016, 28, 115–120. [Google Scholar]
- Efrat, A.; Fan, Q.; Venkatasubramanian, S. Curve matching, time warping, and light fields: New algorithms for computing similarity between Curves. J. Math. Imaging Vis. 2007, 27, 203–216. [Google Scholar] [CrossRef]
- Karlen, W.; Raman, S.; Ansermino, J.M.; Dumont, G.A. Multiparameter respiratory rate estimation from the photoplethysmogram. IEEE Trans. Biomed. Eng. 2013, 60, 1946–1953. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.E.W.; Pollard, T.J.; Shen, L.; Lehman, L.H.; Feng, M.; Ghassemi, M.; Moody, B.; Szolovits, P.; Celi, L.A.; Mark, R.G. MIMIC-III, a freely accessible critical care database. Sci. Data 2016, 3, 160035. [Google Scholar] [CrossRef]
- Zhang, J.; Yi, S.; Guo, L.; Gao, H.; Hong, X.; Song, H. A new bearing fault diagnosis method based on modified convolutional neural networks. Chin. J. Aeronaut. 2020, 33, 439–447. [Google Scholar] [CrossRef]
- Rijnbeek, P.R.; Witteman, J.C.M.; Kors, J.; van Herpen, G.; Bots, M.L.; Man, S.; Verweij, N.; Hofman, A.; Hillege, H.; Numans, M.E.; et al. Normal values of the electrocardiogram for ages 16–90 years. J. Electrocardiol. 2014, 47, 914–921. [Google Scholar] [CrossRef]
- Ding, B.; Qian, H.; Zhou, J. Activation functions and their characteristics in deep neural networks. In Proceedings of the 2018 Chinese Control and Decision Conference (CCDC), Shenyang, China, 9–11 June 2018. [Google Scholar]
- Szandała, T. Review and comparison of commonly used activation functions for deep neural networks. Bio-Inspired Neurocomput. 2021, 903, 203–224. [Google Scholar]
- Vargas, V.M.; Gutiérrez, P.A.; Barbero-Gómez, J.; Hervás-Martínez, C. Activation Functions for Convolutional Neural Networks: Proposals and Experimental Study. IEEE Trans. Neural Netw. Learn. Syst. 2023, 34, 1478–1488. [Google Scholar] [CrossRef] [PubMed]



| Align with | RMSE (mV) | r | (mV) | |
|---|---|---|---|---|
| Experiments I | No | 0.037 ± 0.028 | 0.977 ± 0.029 | 0.010 ± 0.004 |
| Experiments II | Yes | 0.037 ± 0.027 | 0.978 ± 0.026 | 0.010 ± 0.004 |
| Method | Data Used | Alignment Required in Preprocessing | Segment Length | RMSE (mV) | r | (mV) |
|---|---|---|---|---|---|---|
| DCT Model [18] | TBME-RR [36]: 42 Records MIMIC III [37]: 103 Records Self-collected: 2 Records | Yes | Beat | NR | 0.984 0.940 0.904 | NR |
| XDJDL model [19] | MIMIC III [37]: 33 Records | Yes | Beat | NR | 0.88 | NR |
| Bi-LSTM model [20] | MIMIC III [37]: 100 Records | Yes | 1 s 2 s 3 s 4 s | 0.063 0.068 0.063 0.059 | 0.893 0.874 0.891 0.904 | NR |
| This study (PPG2ECGps) | Cuffless [25]: 500 Records | No | 8.192 s | 0.037 | 0.977 | 0.010 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, Q.; Chen, Z.; Ward, R.; Menon, C.; Elgendi, M. PPG2ECGps: An End-to-End Subject-Specific Deep Neural Network Model for Electrocardiogram Reconstruction from Photoplethysmography Signals without Pulse Arrival Time Adjustments. Bioengineering 2023, 10, 630. https://doi.org/10.3390/bioengineering10060630
Tang Q, Chen Z, Ward R, Menon C, Elgendi M. PPG2ECGps: An End-to-End Subject-Specific Deep Neural Network Model for Electrocardiogram Reconstruction from Photoplethysmography Signals without Pulse Arrival Time Adjustments. Bioengineering. 2023; 10(6):630. https://doi.org/10.3390/bioengineering10060630
Chicago/Turabian StyleTang, Qunfeng, Zhencheng Chen, Rabab Ward, Carlo Menon, and Mohamed Elgendi. 2023. "PPG2ECGps: An End-to-End Subject-Specific Deep Neural Network Model for Electrocardiogram Reconstruction from Photoplethysmography Signals without Pulse Arrival Time Adjustments" Bioengineering 10, no. 6: 630. https://doi.org/10.3390/bioengineering10060630
APA StyleTang, Q., Chen, Z., Ward, R., Menon, C., & Elgendi, M. (2023). PPG2ECGps: An End-to-End Subject-Specific Deep Neural Network Model for Electrocardiogram Reconstruction from Photoplethysmography Signals without Pulse Arrival Time Adjustments. Bioengineering, 10(6), 630. https://doi.org/10.3390/bioengineering10060630







_Elgendi.png)

