Quantification of the Monomer Compositions of Poly(3-hydroxybutyrate-co-3-hydroxyvalerate) and Poly(3-hydroxyvalerate) by Alkaline Hydrolysis and Using High-Performance Liquid Chromatography
Abstract
1. Introduction
2. Materials and Methods
2.1. P(3HB-co-3HV) Production
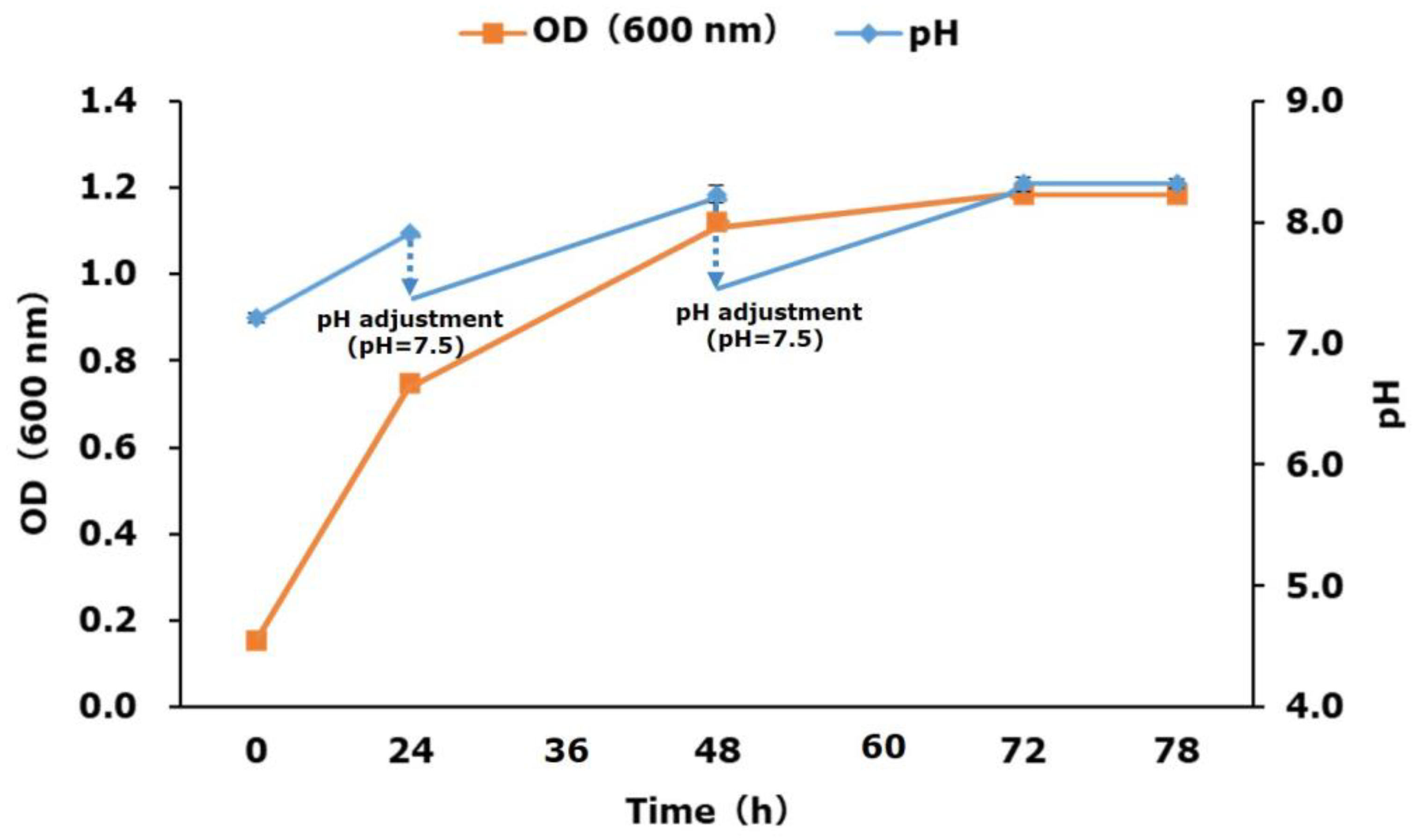
2.2. P(3HV) Production
2.3. Extraction of P(3HB-co-3HV) and P(3HV)
2.3.1. Alkaline Decomposition of Homopolymers P(3HB) and P(3HV)
2.3.2. Alkaline Decomposition of Copolymer P(3HB-co-3HV)
2.4. Analysis
2.4.1. HPLC Analysis
Calibration Curve Preparation Using Standard 2BE, 2PE, and 3HB
2.4.2. GC Analysis
3. Results and Discussion
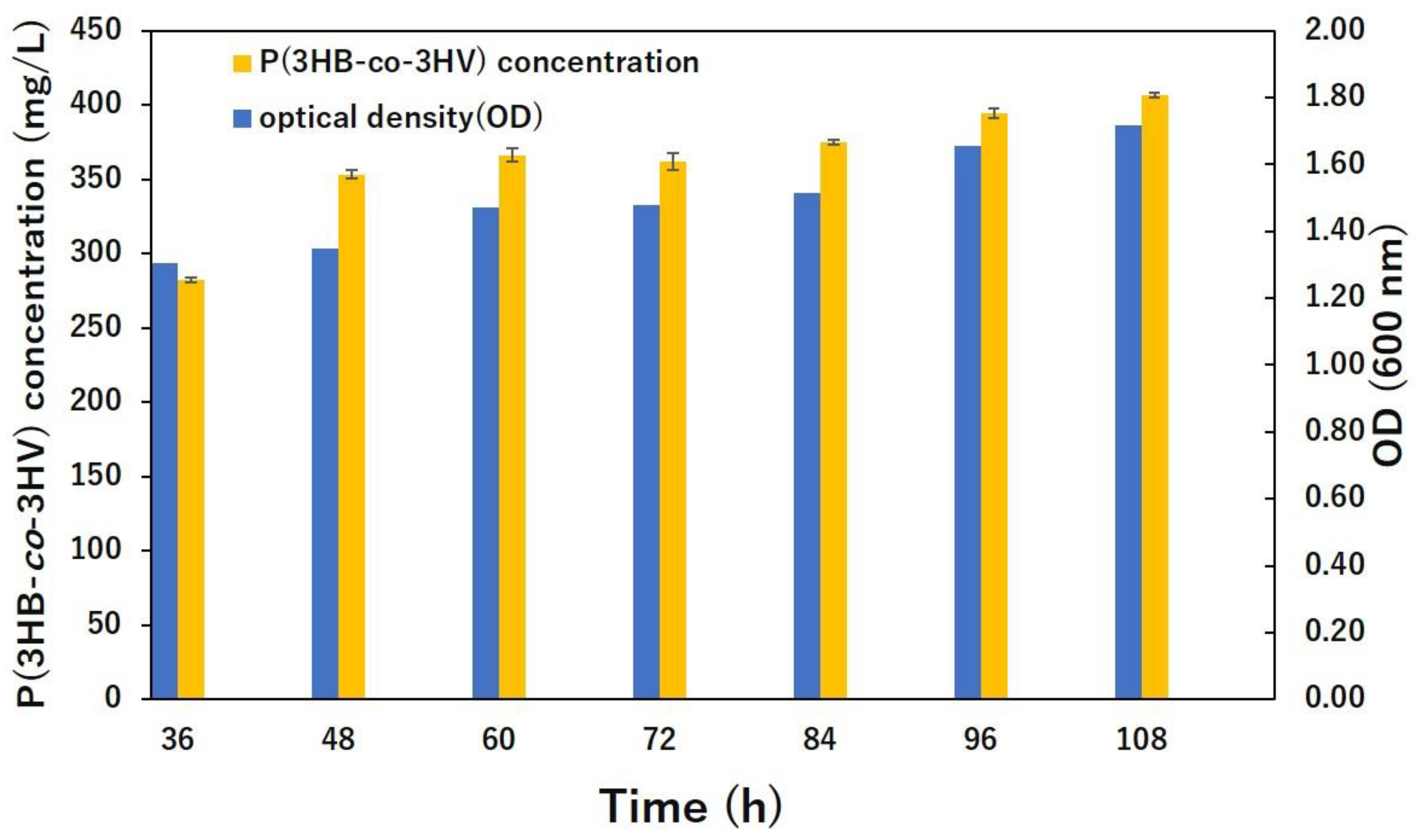
3.1. Bacterial Growth and Polymer Production
3.2. HPLC Results
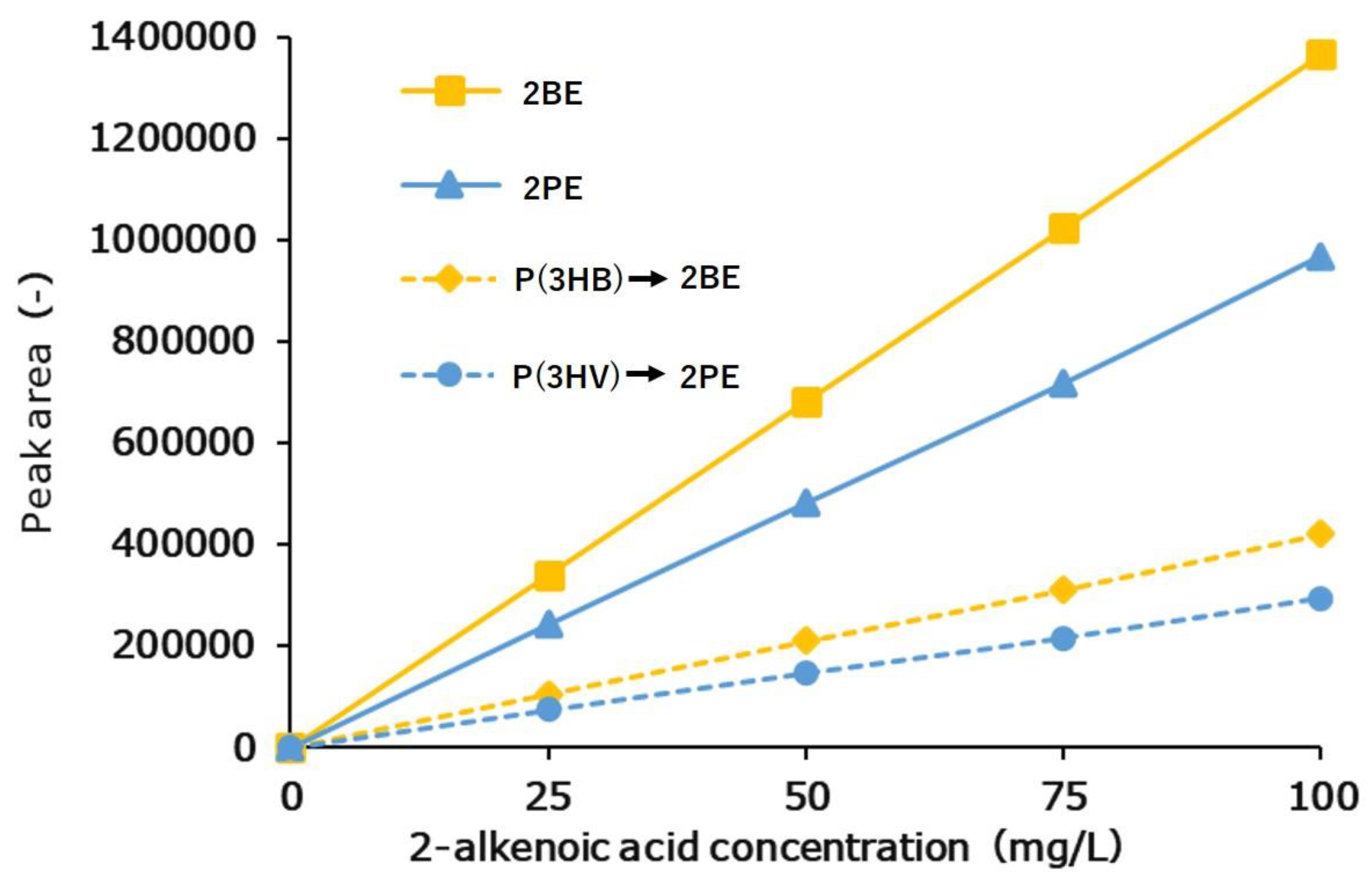
3.2.1. Calibration Curve Results of 2BE, 2PE, and 3HB Standards
3.2.2. Calibration Curve Results of P(3HB) and P(3HV) Homopolymers
3.2.3. Quantification of Copolymer P(3HB-co-3HV)
3.3. GC Results
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rogovina, S.Z.; Prut, E.V.; Berlin, A.A. Composite materials based on synthetic polymers reinforced with natural fibers. Polym. Sci. Ser. A 2019, 61, 417–438. [Google Scholar] [CrossRef]
- Raturi, G.; Shree, S.; Sharma, A.; Panesar, P.S.; Goswami, S. Recent approaches for enhanced production of microbial polyhydroxybutyrate: Preparation of biocomposites and applications. Int. J. Biol. Macromol. 2021, 182, 1650–1669. [Google Scholar]
- Reddy, M.V.; Watanabe, A.; Onodera, R.; Mawatari, Y.; Tsukiori, Y.; Watanabe, A.; Kudou, M.; Chang, Y.C. Polyhydroxyalkanoates (PHA) production using single or mixture of fatty acids with Bacillus sp. CYR1: Identification of PHA synthesis genes. Bioresour. Technol. Rep. 2020, 11, 100483. [Google Scholar] [CrossRef]
- Reddy, M.V.; Mawatari, Y.; Yajima, Y.; Seki, C.; Hoshino, T.; Chang, Y.C. Poly-3-hydroxybutyrate (PHB) production from alkylphenols, mono and poly-aromatic hydrocarbons using Bacillus sp. CYR1: A new strategy for wealth from waste. Bioresour. Technol. 2015, 192, 711–717. [Google Scholar] [CrossRef]
- Chang, Y.C.; Reddy, M.V.; Choi, D. Cometabolic degradation of toxic trichloroethene or cis-1,2-dichloroethene with phenol and production of poly-β-hydroxybutyrate (PHB). Green Chem. 2021, 23, 2729–2737. [Google Scholar] [CrossRef]
- Reddy, M.V.; Yajima, Y.; Mawatari, Y.; Hoshino, T.; Chang, Y.C. Degradation and conversion of toxic compounds into useful bioplastics by Cupriavidus sp. CY-1: Relative expression of the PhaC gene under phenol and nitrogen stress. Green Chem. 2015, 17, 4560–4569. [Google Scholar] [CrossRef]
- Koller, M.; Maršálek, L.; Dias, M.M.de.S.; Braunegg, G. Producing microbial polyhydroxyalkanoate (PHA) biopolyesters in a sustainable manner. New Biotech. 2017, 37, 24–38. [Google Scholar] [CrossRef]
- Li, M.; Wilkins, M.R. Recent advances in polyhydroxyalkanoate production: Feedstocks, strains and process developments. Int. J. Biol. Macromol. 2020, 156, 691–703. [Google Scholar] [CrossRef] [PubMed]
- Strong, P.J.; Laycock, B.S.; Mahamud, N.S.; Jensen, P.D.; Lant, P.A.; Tyson, G.; Pratt, S. The opportunity for high-performance biomaterials from methane. Microorganisms 2016, 4, 11. [Google Scholar] [CrossRef] [PubMed]
- Wong, H.S.J.; Bhubalan, K.; Amirul, A.A. A critical review on the economically feasible and sustainable poly(3-Hydroxybutyrate-co-3-hydroxyvalerate) production from alkyl alcohols. Polymers 2022, 14, 670. [Google Scholar] [CrossRef] [PubMed]
- Reddy, M.V.; Mawatari, Y.; Yajima, Y.; Satoh, K.; Mohan, S.V.; Chang, Y.C. Production of poly-3-hydroxybutyrate(P3HB) and poly(3-hydroxbutyrate-co-3-hydroxyvalerate) P(3HB-co-3HV) from synthetic wastewater using Hydrogenophaga palleronii. Bioresour. Technol. 2016, 215, 155–162. [Google Scholar] [CrossRef]
- Amini, M.; Sobhani, S.; Younesi, H.; Abyar, H.; Salamatinia, B.; Mohammadi, M. Evaluating the feasibility of poly (3-hydroxybutyrate-co-3-hydroxyvalerate) co-biopolymer production from rice wastewater by Azohydromonas lata. Appl. Food Biotechnol. 2020, 7, 73–83. [Google Scholar]
- Wong, H.S.J.; Huong, K.H.; Hani, S.N.A.; Amirul, A.A. Genetic incorporation of oil-utilizing ability in Cupriavidus malaysiensis USMAA2-4 for sustainable polyhydroxyalkanoates production from palm olein and 1-pentanol. J. Biotechnol. 2021, 337, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Kiun, J.T.; Amelia, T.S.M.; Huong, K.H.; Amirul, A.A.; Bhubalan, K. Optimizing the biosynthesis of renewable polyhydroxyalkanoate copolymer containing 3-hydroxyvalerate by Massilia haematophila using statistical modeling. BioTechnologia 2019, 100, 359–371. [Google Scholar] [CrossRef]
- Steinbüchel, A.; Debzi, E.M.; Marchessault, R.H.; Timm, A. Synthesis and production of poly(3-hydroxyvaleric acid) homopolyester by Chromobacterium violaceum. Appl. Microbiol. Biotechnol. 1993, 39, 443–449. [Google Scholar] [CrossRef]
- Steinbüchel, A.; Schmack, G. Large-scale production of poly (3-hydroxyvaleric acid) by fermentation of Chromobacterium violaceum, processing, and characterization of the homopolyester. J. Environ. Polym. Degrad. 1995, 3, 243–258. [Google Scholar] [CrossRef]
- Fukui, T.; Kichise, T.; Yoshida, Y.; Doi, Y. Biosynthesis of poly(3-hydroxybutyrate-co-3-hydroxyvalerate-co-3-hydroxy-heptanoate) terpolymers by recombinant Alcaligenes eutrophus. Biotechnol. Lett. 1997, 19, 1093–1097. [Google Scholar] [CrossRef]
- Benesova, P.; Kucera, D.; Marova, I.; Obruca, S. Chicken feather hydrolysate as an inexpensive complex nitrogen source for PHA production by Cupriavidus necator on waste frying oils. Let. Appl. Microbiol. 2017, 65, 182–188. [Google Scholar] [CrossRef]
- Lemoigne, M. Produit de deshydratation et de polymerisation de l’acide β-oxybutyrique. Bull. Soc. Chem. Biol. 1926, 8, 770–782. [Google Scholar]
- Williamson, D.H.; Wilkinson, J.F. The isolation and estimation of the poly-beta-hydroxybutyrate inclusions of Bacillus species. J. Gen. Microbiol. 1958, 19, 198–209. [Google Scholar] [CrossRef]
- Karr, D.B.; Waters, J.K.; Emerich, D.W. Analysis of poly-beta-hydroxybutyrate in Rhizobium japonicum bacteroids by ion-exclusion high-pressure liquid chromatography and UV detection. Appl. Environ. Microbiol. 1983, 46, 1339–1344. [Google Scholar] [CrossRef]
- Braunegg, G.; Sonnleimer, B.; Lafferty, R.M. A rapid gas chromatographic method for the determination of poly-hydroxybutyric acid in microbial biomass. Eur. J. Appl. Microbiol. Biotechnol. 1978, 6, 29–37. [Google Scholar] [CrossRef]
- Degelau, A.; Scheper, T.; Bailey, J.E.; Guske, C. Fluorometric measurement of poly-β hydroxybutyrate in Alcaligenes eutrophus by flow cytometry and spectrofluorometry. Appl. Microbiol. Biotechnol. 1995, 42, 653–657. [Google Scholar] [CrossRef]
- Lundgen, D.G.; Pfister, R.M.; Merrick, J.M. Structure of poly-beta-hydroxybutyric acid granules. J. Gen. Microbiol. 1964, 34, 441–446. [Google Scholar] [CrossRef]
- Stubbe, J.; Tian, J. Polyhydroxyalkanoate (PHA) hemeostasis: The role of PHA synthase. Nat. Prod. Rep. 2003, 20, 445–457. [Google Scholar] [CrossRef]
- Watanabe, Y.; Ichinomiya, Y.; Shimada, D.; Saika, A.; Abe, H.; Taguchi, S.; Tsuge, T. Development and validation of an HPLC-based screening method to acquire polyhydroxyalkanoate synthase mutants with altered substrate specificity. J. Biosci. Bioeng. 2012, 113, 286–292. [Google Scholar] [CrossRef] [PubMed]
- Duvigneau, S.; Kettner, A.; Carius, L.; Grieh, C.; Findeisen, R.; Kien, A. Fast, inexpensive, and reliable HPLC method to determine monomer fractions in poly(3-hydroxybutyrate-co-3-hydroxyvalerate). Appl. Microbiol. Biotechnol. 2021, 105, 4743–4749. [Google Scholar] [CrossRef]
- Liang, S.Y.; Wan, S.C.; Ho, Y.P.; Horng, Y.T.; Soo, P.C.; Peng, W.P. Rapid quantification of polyhydroxybutyrate polymer from single bacterial cells with mass spectrometry. Anal. Chem. 2022, 94, 11734–11738. [Google Scholar] [CrossRef]
- Koller, M.; Niebelschütz, H.; Braunegg, G. Strategies for recovery and purification of poly[(R)-3-hydroxyalkanoates] (PHA) biopolyesters from surrounding biomass. Eng. Life Sci. 2013, 13, 549–562. [Google Scholar] [CrossRef]
- Tamang, P.; Banerjee, R.; Köster, S.; Nogueira, R. Comparative study of polyhydroxyalkanoates production from acidified and anaerobically treated brewery wastewater using enriched mixed microbial culture. J. Environ. Sci. 2019, 78, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Satoh, H.; Skamoto, T.; Kuroki, Y.; Kudo, Y.; Mino, T. Application of the alkaline-digestion-HPLC method to the rapid determination of polyhydroxyalkanoate in activated sludge. J. Water Environ. Technol. 2016, 14, 411–421. [Google Scholar] [CrossRef]
- Law, J.H.; Slepecky, R.A. Assay of poly-beta-hydroxybutyric acid. J. Bacteriol. 1961, 82, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Slepecky, R.A.; Law, J.H. Synthesis and degradation of poly-β-hydroxybutyric acid in connection with sporulation of Bacillus megaterium. J. Bacteriol. 1961, 82, 37–42. [Google Scholar] [CrossRef]
- Yu, J.; Plackett, D.; Chen, L.X.L. Kinetics and mechanism of the monomeric products from abiotic hydrolysis of poly[(R)-3-hydroxybutyrate] under acidic and alkaline conditions. Polym. Degrad. Stab. 2005, 89, 289–299. [Google Scholar] [CrossRef]
- Zou, Y.; Yang, M.; Tao, Q.; Zhu, K.; Liu, X.; Wan, C.; Harder, M.K.; Yan, Q.; Liang, B.; Ntaikou, I.; et al. Recovery of polyhydroxyalkanoates (PHAs) polymers from a mixed microbial culture through combined ultrasonic disruption and alkaline digestion. J. Environ. Manag. 2023, 326, 116786. [Google Scholar] [CrossRef] [PubMed]

| Prepared Sample Concentration (mg/L) | Measured Concentration (mg/L) | ||
|---|---|---|---|
| 3HB | 3HV | P(3HB-co-3HV) | |
| 0 | 0 | 0 | 0 |
| 25 | 23.1 ± 1.17 | 2.5 ± 0.073 | 25.6 ± 1.23 |
| 50 | 47.8 ± 1.60 | 5.1 ± 0.24 | 52.9 ± 1.82 |
| 75 | 69.3 ± 3.01 | 7.1 ± 0.31 | 76.5 ± 3.31 |
| 100 | 95.0 ± 3.91 | 9.9 ± 0.35 | 104.8 ± 4.23 |
| HPLC Method | GC Method | |||||
|---|---|---|---|---|---|---|
| 3HB | 3HV | P(3HB-co-3HV) | 3HB | 3HV | P(3HB-co-3HV) | |
| Sample concentration (50 mg/L) | 18.3 ± 0.870 | 29.5 ± 1.32 | 47.8 ± 2.11 | 14.7 ± 0.751 | 33.1 ± 1.31 | 47.8 ± 2.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saito, K.; Reddy, M.V.; Sarkar, O.; Kumar, A.N.; Choi, D.; Chang, Y.-C. Quantification of the Monomer Compositions of Poly(3-hydroxybutyrate-co-3-hydroxyvalerate) and Poly(3-hydroxyvalerate) by Alkaline Hydrolysis and Using High-Performance Liquid Chromatography. Bioengineering 2023, 10, 618. https://doi.org/10.3390/bioengineering10050618
Saito K, Reddy MV, Sarkar O, Kumar AN, Choi D, Chang Y-C. Quantification of the Monomer Compositions of Poly(3-hydroxybutyrate-co-3-hydroxyvalerate) and Poly(3-hydroxyvalerate) by Alkaline Hydrolysis and Using High-Performance Liquid Chromatography. Bioengineering. 2023; 10(5):618. https://doi.org/10.3390/bioengineering10050618
Chicago/Turabian StyleSaito, Kyo, M. Venkateswar Reddy, Omprakash Sarkar, A. Naresh Kumar, DuBok Choi, and Young-Cheol Chang. 2023. "Quantification of the Monomer Compositions of Poly(3-hydroxybutyrate-co-3-hydroxyvalerate) and Poly(3-hydroxyvalerate) by Alkaline Hydrolysis and Using High-Performance Liquid Chromatography" Bioengineering 10, no. 5: 618. https://doi.org/10.3390/bioengineering10050618
APA StyleSaito, K., Reddy, M. V., Sarkar, O., Kumar, A. N., Choi, D., & Chang, Y.-C. (2023). Quantification of the Monomer Compositions of Poly(3-hydroxybutyrate-co-3-hydroxyvalerate) and Poly(3-hydroxyvalerate) by Alkaline Hydrolysis and Using High-Performance Liquid Chromatography. Bioengineering, 10(5), 618. https://doi.org/10.3390/bioengineering10050618










